Abstract
Background
State laws in the USA mandate that blood be drawn from all newborn infants to screen for health-threatening conditions. These screening assays consume only a small portion of the blood samples, which are collected on filter paper (‘Guthrie’) cards. Many states archive unused blood spots, often in unrefrigerated storage. Objectives: While individual RNA transcripts have been identified from archived neonatal blood spots, no study to date has performed quantitative analysis of archived blood spot RNA.
Methods
We demonstrate that RNA can be isolated and amplified from newborn blood spots stored unfrozen for as long as 9 years, and can be analyzed by microarray and qPCR.
Results
Microarray assays of archived neonatal blood spots consistently detected 3,000–4,000 expressed genes with correlations of 0.90 between replicates. Blood spot mRNA is amenable to qPCR and we detected biologically relevant expression levels of housekeeping and immune-mediating genes.
Conclusions
These experiments demonstrate the feasibility of using blood spots as a source of RNA which can be analyzed using quantitative microarray and qPCR assays. The application of these methods to the analysis of widely collected biological specimens may be a valuable resource for the study of perinatal determinants of disease development.
Key Words: Guthrie, Blood spot, RNA, Microarray, qPCR, Real-time PCR, Neonatal screening
Introduction
Newborns are routinely screened for a variety of conditions – predominantly inborn errors of metabolism – for which early diagnosis can lead to therapy and subsequent prevention of sequelae, including permanent neurological damage [1]. Such screening programs are in place throughout the world. In the USA, mandatory screening programs exist in all 50 states; the number of conditions screened for in each state varies from 4 to 30 [2]. Within 24–48 h after birth, newborn blood is collected on filter paper cards (‘Guthrie cards’) and submitted for laboratory analysis using principally tandem mass spectrometry, high-performance liquid chromatography, and antibody-based assays. These assays generally consume only a small portion of the available blood on the cards. The portion of the cards with remaining blood is then archived under varying conditions, and for varying durations. A recent survey found that 14 of the 51 US newborn screening programs retain newborn blood for 21 years or more [3]. These population-wide archives of biologic specimens represent a unique resource for the retrospective study of diseases of perinatal origin.
To be of use in health research, informative material in dried blood spots must persist after years of dry, room temperature storage. Proteins in blood spots are beyond detection in a matter of days [4], but nucleic acids show evidence of long-term stability. The published literature shows extensive examples of recovery and analysis of nucleic acids from dried blood spots. Many studies have involved screening for viral and bacterial DNA and RNA in dried blood spots [5,6,7,8,9]. The presence of infectious agents at birth may be very relevant to understanding diseases of perinatal origin, but since analysis of nucleic acids from infectious agents is relatively well documented, we focus our attention on DNA and RNA from the neonate.
Most reports on the analysis of neonatal nucleic acids from archived blood spots have focused on DNA, and several procedures for extracting DNA from filter paper have been published [10,11,12,13]. Subsequent analyses of blood spot DNA have included on-chip PCR-based microarray screening for specific gene polymorphisms [14], screening for disease markers and mutations [15,16,17,18] and post-mortem diagnosis of a disease-causing mutation [19]. Another interesting use of archived neonatal blood DNA has been to use PCR and sequencing to identify leukemic cells present at birth and other diagnostic markers in infants who subsequently developed leukemia [20,21,22,23].
As RNA is known to be more labile than DNA, it is reasonable to doubt the stability of RNA in long-term unfrozen storage. Yet peer-reviewed research shows that RNA in dried blood spots is stable after years of storage, contains transcripts indicative of specific diseases, and mirrors the gene expression in fresh blood. Blood spot RNA analysis was first published by Matsubara et al. [24] and Zhang et al. [23], who performed mutation and restriction enzyme analysis of expressed genes from spots stored for several years. Long-term RNA stability has been most strongly demonstrated by Swedish researchers who have recovered intact RNA transcripts from blood spots archived up to 21 years [25]. Several researchers show that genes expressed in archived blood samples reflect disease states. Suttorp et al. [26] detected RNA for the bcr-abl rearrangement indicative of the Philadelphia chromosome in dried blood spots. Pai et al. [27] showed that patients who lack expression of the FMR-1 gene due to fragile-X syndrome lack FMR-1 RNA in their neonatal blood spot. Both Pai et al. and Suttorp et al. verified their blood spot findings against RNA from fresh blood. We found one paper that compared RNA from a sample of fresh blood against RNA from identical blood spotted, dried, and stored for several months [28]. Maeno et al. confirmed that, after several months of room temperature storage, infection-induced cytokine mRNA in the blood spot precisely mirrored the genes expressed in the original fresh blood sample. While these publications all support the stability of blood spot RNA and its ability to reflect disease states, they only examined from 1 to 13 RNA transcripts at a time.
Researchers have yet to apply the latest tools in RNA technology to measure blood spot gene expression on a larger scale. We can find no published reports of microarray-based gene expression analysis of RNA from archived neonatal blood spots. If whole-expressed genome quantitative RNA expression can be applied to archived newborn blood spots, these specimens would constitute a remarkable resource for clinicians and researchers. More so than DNA, blood spot RNA would provide a temporal molecular snapshot of physiologic and metabolic function shortly after birth. These ‘expressed genetic profiles’ could then be retrospectively studied to identify markers and potential causal factors of childhood (and perhaps adult-onset) diseases from the perinatal period. RNA data could add further value to the already universally collected (and in some states research-available) neonatal blood spot.
In this report, we describe the results of parallel microarray-based gene expression and quantitative PCR analyses of RNA extracted from 12 archived neonatal blood spots.
Materials and Methods
Selection of Neonatal Blood Spots
In Michigan, 5 circles of 1.25 cm diameter on Guthrie cards are filled with blood obtained from a heel stick blood draw obtained between 24 and 72 h after birth. In most instances, only a portion of the first two spots are used for clinical testing. By state law in effect since 1986, the remaining blood is archived for the next 21.5 years. The Michigan Department of Community Health stores the Guthrie cards in boxes on shelves in rooms at ambient temperature. Michigan state law governing the use of these archived blood spots explicitly encourages their use for medical research, and anonymous specimens can be obtained for research purposes with IRB approval [MI Public Health Code 333.5431]. Individually identified spots can be obtained with parental informed consent.
After receiving IRB approvals from MSU and the Michigan Department of Community Health (MDCH), we obtained from MDCH an anonymous stratified random sample of newborn blood spots collected from 1996 to 2004. All blood spots were stored in boxes corresponding to the date of laboratory testing of the samples. Within the boxes, which are filed by birth date in the Michigan Department of Laboratories, blood spot cards are bundled in groups of 70, with most boxes containing between 8 and 20 bundles. A stratified sampling procedure was used to randomly select boxes, bundles, and cards within bundles. We selected only cards with three unused spots, or two unused spots and a third spot with only a single punch taken. If the randomly selected card was not eligible, we alternated examining cards on either side of the selected card until we found a usable card. This procedure was required less than 5% of the time. We then excised one unused 1.25-cm diameter blood spot and placed it in a plastic envelope. We aimed to obtain 10 samples per year for each of the 9 years, and using this method were able to obtain 85 archived specimens.
Isolation, Quantification, and Amplification of RNA from Neonatal Blood Spots
We randomly selected 12 of the 85 blood spots for RNA testing; three blood spots (each representing a different neonate) from each of the years 1998, 2000, 2003, and 2004. The 73 remaining spots were set aside for future experiments. We collected eight 3-mm punches from each blood spot. Punches were soaked in 1 ml TRIzol reagent (Invitrogen) for 15 min and agitated until all blood was dissolved from the spot. Tubes were then spun in a microcentrifuge to pellet the filter paper. TRIzol reagent containing the lysed blood cells was transferred to a fresh tube with 200 μl chloroform (Omnipur), vortexed, and centrifuged at 20,100 RCF, 4°C for 15 min. The upper aqueous portion was transferred to a fresh tube containing 500 μl isopropanol (Tedia) and 30 μg glycogen (Ambion), and vortexed. We precipitated RNA by centrifugation at 20,100 RCF, 4°C for 30 min. RNA pellets were rinsed with 75% ethanol/25% nuclease-free water (Aaper, Ambion) and eluted in 15 μl nuclease-free water. 2 μl of each sample were used to determine RNA concentration by fluorescent RNA detection (Quant-iT™ RNA Assay Kit; Invitrogen); a 5-μl sample was reserved for future assays, and the remaining 8 μl were used as template for two rounds of RNA amplification using the Message Amp 2 standard protocol (Ambion). 2 μl of total cDNA for each sample were set aside during the first round of amplification for quantitative PCR analysis. Amplified RNA (aRNA) was quantified using the Nanodrop ND1000 spectrophotometer.
DNA Microarray Analysis of Neonatal Blood Spot RNA
For each of our 12 samples, we performed the following procedure in triplicate. For each microarray, reverse transcription using SuperScript II (Invitrogen) was performed on 20 μg of aRNA primed with random hexamers (Invitrogen) in the presence of cyanine-3 dCTP (PerkinElmer). To enable comparison within and across all replicate samples, each individual sample was compared against a common reference; equal amounts of aRNA from all 12 neonatal samples were pooled to form the common reference. This reference pool was labeled with cyanine-5 dCTP and divided for use across all arrays. Microarrays were synthesized at the Van Andel Institute and had passed internal quality assurance/quality control standards. Labeled cDNA cleanup, hybridization to 20,000 feature human cDNA arrays, and post-hybridization array washes were performed in triplicate for each sample as previously described [29, 30]. Arrays were scanned at 532 and 635 nm using standard settings in an Agilent Microarray Scanner (Agilent Technologies, Palo Alto, Calif., USA). Images were analyzed using GenePix Pro 5.0 (Axon, Union City, Calif., USA). Spots with signal lower than three times the standard deviation of the global array background in either channel were excluded from analysis. The array intensities were adjusted (normalized) using the within print-tip normalization methods as implemented in the limma package [31] for the R statistical environment [32].
Quantitative PCR Analysis of Neonatal Blood Spot RNA
The 2-μl aliquot of double-stranded cDNA, representing mRNA extracted from each blood spot, was divided into 12 aliquots to permit study of four genes in triplicate from each spot. These four genes were two housekeeping/metabolic genes: mitochondrial ribosomal protein L11 (MRPL11) and hexokinase 1 (HK1); and two inflammatory genes: nuclear factor kappa B1 (NFκB1) and alpha × integrin (ITGAX). The primers used for amplification were specifically selected to amplify only RNA. In each pair of primers, either one or both were exon-spanning, and the primers were always located on different exons. The introns present in genomic DNA prevent primer binding and thus the PCR reaction cannot be initiated by contaminating DNA. Two additional steps were then used to confirm that we had amplified mRNA. First, we used commercially available genomic DNA in each experimental array and showed that amplification did not occur with DNA. Second, we applied RNAse to our experimental arrays and showed that after this treatment, amplification did not occur, indicating that we were detecting mRNA in our experiments. To remove any further uncertainty, all of our primer sets were purchased from Super Array Bioscience Corp. (Frederick, Md., USA), a company that uses stringent criteria for primer design and validation. All primers were subjected to standard curve analysis using known mRNA samples [33]. Only the primers that gave a slope of standard curve of −3.32 ± 0.1 (−3.32 represents 100% efficiency) were used in this study.
The SYBR Green dye, which binds to the minor groove of double-stranded DNA, was used to detect amplicons whose quantity is indicated by the intensity of fluorescent emissions. The SYBR Green method is as effective as the TaqMan method for validation of microarray gene expression data, is less expensive, and uses a more straightforward protocol [34,35,36]. Each 2-μl sample of total cDNA (after dilution and separation into 12 aliquots) was mixed with 2× SYBR Green PCR Master Mix (Applied Biosystems) and the qPCR analysis was performed at the Research Technology Support Facility (RTSF) at MSU on an ABI Prism 7900HT Sequence Detection System. Hexokinase 1, a stably and highly expressed neonatal blood transcript, was used as a loading control in qPCR analysis. We monitored the amount of DNA produced in each PCR cycle and counted the fold changes required to obtain satisfactory binding to SYBR Green.
Results
Isolation, Quantification, and Amplification of RNA from Neonatal Blood Spots
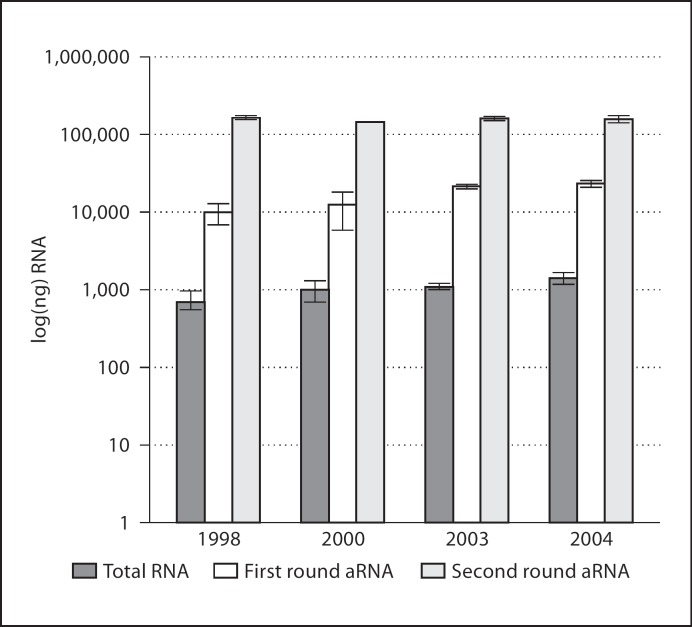
Each column in figure 1 depicts the average yield of RNA from three different samples within a given year. Although one sees a slight decrease in total RNA and first round aRNA when comparing older versus newer blood spots, differences in yield are negligible by the second round of amplification. The 12 samples averaged 160 ± 13 μg aRNA. First round amplification products should meet the requirements for most commercial microarray platforms (9, 11, 21 and 23 μg, from oldest to newest spots), while the second round of RNA amplification provides enough aRNA to perform multiple microarrays.
Fig. 1.
Average neonatal blood spot RNA by year. For each sample year we show the average RNA yield from three key points in the microarray process: (1) initial total RNA yield, (2) aRNA yield after the first round of amplification, and (3) aRNA yield after the second round of amplification.
DNA Microarray Analysis of Neonatal Blood Spot RNA
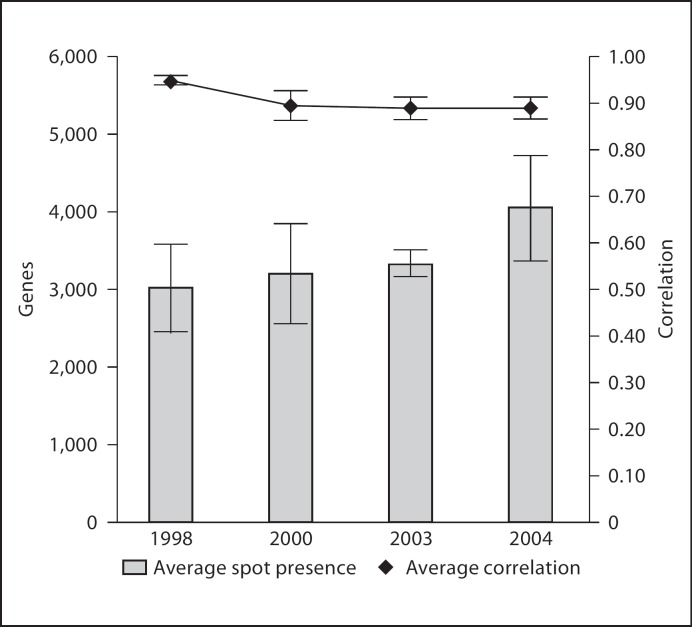
As shown in figure 2, the average number of expressed genes detected between all years and all neonatal blood spots was 3,480 of 20,000 total. This ranged from an average of 3,017 genes for blood spots from 1998 and 4,067 genes for blood spots from 2004. The average correlation within triplicate assays for all neonatal blood samples was 0.904 ± 0.042.
Fig. 2.
Average gene presence and average correlation across triplicate arrays. Left axis - average gene presence: each column represents the average number of genes measured as ‘present’ across 9 microarrays; 3 different blood spots for each sample year, each spot assayed in triplicate. Right axis - average correlation: we calculated Pearson correlation coefficients between each replicate blood spot array. We averaged these coefficients within each sample year; each point represents aggregate average correlations for 9 microarrays.
Quantitative PCR Analysis of Neonatal Blood Spot RNA
We amplified four genes in triplicate from each of our 12 blood spots, for a total of 36 assays per gene and 144 total qPCR reactions. Because of limited starting material, some samples did not have enough material for triplicates. Standard deviations were not calculated for these samples.
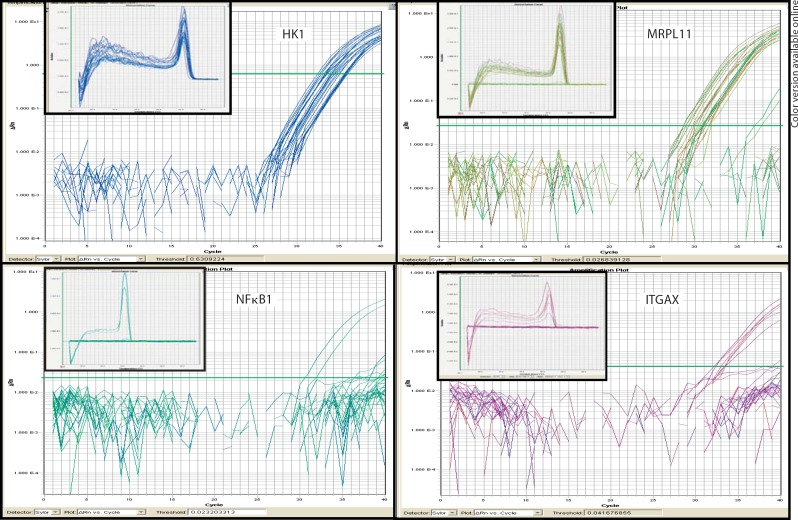
Figure 3 (main window) shows saturation curves for individual assays. HK1, a metabolic gene expressed in higher number in neonatal erythrocytes [37], gave Ct values before 35 cycles for 26/36 assays (72%) and was present in all (12/12) samples. MRPL11 gave Ct values before 35 cycles for 24/36 assays (67%, 11/12 samples). ITGAX, also known as leukocyte adhesion receptor, reached Ct before 35 cycles in 13/36 assays (36%, 9/12 samples). As ITGAX can mediate cell-cell interactions during inflammation [38], we would not expect it to be expressed in all subjects. The same is true of qPCR assays on the pro-inflammatory cytokine NFκB1; only five of our twelve blood spot samples (6/36 aliquots) reached Ct for this cytokine between 20 and 35 cycles.
Fig. 3.
Neonatal blood spot qPCR results. Main window: saturation curves representing abundance of transcripts within each sample, as measured by fluorescence. Inset: dissociation curves confirming the amplification of a single fragment of expected melting temperature for each assay.
The dissociation curves (fig. 3; inset) for the amplification products of each assay show dissociation at only one temperature, indicating that – where there was amplification – our primers amplified a single transcript with a uniform melting point. This shows a lack of non-specific amplification and further validates the specificity of our pPCR measurements.
In table 1, values with standard deviations indicate there was adequate starting material for triplicate assays. Relative expression levels, after normalization to HK1, show biologically plausible results. As is expected, NFκB and ITGAX were not expressed in all samples and relative expression levels were lower for these inflammatory genes than for MRPL11. One of our randomly selected blood spots (1998_B) showed relatively higher levels of NFκB1 when compared to the other four NFκB1 expressing blood spots.
Table 1.
Neonatal blood spot qPCR expression levels relative to hexokinase 1
| Year | Sample No. | 2 Δct compared to hexokinase l |
||
|---|---|---|---|---|
| MRPL11 | ITGAX | NFkBI | ||
| 1998 | 1998_A | ND | ND | ND |
| 1998_B | 1.94±1.63 | ND | 1.27 | |
| 1998_C | 0.66±0.01 | ND | ND | |
| 2000 | 2000 A | 1.36±1.91 | 0.003 | ND |
| 2000 B | 0.69±0.54 | 0.57±0.60 | 0.002 | |
| 2000_C | 0.81±0.39 | 0.02±0.01 | ND | |
| 2003 | 2003_A | 0.29±0.40 | 0.16±0.10 | 0.004 |
| 2003_B | 1.61 | 0.16 | ND | |
| 2003_C | 0.56±0.04 | 0.02 | ND | |
| 2004 | 2004_A | 1.92±0.70 | 0.24±0.31 | ND |
| 2004_B | 0.22±0.11 | 0.15 | 0.10 | |
| 2004_C | 0.59±0.67 | 0.08±0.02 | 0.001 | |
Concordance between Microarray and Quantitative PCR for Blood Spot RNA
Our qPCR experiment was not designed to validate the microarray results, but rather was performed in parallel as a proof-of-principle study. Because of this we used different sets of primers for microarray and qPCR analysis, and were unable to target qPCR to measure the genes reported as most highly misregulated by microrarray. However, even with this caveat, we observed a very high (79 ± 22%) concordance between genes measured as present by microarray and genes measured as present by qPCR. Concordance of RNA transcript presence was highest for the housekeeping genes HXK and MRPL11 (100 and 92% agreement, respectively) and lower for ITGAX and NFκB (75 and 50% agreement, respectively). To enable accurate microarray validation in the future we will use the exact same sets of primers, based on microarray sequences, for qPCR assays.
Discussion
In this study we used two different laboratory methods – DNA microarray and quantitative PCR – to demonstrate the feasibility of quantitative analysis of RNA from archived blood spots. Both methods demonstrated that the RNA obtained from archived blood spot samples is usable for quantitative gene expression analysis. Our results show that consistent, quantitative RNA measurements can be obtained from neonatal blood spot samples archived at room temperature for up to 9 years.
Further investigation is warranted to determine if improvements in RNA isolation and amplification will increase the quantity and quality of data from blood spots. Targeted removal of globin RNA could unmask more genes in the samples [39]. We realize the threat that DNA contamination can play when using small amounts of RNA and have performed experiments showing the efficacy of treatment with RNAse-free DNAse (data not shown). We plan to apply this treatment to all future experiments. Future experiments will also utilize commercially available Agilent arrays; these arrays require less input RNA, have inherent QC and QA considerations, and provide more standardized measurements that will facilitate across-laboratory comparisons. Agilent microarrays will also enable us to design qPCR primers that correspond to the oligonucleotide probes used on the array, enabling qPCR validation of our microarray results. As we only consumed a portion of the available blood spots (8 of 14 3 mm punches), we will preserve at least part of the spot for future assays, including viral and bacterial screening by PCR.
Newborn metabolic screening continues to be a vital and successful clinical assay of neonates. By teaming with state health departments and utilizing existing blood spot archives, we hope to improve our understanding of diseases that are not immediately apparent at birth, but have roots in the perinatal period. In conclusion, these findings correlate with previously published research showing that, in addition to being a source of informative DNA, RNA from archived neonatal blood spots is amenable to quantitative gene expression measurement. Measuring the relative abundance of thousands of expressed genes from universally collected neonatal blood spots may open new avenues of research into perinatal markers, determinants of disease development, and add value to an underutilized biologic archive.
Acknowledgements
The authors would like to thank Todd Lydic and Shirali Gaurang Patel for RNA work, and Kyle Furge and Karl Dykema of the VARI Bioinformatics core for their analysis contributions and guidance. We also appreciate the efforts of Madeleine Lenski at Michigan State University and Harry Hawkins at the Michigan Department of Community Health for neonatal sample acquisition.
Partial support for this work was provided by the Office of Research, College of Human Medicine, Michigan State University, as well as generous support from the Jay and Betty Van Andel Foundation (J.H.R., E.J.K., and P.T.H.). P.T.H. and E.J.K. are also supported through a T32 training grant in perinatal epidemiology from the NIH.
References
- 1.Raghuveer TS, Garg U, Graf WD. Inborn errors of metabolism in infancy and early childhood: an update. Am Fam Physician. 2006;73:1981–1990. [PubMed] [Google Scholar]
- 2.Green NS, Pass KA. Neonatal screening by DNA microarray: spots and chips. Nat Rev Genet. 2005;6:147–151. doi: 10.1038/nrg1526. [DOI] [PubMed] [Google Scholar]
- 3.Therrell BL, Johnson A, Williams D. Status of newborn screening programs in the United States. Pediatrics. 2006;117:S212–S252. doi: 10.1542/peds.2005-2633C. [DOI] [PubMed] [Google Scholar]
- 4.Miller AA, Sharrock KC, McDade TW. Measurement of leptin in dried blood spot samples. Am J Hum Biol. 2006;18:857–860. doi: 10.1002/ajhb.20566. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Muñoz MT, Zaragoza-Rodríguez S, Rojas-Montes O, Palacios-Saucedo G, Vázquez-Rosales G, Gómez-Delgado A, Torres J, Muñoz O. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch Med Res. 2005;36:382–386. doi: 10.1016/j.arcmed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J Acquir Immune Defic Syndr. 2005;38:615–617. doi: 10.1097/01.qai.0000143604.71857.5d. [DOI] [PubMed] [Google Scholar]
- 7.Lewensohn-Fuchs I, Osterwall P, Forsgren M, Malm G. Detection of herpes simplex virus DNA in dried blood spots making a retrospective diagnosis possible. J Clin Virol. 2003;26:39–48. doi: 10.1016/s1386-6532(02)00019-7. [DOI] [PubMed] [Google Scholar]
- 8.Solmone M, Girardi E, Costa F, Pucillo L, Ippolito G, Capobianchi MR. Simple and reliable method for detection and genotyping of hepatitis C virus RNA in dried blood spots stored at room temperature. J Clin Microbiol. 2002;40:3512–3514. doi: 10.1128/JCM.40.9.3512-3514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCabe KM, Zhang YH, Huang BL, Wagar EA, McCabe ER. Bacterial species identification after DNA amplification with a universal primer pair. Mol Genet Metab. 1999;66:205–211. doi: 10.1006/mgme.1998.2795. [DOI] [PubMed] [Google Scholar]
- 10.Rubin EM, Andrews KA, Kan YW. Newborn screening by DNA analysis of dried blood spots. Hum Genet. 1989;82:134–136. doi: 10.1007/BF00284045. [DOI] [PubMed] [Google Scholar]
- 11.Carducci C, Ellul L, Antonozzi I, Pontecorvi A. DNA elution and amplification by polymerase chain reaction from dried blood spots. Biotechniques. 1992;13:735–737. [PubMed] [Google Scholar]
- 12.Mackey K, Steinkamp A, Chomczynski P. DNA extraction from small blood volumes and the processing of cellulose blood cards for use in polymerase chain reaction. Mol Biotechnol. 1998;9:1–5. doi: 10.1007/BF02752692. [DOI] [PubMed] [Google Scholar]
- 13.Sørensen KM, Jespersgaard C, Vuust J, Hougaard D, Nørgaard-Pedersen B, Andersen PS. Whole genome amplification on DNA from filter paper blood spot samples: an evaluation of selected systems. Genet Test. 2007;11:65–71. doi: 10.1089/gte.2006.0503. [DOI] [PubMed] [Google Scholar]
- 14.Mitterer G, Bodamer O, Harwanegg C, Maurer W, Mueller MW, Schmidt WM. Microarray-based detection of mannose-binding lectin 2 (MBL2) polymorphisms in a routine clinical setting. Genet Test. 2005;9:6–13. doi: 10.1089/gte.2005.9.6. [DOI] [PubMed] [Google Scholar]
- 15.Chow JC, Chen DJ, Lin CN, Chiu CY, Huang CB, Chiu PC, Lin CH, Lin SJ, Tzeng CC. Feasibility of blood spot PCR in large-scale screening of fragile X syndrome in southern Taiwan. J Formos Med Assoc. 2003;102:12–16. [PubMed] [Google Scholar]
- 16.Dobrowolski SF, Angeletti J, Banas RA, Naylor EW. Real-time PCR assays to detect common mutations in the biotinidase gene and application of mutational analysis to newborn screening for biotinidase deficiency. Mol Genet Metab. 2003;78:100–107. doi: 10.1016/s1096-7192(02)00231-7. [DOI] [PubMed] [Google Scholar]
- 17.Majumdar R, Rehana Z, Al Jumah M, Fetaini N. Spinal muscular atrophy carrier screening by multiplex polymerase chain reaction using dried blood spot on filter paper. Ann Hum Genet. 2005;69:216–221. doi: 10.1046/j.1529-8817.2004.00149.x. [DOI] [PubMed] [Google Scholar]
- 18.Olshan AF, Shaw GM, Millikan RC, Laurent C, Finnell RH. Polymorphisms in DNA repair genes as risk factors for spina bifida and orofacial clefts. Am J Med Genet A. 2005;135:268–273. doi: 10.1002/ajmg.a.30713. [DOI] [PubMed] [Google Scholar]
- 19.Skinner JR, Chong B, Fawkner M, Webster DR, Hegde M. Use of the newborn screening card to define cause of death in a 12-year-old diagnosed with epilepsy. J Paediatr Child Health. 2004;40:651–653. doi: 10.1111/j.1440-1754.2004.00498.x. [DOI] [PubMed] [Google Scholar]
- 20.Taub JW, Konrad MA, Ge Y, Naber JM, Scott JS, Matherly LH, Ravindranath Y. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood. 2002;99:2992–2996. doi: 10.1182/blood.v99.8.2992. [DOI] [PubMed] [Google Scholar]
- 21.Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, Greaves MF. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagi T, Hibi S, Tabata Y, Kuriyama K, Teramura T, Hashida T, Shimizu Y, Takimoto T, Todo S, Sawada T, et al. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood. 2000;96:264–268. [PubMed] [Google Scholar]
- 23.Wiemels JL, Xiao Z, Buffler PA, Maia AT, Ma X, Dicks BM, Smith MT, Zhang L, Feusner J, Wiencke J, et al. In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood. 2002;99:3801–3805. doi: 10.1182/blood.v99.10.3801. [DOI] [PubMed] [Google Scholar]
- 24.Matsubara Y, Ikeda H, Endo H, Narisawa K. Dried blood spot on filter paper as a source of mRNA. Nucleic Acids Res. 1992;20:1998. doi: 10.1093/nar/20.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlsson H, Guthenberg C, von Döbeln U, Kristenssson K. Extraction of RNA from dried blood on filter papers after long-term storage. Clin Chem. 2003;49:979–981. doi: 10.1373/49.6.979. [DOI] [PubMed] [Google Scholar]
- 26.Suttorp M, Ritgen M, von Neuhoff N, Schoch R, Schmitz N. Blood on filter paper as a readily available source of bcr-abl rearranged mRNA. Blood. 1997;90:1713–1715. [PubMed] [Google Scholar]
- 27.Pai JT, Tsai SF, Horng CJ, Chiu PC, Cheng MY, Hsiao KJ, Wuu KD. Absence of FMR-1 gene expression can be detected with RNA extracted from dried blood specimens. Hum Genet. 1994;93:488–493. doi: 10.1007/BF00202810. [DOI] [PubMed] [Google Scholar]
- 28.Maeno Y, Nakazawa S, Nagashima S, Sasaki J, Higo KM, Taniguchi K. Utility of the dried blood on filter paper as a source of cytokine mRNA for the analysis of immunoreactions in Plasmodium yoelii infection. Acta Trop. 2003;87:295–300. doi: 10.1016/s0001-706x(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 29.Whitwam T, Vanbrocklin MW, Russo ME, Haak PT, Bilgili D, Resau JH, Koo HM, Holmen SL. Differential oncogenic potential of activated RAS isoforms in melanocytes. Oncogene. 2007;26:4563–4570. doi: 10.1038/sj.onc.1210239. [DOI] [PubMed] [Google Scholar]
- 30.Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, Werts A, Haak P, Vande Woude GF. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 32.Ihaka R, Gentleman R. A language for data analysis and graphics. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
- 33.Applied Biosystems User bulletin #2: ABI PRISM 7700 Sequence Detection System, 2001.
- 34.Hembruff SL, Villeneuve DJ, Parissenti AM. The optimization of quantitative reverse transcription PCR for verification of cDNA microarray data. Anal Biochem. 2005;345:237–249. doi: 10.1016/j.ab.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ, et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.al-Naama MM, al-Naama LM, al-Sadoon TA. Glucose-6-phosphate dehydrogenase, hexokinase and pyruvate kinase activities in erythrocytes of neonates and adults in Basrah. Ann Trop Paediatr. 1994;14:195–200. doi: 10.1080/02724936.1994.11747717. [DOI] [PubMed] [Google Scholar]
- 38.Ugarova TP, Yakubenko VP. Recognition of fibrinogen by leukocyte integrins. Ann NY Acad Sci. 2001;936:368–385. doi: 10.1111/j.1749-6632.2001.tb03523.x. [DOI] [PubMed] [Google Scholar]
- 39.Field LA, Jordan RM, Hadix JA, Dunn MA, Shriver CD, Ellsworth RE, Ellsworth DL. Functional identity of genes detectable in expression profiling assays following globin mRNA reduction of peripheral blood samples. Clin Biochem. 2007;40:499–502. doi: 10.1016/j.clinbiochem.2007.01.004. [DOI] [PubMed] [Google Scholar]