Abstract
DLX1 and DLX2 transcription factors are necessary for forebrain GABAergic neuron differentiation, migration, and survival. We generated transgenic mice that express Cre-recombinase under the control of two ultra-conserved DNA elements near the Dlx1&2 locus termed I12b and URE2. We show that Cre-recombinase is active in a “Dlx-pattern” in the embryonic forebrain of transgenic mice. I12b-Cre is more active than URE2-Cre in the medial ganglionic eminences and its derivatives. Fate-mapping of EGFP+ cells in adult Cre;Z/EG animals demonstrated that GABAergic neurons, but not glia, are labeled. Most NPY+, nNOS+, parvalbumin+, and somatostatin+ cells are marked by I12b-Cre in the cortex and hippocampus, while 25-40% of these interneuron subtypes are labeled by URE2-Cre. Labeling of neurons generated between E12.5 to E15.5 indicated differences in birth-dates of EGFP+ cells that populate the olfactory bulb, hippocampus, and cortex. Finally, we provide the first in vivo evidence that both I12b and URE2 are direct targets of DLX2 and require Dlx1 and Dlx2 expression for proper activity.
Introduction
Gamma-amino butyric acid (GABA)-ergic neurons comprise ∼20% of all neurons within the cerebral cortex and hippocampus and ∼95% of neurons within the striatum (Danglot et al., 2006; Rymar et al., 2004; Tepper et al., 2004; Wonders and Anderson, 2006). As inhibitory local circuit neurons within the cortex, GABAergic interneurons modulate neuronal activity and synaptic plasticity; as projection neurons within the striatum and pallidum, they perform key inhibitory functions within neural systems. Disruption of GABAergic neuron function has been associated with several disorders such as autism, schizophrenia, Tourette's syndrome, bipolar depression, and epilepsy (Benes and Berretta, 2001; Cossart et al., 2005; Kalanithi et al., 2005; Lewis et al., 2005; Rubenstein and Merzenich, 2003).
In the rodent, GABAergic telencephalic interneurons are generated in the subpallial ganglionic eminences and tangentially migrate to populate the olfactory bulb, cortex, and hippocampus (Metin et al., 2006; Wonders and Anderson, 2005). GABAergic interneurons are remarkably diverse and can be subdivided by morphology, connectivity, electrophysiology, and molecular markers (Markram et al., 2004). GABAergic neuron markers include Ca2+- binding proteins, such as calbindin (CB), calretinin (CR), and parvalbumin (PV), neuropeptides, such as somatostatin (SOM) and neuropeptide Y (NPY), and neurotransmitters such as neuronal nitric oxide synthase (nNOS). Expression of these markers within GABAergic interneuron subtypes varies between regions, yet ∼85% of interneurons can be classified by largely non-overlapping expression of PV, CR, and SOM in the cortex and PV, CR, and CB in the hippocampus (Freund and Buzsaki, 1996; Gonchar, 2008; Gonchar and Burkhalter, 1997; Jinno and Kosaka, 2006; Kubota and Kawaguchi, 1994; Miyoshi et al., 2007). GABAergic interneuron diversity arises during embryogenesis and is influenced by both the place and time of their specification. For example, PV and SOM expressing interneurons are generated first and arise primarily from the medial ganglionic eminence (MGE), while CR+/SOM- interneurons are born later and arise in the caudal ganglionic eminence (CGE) (Butt et al., 2005; Fogarty et al., 2007; Wonders et al., 2008).
The DLX family of homeobox transcription factors are key molecular regulators of GABAergic neuron differentiation, migration, and survival. Dlx genes are expressed during development in several tissues, including the forebrain, limbs, face, and tail (Jeong et al., 2008; Kraus and Lufkin, 2006; Panganiban and Rubenstein, 2002). In the CNS, four of the Dlx family members, Dlx1, Dlx2, Dlx5, and Dlx6, are expressed in the ventral telencephalon in regions that coincide with GABAergic neuron specification or differentiation (Stühmer et al., 2002a; Stühmer et al., 2002b); in general, expression of Dlx1 and Dlx2 precedes Dlx5 and Dlx6 (Eisenstat et al., 1999). Dlx1 and Dlx2 double mutant mice (Dlx1&2 mutants) lose the majority of GABAergic neocortical, hippocampal, and olfactory bulb interneurons due to defects in neuronal maturation and migration (Anderson et al., 1997a; Cobos et al., 2007; Cobos et al., 2005; Long et al., 2007; Long et al., 2008). Dlx1&2 mutants also have a block in the differentiation of basal ganglia GABAergic projection and interneurons (Anderson et al., 1997b; Marin et al., 2000; Yun et al., 2002). Thus DLX1 and DLX2 are essential for GABAergic neuron formation throughout the forebrain.
Directed gene targeting in ES cells and generation of transgenic mice offer researchers powerful tools to explore the mechanisms of development and disease (Gossen and Bujard, 2002). The Cre/loxP system enables tissue and temporal restricted disruption of genes or reporter expression through Cre-recombinase mediated recombination of loci that contain flanking loxP sites (“floxed”) (Gaveriaux-Ruff and Kieffer, 2007). The utility of this system is limited by the availability of mice that express Cre-recombinase in specific temperospatial patterns. The few Cre-expressing transgenic mouse lines that target forebrain GABAergic neurons use an enhancer element from the Dlx5 and Dlx6 locus (Kohwi et al., 2007; Monory et al., 2006; Stenman et al., 2003; Zerucha et al., 2000). However, these mouse lines have some disadvantages as there is incomplete recombination in GABAergic neurons and ectopic expression of Cre-recombinase sometimes occurs in the caudal-ventral cortex (unpublished observations). Therefore, we wished to generate mouse lines that express Cre-recombinase specifically in a large complement of forebrain GABAergic neurons, as well as in specific subsets of forebrain GABAergic neurons. Since Dlx1 and Dlx2 are expressed earlier and in more GABAergic neurons than Dlx5 and Dlx6, we decided to generate transgenic mice that utilized recently described enhancer elements from the Dlx1 and Dlx2 locus (Ghanem et al., 2007).
We report the generation and characterization of two Cre transgenic mouse lines using distinct Dlx1 and Dlx2 elements termed I12b and URE2. I12b-Cre and URE2-Cre mice show strong activity in the telencephalon and diencephalon in a pattern matching Dlx1 and Dlx2 expression where GABAergic neurons are formed and differentiate (Stühmer et al., 2002a; Stühmer et al., 2002b). We show that I12b-Cre is expressed by nearly all cortical and hippocampal GABAergic interneurons, while URE2-Cre induced recombination in a subset of these interneurons. We use lineage-tracing with floxed EGFP reporter mice and thymidine-analog labeling to show that GABAergic neurons destined for the olfactory bulb, cortex, or hippocampus are born with different time-courses and that I12b-Cre and URE2-Cre labeled cells show differences in their birthdates. Finally, we provide the first in vivo evidence that DLX binds directly to I12b and URE2 and that DLX1 and DLX2 function are necessary for I12b and URE2 activity.
Results
DLX1 and DLX2 are lineage-specific transcription factors required for forebrain GABAergic neuron differentiation, migration, and survival (Anderson et al., 1997a; Anderson et al., 1997b; Cobos et al., 2005; Long et al., 2007; Panganiban and Rubenstein, 2002; Petryniak et al., 2007; Yun et al., 2002). Two distinct, conserved noncoding DNA elements near the Dlx1 and Dlx2 (Dlx1&2) locus drive transgene expression in the developing forebrain in a pattern similar to endogenous DLX1 and DLX2 (Ghanem et al., 2007). The ∼450 bp I12b element is located in the intragenic region of Dlx1&2 and the ∼1400 bp URE2 element is located approximately 12 kbp 5′ of the Dlx1 ATG start site in the mouse (Fig. 1A). Both of these elements are active in the embryonic forebrain, with high telencephalic activity in the lateral, medial, and caudal ganglionic eminences (LGE, MGE, CGE), and their derivatives, and in the diencephalon (Ghanem et al., 2007). To generate transgenic mice that express Cre-recombinase in the developing forebrain, we placed the I12b or URE2 elements upstream of a β-globin minimal promoter and a Cre-recombinase with a nuclear localization signal (NLS) (Fig. 1B, 1C). These constructs were linearized and injected into oocytes to generate transgenic animals. Transgenic mice were screened by PCR and Southern blot analysis. Eleven I12b-Cre and seven URE2-Cre were identified by PCR, of which four I12b-Cre and four URE2-Cre had single integration sites (Supplemental Fig. S1). To determine which transgenic mice express Cre-recombinase capable of catalyzing recombination of loxP sites in vivo, animals with single integration sites were mated with Z/EG Cre-reporter mice, which have a floxed enhanced green fluorescent protein (EGFP) allele, and their progeny analyzed for EGFP fluorescence at birth. All I12b-Cre;Z/EG mice, and all URE2-Cre;Z/EG mice, had similar patterns of EGFP expression at birth, indicating that the pattern of Cre-recombinase activity was not strongly affected by the insertion-sites or transgene copy number. Analysis of histological sections at birth from two different I12b-Cre and URE2-Cre transgenic mice gave nearly identical results (data not shown); thus we focused our analysis on one line of I12b-Cre and one line of URE2-Cre mice.
Figure 1. Generation of I12b-Cre and URE2-Cre transgenic mice.
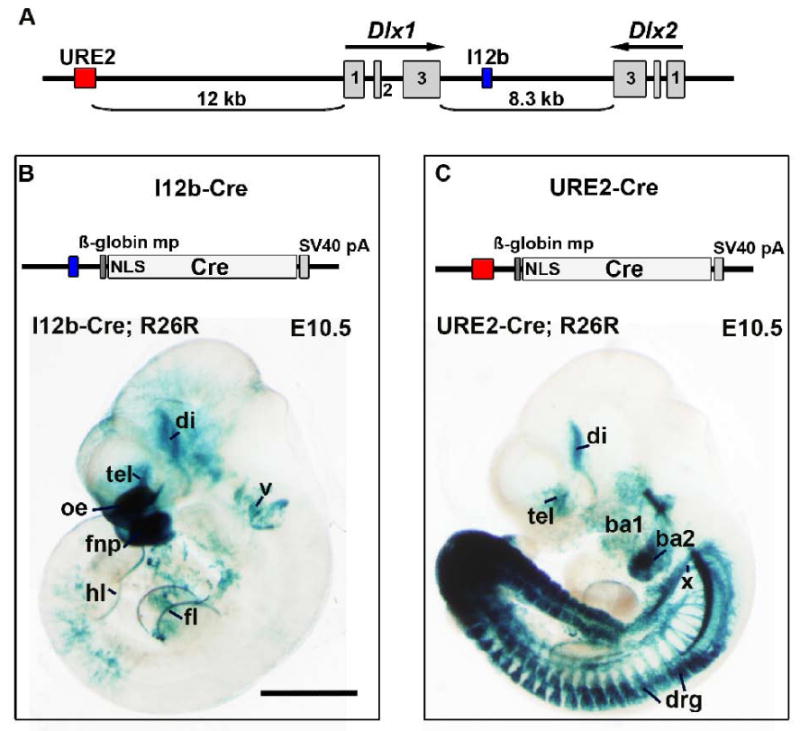
(A) Schematic of the Dlx1 and Dlx2 genomic locus showing the location of the I12b and URE2 regulatory elements.
(B) Top, Schematic of the I12b-Cre transgene; Bottom, Wholemount X-gal staining of an E10.5 I12b-Cre embryo crossed to the ROSA26-lacZ reporter (R26R).
(C) Top, Schematic of the URE2-Cre transgene; Bottom, Wholemount X-gal staining of an E10.5 URE2-Cre embryo crossed to the ROSA26-lacZ reporter (R26R).
ba1, first branchial arch; ba2, second branchial arch; di, diencephalon; drg, dorsal root ganglia; fl, forelimb; fnp, frontonasal prominence; hl, hindlimb; mp, minimal promoter; NLS, nuclear localization signal; oe, olfactory epithelium; pA, poly A sequence; tel, telencephalon; v, trigeminal ganglion; x, vagus ganglion. Scale bar = 1 mm.
I12b-Cre and URE2-Cre expression during development
Dlx genes are expressed in several developing tissues, including craniofacial components, limbs, tail and forebrain (Jeong et al., 2008; Kraus and Lufkin, 2006; Panganiban and Rubenstein, 2002). To determine the location of I12b or URE2 activity during embryogenesis, we analyzed β-galactosidase (β-gal) expression in double-transgenic I12b-Cre; ROSA26-lacZ or URE2-Cre;ROSA26-lacZ mice at embryonic day (E)10.5. ROSA26-lacZ mice express β-gal after Cre-mediated recombination of a floxed STOP cassette that has been targeted to the ubiquitously express ROSA26 locus (Soriano, 1999). I12b-Cre activity was detected in the telencephalon and diencephalon, the anterior ectodermal ridge (AER) of the forelimb and hindlimb, the frontonasal prominence and olfactory epithelium, and in the trigeminal ganglion (Fig.1B). Similar to I12b-Cre, URE2-Cre activity was detected in the forebrain and AER of the forelimb and hindlimb. However, activity was also detected in the first and second branchial arch, vagus ganglion, tail, and dorsal root ganglion (Fig. 1C).
To determine the expression pattern of the regulatory elements during embryonic development in more detail, we examined the progeny of transgenic I12b-Cre or URE2-Cre mice that had been crossed to the Z/EG reporter (Novak et al., 2000). EGFP fluorescence was readily detected in the forebrain by E10.5 using a fluorescent dissecting scope. At E10.5 and other time points, expression of the EGFP reporter matched the expression pattern of the ROSA26-lacZ reporter (data not shown). At E12.5, EGFP expression in I12b-Cre;Z/EG embryos was detected in the LGE, MGE, and CGE, the olfactory epithelium and olfactory bulb, prethalamus, preoptic area, the hypothalamus, and in the skin overlying the head (Fig. 2A, 2C). In addition, EGFP+ cells, which are most likely tangentially migrating immature interneurons, can be seen streaming from the ganglionic eminences into the cortex (arrowheads in Fig. 2C, Fig. 9). Expression of EGFP in URE2-Cre;Z/EG embryos was detected in the MGE, LGE, hypothalamus, and prethalamus, although the level of expression was lower compared to I12b-Cre;Z/EG embryos (Fig. 2B, 2D). Reporter expression was also found in the trigeminal nerve, dorsal root ganglia, gut, and tail (Fig. 2B). In contrast to I12b-Cre, EGFP expression was not detected in the CGE, POA or in tangentially migrating immature interneurons in URE2-Cre animals at this time point.
Figure 2. EGFP labeling at E12.5 in I12b-Cre;Z/EG and URE2-Cre;Z/EG reveals strong forebrain expression.
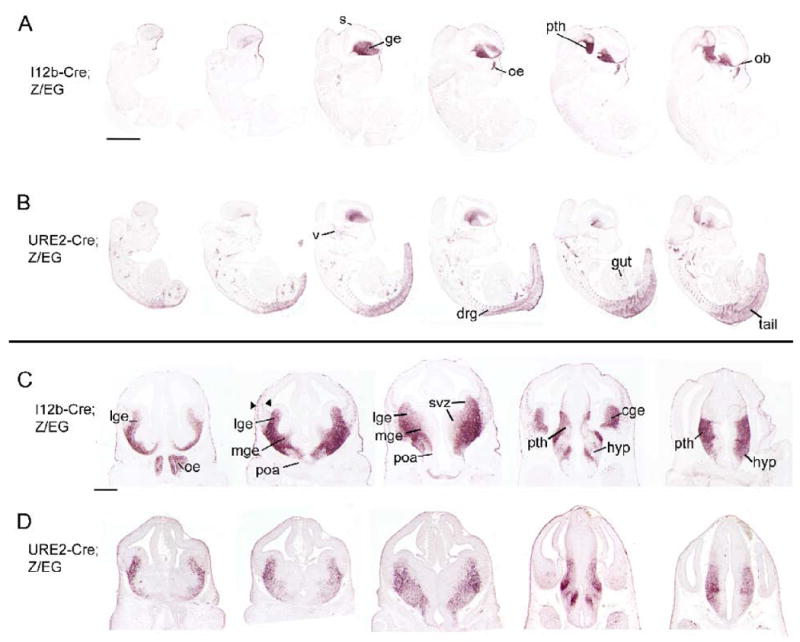
(A-B) Immunohistochemistry for EGFP on parasagittal sections through an E12.5 (A) I12b-Cre;Z/EG or (B) URE2-Cre;Z/EG animal.
(C-D) Immunohistochemistry for EGFP on coronal sections through the E12.5 forebrain of (C) I12b-Cre;Z/EG or (D) URE2-Cre;Z/EG mice. EGFP expression is present in the CGE in I12b-Cre;Z/EG but not URE2-Cre;Z/EG. Also note that tangentially migrating cells (arrowhead) are readily detected in I12b-Cre;Z/EG forebrain.
cge, caudal ganglionic eminence; drg, dorsal root ganglion; ge, ganglionic eminences; hyp, hypothalamus; lge, lateral ganglionic eminence; mge, medial ganglionic eminence; ob, olfactory bulb; oe, olfactory epithelium; poa, preoptic area; pth, prethalamus; s, skin; svz, subventricular zones; v, trigeminal ganglion. Scale bar in A = 1mm and in C = 500 μm.
Figure 9. CRE and EGFP expression in embryonic and adult I12b-Cre;Z/EG mice.
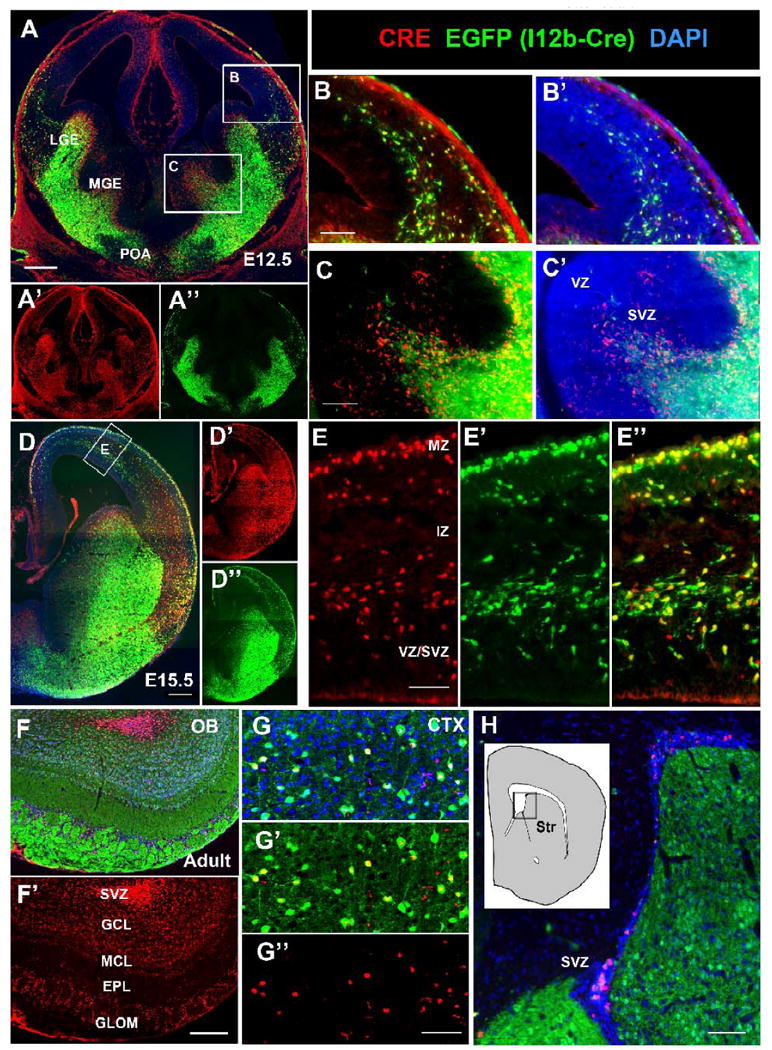
(A-C) Immunofluorescence on E12.5 forebrain sections from I12b-Cre;Z/EG mice for CRE (red), EGFP (green), and DAPI (blue). Boxed regions in (A) are shown at higher magnification in (B) and (C). LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; POA, preoptic area.
(B) Overlapping CRE and EGFP expression is detectable in tangentially migrating interneurons
(C) CRE expression precedes EGFP expression within the SVZ of the MGE. (D-E) Immunofluorescence on E15.5 forebrain sections from I12b-Cre;Z/EG mice for CRE (red), EGFP (green), and DAPI (blue). Boxed regions in (D) is shown at higher magnification in (E).
(E-E″) CRE and EGFP are co-expressed in tangentially migrating interneurons within the marginal zone (MZ) and between the intermediate zone (IZ) and subventricular zone (SVZ).
(F-H) Immunofluorescence on adult forebrain sections from I12b-Cre;Z/EG mice for CRE (red), EGFP (green), and DAPI (blue) in the olfactory bulb (F-F′), cortex (G-G″), and subventricular zone (H).
(F) CRE and EGFP expression within the adult olfactory bulb. Note that migrating neuroblasts in the SVZ express CRE, but not EGFP. In contrast, the granule cell layer (GCL), mitral cell layer (MCL), external plexiform layer (EPL) and glomerular layer (GL) contain cells that co-express EGFP and CRE.
(G) CRE is expressed in a subset of EGFP+ cells within the adult somatosensory cortex.
(H) CRE is expressed within neurogenic niche of SVZ cells of the lateral ventricles, but is not detectable within the striatum (Str).
Scale bars in E, G = 100 μm; B, C, H = 200 μm; A, D, F = 500 μm.
By E15.5, strong EGFP expression was detected in the projection neurons of the caudate-putamen (a derivative of the LGE) of both I12b-Cre;Z/EG and URE2-Cre;Z/EG embryos (Fig. 3A-A′, 3B-B′). In addition, both transgenic mice generated reporter expression within tangentially migrating immature interneurons, the olfactory bulb, septum, hypothalamus, thalamic reticular nucleus, and zona incerta. I12b-Cre was more active than URE2-Cre in subpallial MGE and POA derivatives, including the globus pallidus and ventral pallidum. The enhancers also showed activity outside of regions known to express Dlx1 and Dlx2; I12b-Cre labeled cells and projections of olfactory neurons (which express Dlx5), pretectal nucleus, and the tegmental nucleus, while URE2-Cre was active in inferior olive cells.
Figure 3. Comparison of EGFP expression between E15.5 I12b-Cre;Z/EG and URE2-Cre;Z/EG mice highlights similarities and differences in forebrain expression.
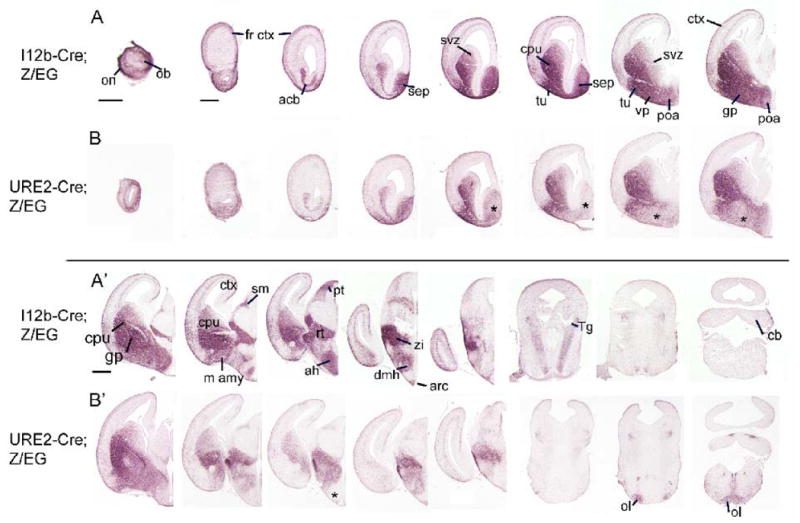
(A-A′, B-B′) Rostral to caudal coronal sections through E15.5 I12b-Cre;Z/EG (A-A′) and URE2-Cre;Z/EG (B-B′) were labeled by immunohistochemistry for EGFP. Note the reduced expression in specific ventral forebrain regions (asterisks) in URE2-Cre;Z/EG embryos.
acb, accumbens; ah, anterior hypothalamus; arc, arcuate nucleus; cb, cerebellum; cpu, caudate-putamen; dmh, dorsomedial hypothalamus; fr ctx, frontal cortex; gp, globus pallidus; lh, lateral hypothalamus; m amy, medial amygala; ob, olfactory bulb; ol, inferior olive; on, olfactory nerve layer; pt, pretectal nucleus; pa, paraventricular hypothalamic nucleus; poa, preoptic area; rt, thalamic reticular nucleus; sep, septum; sf, superior fiber bundle; sm, stria medullaris of thalamus; svz, subventricular zone; Tg, tegmental nucleus; tu, olfactory tubercle; vp, ventral pallidum; zi, zona incerta. Scale bars = 500 μm.
At P0, both I12b-Cre;Z/EG and URE2-Cre;Z/EG pups showed robust reporter expression within the olfactory bulb, caudate-putamen, accumbens, amygdala, cortex, hippocampus, olfactory tubercle, hypothalamus, and in axonal projections through the basal ganglia to the substantia nigra (Fig. 4). I12b-Cre strongly labeled olfactory sensory neurons and produced much higher labeling of the septum, globus pallidus, ventral pallidum, and anterior hypothalamus compared to URE2-Cre (compare asterisk-labeled regions in Fig. 4B-B″ to 4A-A″).
Figure 4. Activity of I12b and URE2 revealed by EGFP immunohistochemistry of sections through newborn I12b-Cre;Z/EG and URE2-Cre;Z/EG mice.
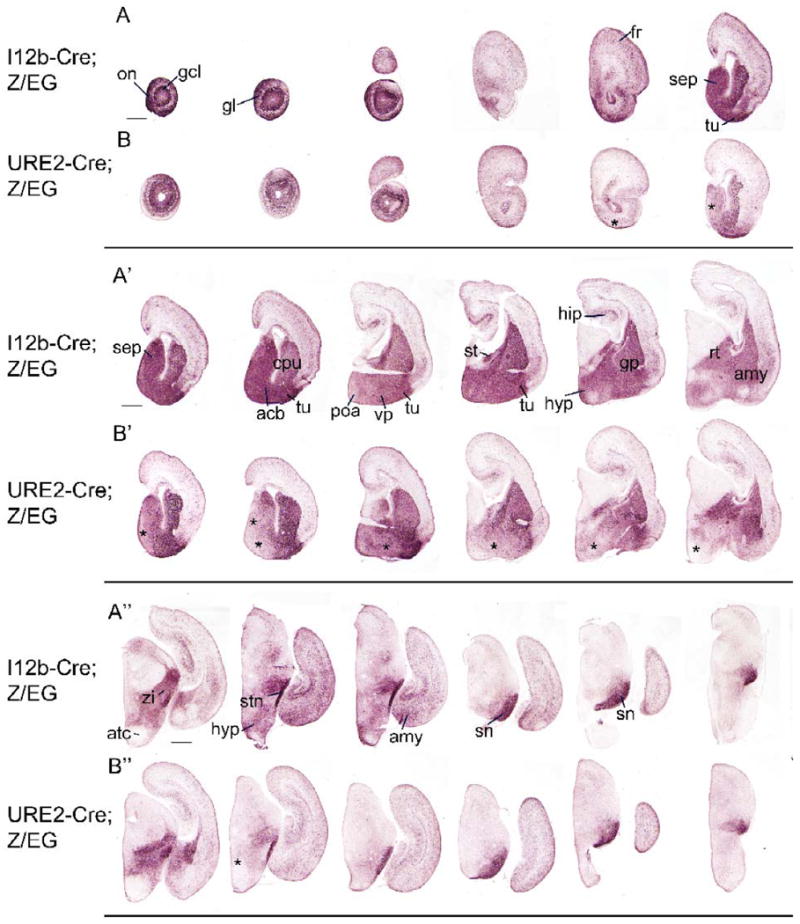
(A-A″, B-B″) Rostral to caudal coronal sections through P0 I12b-Cre;Z/EG (A-A″) and URE2-Cre;Z/EG (B-B″) mice were labeled for EGFP. Expression in URE2-Cre;Z/EG mice is lower in specific regions (asterisks) compared to I12b-Cre;Z/EG.
acb, accumbens; amy, amygdala; atc, anterior hypothalamic area; cpu, caudate-putamen; fr, frontal cortex; gcl, granule cell layer; gl, glomerular layer; hip, hippocampus; hyp, hypothalamus; on, olfactory nerve layer; poa, preoptic area; rt, thalamic reticular nucleus; sep, septum; sn, substantia nigra; stm, stria terminalis; stn, subthalamic nucleus; tu, olfactory tubercle; vp, ventral pallidum; zi, zona incerta. Scale bars = 500 μm.
Adult expression of I12b-Cre and URE2-Cre labeled cells
To determine the adult pattern of I12b-Cre and URE2-Cre lineage cells, we analyzed EGFP expression in adult Cre;Z/EG animals. In the olfactory bulb of both Cre;/ZEG animals, strong expression of the reporter was detected in the granule cells, the external plexiform layer, and periglomerular cells (Fig. 5A). As previously noted, EGFP was strongly expressed within olfactory sensory neuron axons that project to the glomerular layer in I12b-Cre;Z/EG animals. Both I12b-Cre and URE2-Cre labeled cells that populate the caudate-putamen, globus pallidus, preoptic area, accumbens, amygdala, cortex, hippocampus, hypothalamus and thalamic reticular nucleus and axonal projections from the basal ganglia to the substantia nigra (Fig. 5A, 5B and Supplemental Fig. S2 and S3). EGFP+ cells were found in all cortical areas.
Figure 5. Characterization of EGFP expression in adult I12b-Cre;Z/EG and URE2-Cre;Z/EG forebrain.
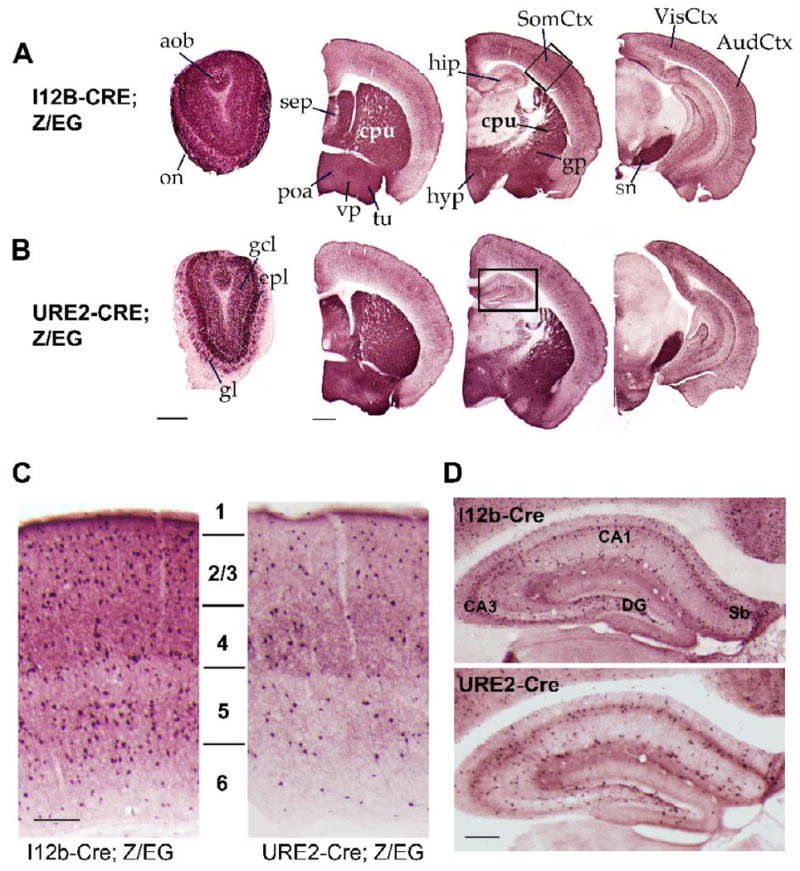
(A-B) EGFP+ cells were identified in coronal sections through adult I12b-Cre;Z/EG (A) or URE2-Cre;Z/EG (B) mice. The boxes denote similar regions of the cortex and hippocampus shown at higher magnification in (C) and (D).
(C) EGFP+ cells in the adult somatosensory cortex of I12b-Cre;Z/EG (left) and URE2-Cre;Z/EG (right) mice populate cortical layers 1 to 6. I12b-Cre labels about 2.3× more cells than URE2-Cre.
(D) EGFP+ cells are found throughout the adult hippocampus of I12b-Cre;Z/EG (top) and URE2-Cre;Z/EG (bottom) mice. I12b-Cre labels about 1.6× more cells than URE2-Cre.
amy, amygdala; aob, accessory olfactory bulb; AudCtx, auditory cortex; cpu, caudate-putamen; DG, dentate gyrus; epl, external plexiform layer; gcl, granule cell layer; gl, glomerular layer; gp, globus pallidus; hip, hippocampus; hyp, hypothalamus; on, olfactory nerve layer; poa, preoptic area; Sb, subiculum; sep, septum; snr, substania nigra reticulata; SomCtx, somatosensory cortex; tu, olfactory tubercle; VisCtx, visual cortex; vp, ventral pallidum. Scale bar in A, B = 500 μm; C = 100 μm; D= 150 μm.
We quantified the distribution of EGFP+ cells within the adult somatosensory cortex of I12b-Cre;Z/EG and URE2-Cre;Z/EG mice. Labeled cells were found in all layers. I12b-Cre generated ∼2.3-fold more EGFP+ cells than URE2-Cre (n=4; 24,723 vs. 10,562 EGFP+ cells in I12b-Cre;Z/EG and URE2-Cre;Z/EG cortical sections). I12b-Cre;Z/EG (n=4) labeling resulted in the following laminar distribution of EGFP+ cells: layer 1: 6.2%, layer 2/3: 26.1%, layer 4: 21.9%, layer 5: 21.5%, and layer 6: 24.3%. URE2-Cre;Z/EG (n=4) labeling resulted in the following laminar distribution of EGFP+ cells: layer 1: 5.6%, layer 2/3: 29.3%, layer 4: 23.8%, layer 5: 22.5%, and layer 6: 18.9%. Thus, relatively more I12b-Cre labeled cells (compared to URE2-Cre) were found in layer 6, and relatively less were in layers 2/3. Furthermore, strong EGFP expression was detected in the neuropile of layer 4 in both I12b-Cre and URE2-Cre, but was more prominently labeled by URE2-Cre.
We next examined the distribution of labeled cells within the adult hippocampus. EGFP+ cells were found in the molecular and cellular layers of CA1-3 and dentate gyrus (DG) regions of the hippocampus (Fig. 5D). Approximately 1.6 fold more cells were labeled in I12b-Cre;Z/EG compared to URE2-Cre;Z/EG (n=4; 12,939 vs. 8,226 EGFP+ cells in comparable I12b-Cre;Z/EG and URE2-Cre;Z/EG sections). I12b-Cre;Z/EG labeling resulted in the following distribution: DG: 19.6%, CA3: 31.3%, and CA1/2: 49.0% (n=4) (Fig. 5D). URE2-Cre;Z/EG labeling resulted in the following distribution: DG: 18.4%, CA3: 28.1%, and CA1/2: 53.5% (n=4). Thus, proportionally more URE2-Cre lineage cells populated the CA1/2 region, while proportionally more I12b-Cre lineage cells populated DG and CA3.
I12b-Cre and URE2-Cre label neurons and not glia in the forebrain
In the embryonic forebrain, the expression pattern of DLX transcription factors is almost identical to that of the glutamic acid decarboxylases (GAD65 and GAD67), enzymes that synthesize the neurotransmitter GABA (Stühmer et al., 2002a). Ectopic expression of Dlx2 and Dlx5 can induce GAD expression in cortical progenitors (Stühmer et al., 2002a). Dlx genes are expressed in all migrating GABAergic neurons, and differentiation, migration, and survival are dependant upon Dlx1 and/or Dlx2 function (Anderson et al., 1997a; Cobos, 2006; Cobos et al., 2007; Cobos et al., 2005). The expression pattern of EGFP in i12b-Cre;Z/EG and URE2-Cre;Z/EG animals during embryonic development and in the adult closely resembles forebrain GABA expression (Fig. 2-5 and (Katarova et al., 2000; Stühmer et al., 2002a). Thus, we hypothesized that I12b-Cre and URE2-Cre activity would label GABAergic neurons. To test this idea, we analyzed co-expression of EGFP and GABA in the somatosensory cortex and hippocampus of I12b-Cre;Z/EG and URE2-Cre;Z/EG mice (Fig. 6A-D). Greater than 95% of GABA+ cells co-expressed EGFP in the somatosensory cortex and hippocampus of I12b-Cre;Z/EG animals (Fig. 6A, 6C, and data not shown) while approximately 43% of GABA+ cells co-labeled for EGFP in the somatosensory cortex and hippocampus of URE2-Cre;Z/EG animals (Fig. 6B, 6C, and data not shown). This indicates that nearly all GABAergic neurons in the cortex and hippocampus were captured by lineage-mapping using I12b-Cre, but not with URE2-Cre.
Figure 6. I12b-Cre and URE2-Cre label GABAergic neurons and not glia in the adult brain.
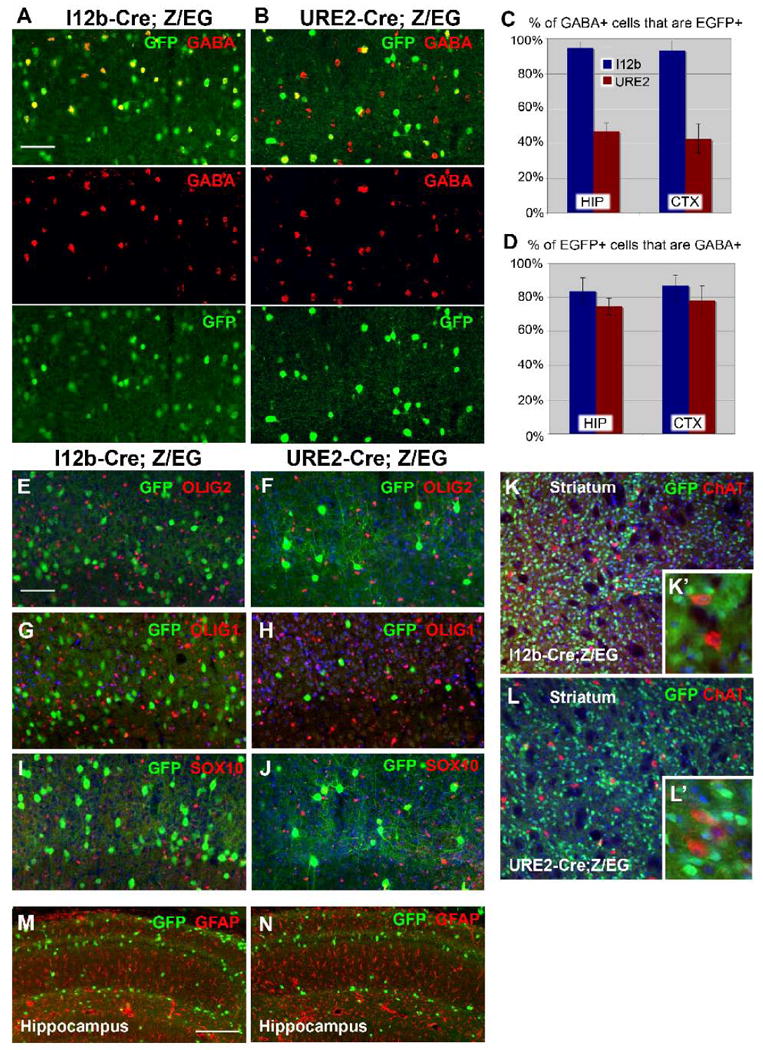
(A-B) Double labeling for EGFP (green) and GABA (red) in the somatosensory cortex of I12b-Cre;Z/EG (A) and URE2-Cre;Z/EG (B) mice.
(C) Graph of the percentage of GABA+ cells that are EGFP+ in the adult hippocampus (HIP) and somatosensory cortex (CTX). I12b-Cre;Z/EG in blue and URE2-Cre;Z/EG in red. Bars are mean ± SD (n=3).
(D) Graph of the percentage of EGFP+ cells that are GABA+ in the adult hippocampus (HIP) and somatosensory cortex (CTX). I12b-Cre;Z/EG in blue and URE2-Cre;Z/EG in red. Bars are mean ± SD (n=3).
(E-F, G-H, I-J) Double labeling for EGFP and the oligodendrocyte markers OLIG2 (E-F), OLIG1 (G-H), and SOX10 (I-J) in I12b-Cre;Z/EG (E,G,I) or URE2-Cre;Z/EG (F,H,J) cortex.
(K-L) Double labeling for EGFP and the cholinergic neuron marker choline acetyltransferase (ChAT) in I12b-Cre;Z/EG (K) and URE2-Cre;Z/EG (L) striatum. No co-expression was detected. The inserts depict higher magnification images of the striatum (K′, L′).
(M-N) Double labeling for EGFP and the astrocytic marker GFAP in I12b-Cre;Z/EG (M) or URE2-Cre;Z/EG (N) hippocampus.
Scale bars = 100 μm.
We next examined the percentage of EGFP+ cells that co-express GABA. In the cortex and hippocampus, approximately 85% and 75% of EGFP+ cells in I12b-Cre;Z/EG and URE2-Cre;Z/EG mice, respectively, expressed detectable levels of GABA. The lack of 100% overlap probably arises from a well-known limitation in GABA-detection (Pow, 1997).
Basal forebrain cholinergic neurons, which express choline acetyltransferase (ChAT), are generated in the MGE, septal area, and anterior POA (Furusho et al., 2006). To determine if I12b-Cre or URE2-Cre are active in this forebrain population, we double-labeled sections for ChAT and EGFP from adult I12b-Cre;Z/EG and URE2-Cre;Z/EG mice. Strongest ChAT immunoreactivity was detected in the striatum, septal nucleus, substantia innominata, diagonal band, and hypothalamus. In the adult striatum, EGFP was not co-localized with ChAT-expressing neurons in both I12b-Cre;Z/EG and URE2-Cre;Z/EG mice (Figure 6K and 6L) indicating that I12b and URE2 are not active in striatal cholinergic neurons. Interestingly, a small percentage of basal forebrain cholinergic neurons in the diagonal band, magnocellular preoptic area, substantia innominata, and hypothalamus co-labeled for EGFP and ChAT in i12b-Cre;Z/EG animals (n=3), while no co-localization of EGFP and ChAT was detected in similar regions in URE2-Cre;Z/EG animals (n=3) (Supplemental Figure S4). Thus, I12b, but not URE2, marks a subset of basal forebrain cholinergic neurons.
Progenitors within the mouse MGE and LGE give rise to both GABAergic neurons and oligodendrocytes (He et al., 2001; Petryniak et al., 2007; Yung et al., 2002). Transgenic mice that express Cre in the ventricular zone of the LGE (i.e. Gsh2-Cre) or MGE (i.e. Nkx2.1-Cre) label both GABAergic interneurons and oligodedendrocytes (Fogarty et al., 2007; Kessaris et al., 2006; Xu et al., 2008). We demonstrated that DLX2 is expressed in uncommitted progenitors of the MGE that generate both oligodendrocytes and GABAergic interneurons; continued expression of DLX restricts progenitors to a neuronal fate (Petryniak et al., 2007). Thus, we predict, unlike other transgenic mice that express CRE in the ganglionic eminences, that I12b-Cre and URE2-Cre would label GABAergic neurons, but not glia, if expression of Cre-recombinase occurs after the neuron-glia fate decision.
To determine whether i12b-Cre and URE2-Cre label oligodendrocytes in addition to GABAergic neurons, we examined the co-expression of EGFP and oligodendrocyte markers OLIG2, OLIG1, and SOX10 in the forebrain of I12b-Cre;Z/EG and URE2-Cre;Z/EG mice (Fig. 6E-6J). We detected less than 0.5% of co-expression between all oligodendrocyte-markers and EGFP in both adult and newborn I12b-Cre;Z/EG and URE2-Cre;Z/EG sections (Fig. 6E-6J and data not shown). It has been reported that Dlx2-lineage cells might give rise to perinatal astrocytes (Marshall and Goldman, 2002). To determine if I12b-lineage or URE2-lineage cells gave rise to forebrain astrocytes, we analyzed the expression of the astrocytic marker GFAP. Numerous GFAP-expressing cells were detected in the hippocampus and white matter of adult mice, while fewer GFAP+ cells were observed in the neocortex. We did not detect any overlap between EGFP and GFAP in the neonatal or adult hippocampus or neocortex of I12b-Cre;Z/EG or URE2-Cre;Z/EG mice (Fig. 6M and 6N and data not shown). Overall this data indicates that I12b-Cre and URE2-Cre are lineage-restricted to neurons within the telencephalon.
Given that DLX2 is expressed in multi-potent progenitors but I12b-Cre and URE2-Cre specifically label neurons but not glia, our data suggests that CRE is expressed after endogenous DLX2 and in progenitors committed to a neuronal fate. To address this directly, we analyzed the expression of DLX2 and CRE in the ganglionic eminence at E12.5, E15.5 and P0 of I12b-Cre and URE2-Cre animals (Fig. 7 and data not shown). Despite functional CRE activity in URE2-Cre animals (Figs. 1-5), we did not detect CRE protein by immunolabeling in URE2-Cre forebrain sections at these ages (data not shown), indicating CRE expression is low in URE2-Cre animals. In I12b-Cre forebrain sections, CRE was highly expressed within the ganglionic eminences and tangentially migrating immature interneurons (Fig. 7). Co-labeling showed numerous DLX2+ cells and few CRE+ cells within the VZ, DLX2/CRE co-expression at the VZ/SVZ border, and many more CRE+ than DLX2+ cells within the MZ. Thus, this supports a model that the neural/glial fate switch takes place either in the VZ or in the layer of the SVZ (SVZ1) that is rich in DLX2+ cells, but has few I12b-CRE+ cells.
Figure 7. DLX2 and CRE expression in embryonic and newborn I12b-Cre;Z/EG mice.
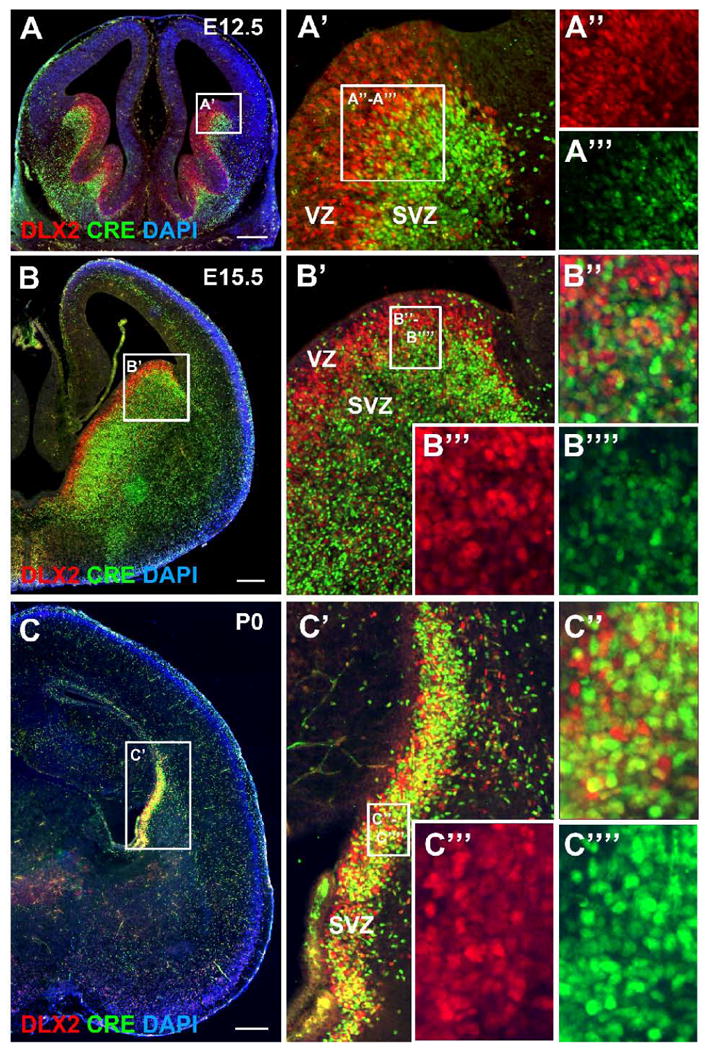
(A - C) Coronal forebrain sections from I12b-Cre;Z/EG animals at E12.5 (A), E15.5 (B), and P0 (C) were double-labeled for DLX2 (red), CRE (green) and counterstained for cell nuclei with DAPI (blue). The boxed regions in (A-C) are shown at higher magnification in (A′-C′). In A′-C′, the boxed regions are shown at higher magnification in A″-A‴, B″-B″″, and C″-C″″. DLX2-only expression is shown in A″, B‴, and C‴, CRE-only expression is shown in A‴, B″″, and C″″, and both DLX2 and CRE expression is shown in A′, B″ and C″. In contrast to CRE expression, DLX2 expression is high in the ventricular zone (VZ). CRE and DLX2 are co-expressed within the subventricular zone (SVZ) at all ages examined. Scale bars in A, B, C = 500 μm.
I12b-Cre and URE2-Cre mark different GABAergic interneuron populations
GABAergic interneurons are diverse and can be classified into distinct subtypes based upon molecular markers, electrophysiological properties, and connectivity (Gonchar, 2008; Markram et al., 2004). The majority of cortical interneurons can be classified by largely non-overlapping expression of PV, CR, and SOM (Gonchar, 2008; Gonchar and Burkhalter, 1997; Kubota and Kawaguchi, 1994; Miyoshi et al., 2007). Analysis of i12b-lacZ and URE2-lacZ transgenic mice suggested that i12b and URE2 were differentially active in cortical SOM, CB, and nNOS expressing neurons (Ghanem et al., 2007). To determine if GABAergic interneuron subtypes were differentially labeled by I12b-Cre or URE2-Cre, we analyzed the co-expression of EGFP and the GABAergic markers CB, CR, nNOS, NPY, PV, and SS in the adult somatosensory cortex and hippocampus of I12b-Cre;Z/EG and URE2-Cre;Z/EG mice (Fig. 8 and Fig. 10).
Figure 8. Co-expression of I12b-Cre and URE2-Cre lineage cells with GABAergic interneuron subtypes in the adult somatosensory cortex.
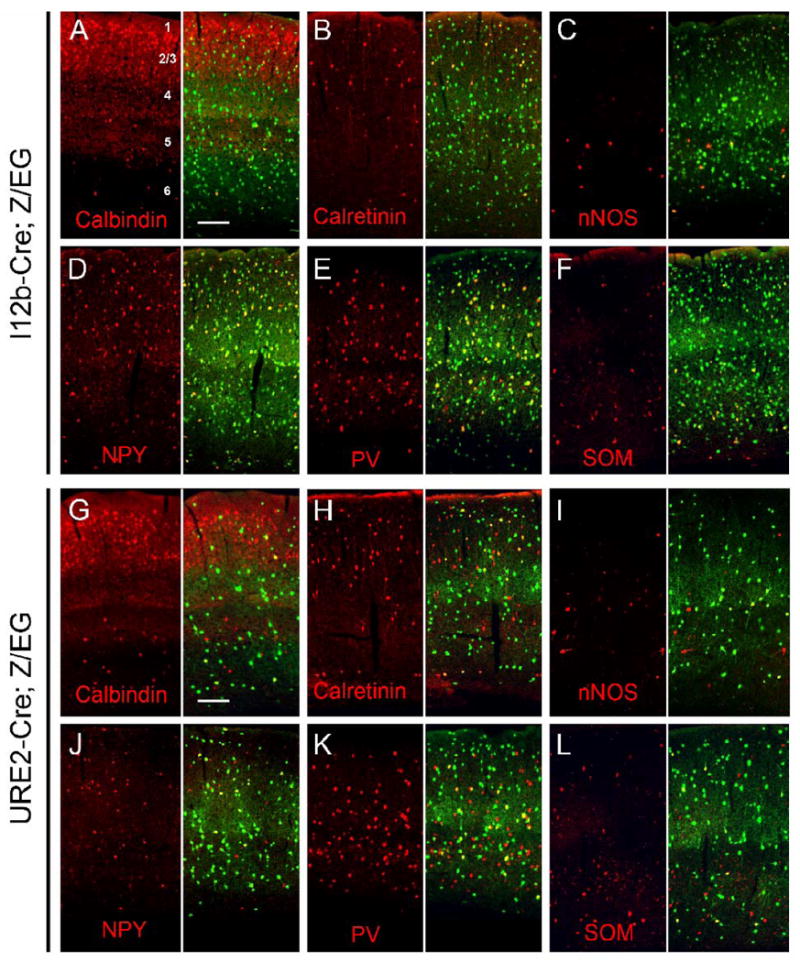
Representative images of double-labeling immunofluorescence with EGFP (green) and interneuron markers (red) in the adult somatosensory cortex of I12b-Cre;Z/EG (A-F) and URE2-Cre;Z/EG (G-L) mice. Quantification of co-expression is presented in Table 1.
(A, G) Calbindin; (B, H) Calretinin; (C,I) nNOS; (D,J) NPY; (E,K) Parvalbumin; (F,L) Somatostatin.
Scale bar= 150 μm.
Figure 10. Co-expression of I12b-Cre and URE2-Cre labeled cells with GABAergic interneuron subtypes in the adult hippocampus.

Representative images of double-labeling immunofluourescence with EGFP (green) and interneuron markers (red) in the adult hippocampus of I12b-Cre;Z/EG (A-F) and URE2-Cre;Z/EG (G-L) mice. Quantification of co-expression is presented in Table 2.
(A, G) Calbindin; (B, H) Calretinin; (C, I) nNOS; (D, J) NPY; (E, K) Parvalbumin; (F, L) Somatostatin.
Scale bar= 200 μm.
Somatosensory Cortex Labeling
GABAergic neuronal markers vary between cortical layers; thus we quantified the overlap in expression in layers 1, layers 2/3, layers 4, layer 5, and layer 6 (Fig. 8 and Table 1). CB was expressed in 80-93% of I12b-Cre labeled cells and 17-20% of URE2-Cre labeled cells in layers 5 and 6. Numerous CB+ cells were found in layers 2/3 and 4 where CB is expressed by both pyramidal and non-pyramidal cells (DeFelipe, 1997; DeFelipe and Jones, 1992; Demeulemeester et al., 1991). We found that CB was co-expressed with fewer I12b-Cre (∼3-11%) and URE2-Cre (∼1-2%) labeled cells in the superficial layers.
TABLE 1. Characterization of labeled cells in Adult Somatosensory Cortex.
Ratio of number of I12b-lineage to URE2-lineage cells in cortex: ∼2.3; ratios significantly different are highlighted
| Percentage of marker+ cells per layer that are EGFP+ | ||||||
|---|---|---|---|---|---|---|
| Mean % ± SD (n=3-4); Numbers in parenthesis are total number of marker+ cells counted; NA, not applicable. | ||||||
| I12b-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| Layer 1 | NA (0) | 61.7 ± 12.6 (20) | 100.0 ± 0.0 (1) | 67.1 ± 16.3 (27) | NA (0) | NA (0) |
| Layer 2/3 | 11.2 ± 3.5 (1852) | 70.0 ± 12.9 (174) | 65.5 ± 37.9 (23) | 86.6 ± 6.3 (253) | 78.7 ± 16.6 (198) | 92.2 ± 6.8 (160) |
| Layer 4 | 3.1 ± 3.6 (1246) | 82.3 ± 11.5 (111) | 100.0 ± 0.0 (5) | 88.3 ± 5.1 (132) | 83.6 ± 10.6 (355) | 92.5 ± 7.6 (155) |
| Layer 5 | 80.2 ± 1.6 (229) | 78.2 ± 13.4 (42) | 88.0 ± 12.5 (22) | 92.3 ± 4.0 (100) | 81.6 ± 14.0 (225) | 88.4 ± 7.4 (207) |
| Layer 6 | 93.1 ± 4.0 (211) | 58.2 ± 18.3 (59) | 90.5 ± 16.5 (42) | 85.6 ± 2.2 (185) | 74.3 ± 8.1 (292) | 86.8 ± 11.6 (315) |
| URE2-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| Layer 1 | NA (0) | 56.3 ± 8.8 (30) | 0.0 ± 0.0 (1) | 23.3 ± 40.4 (22) | NA (0) | NA (0) |
| Layer 2/3 | 1.7 ± 0.8 (2291) | 33.8 ± 3.1 (312) | 14.8 ± 17.0 (15) | 32.8 ± 5.8 (255) | 19.5 ± 9.0 (155) | 25.4 ± 4.7 (145) |
| Layer 4 | 1.0 ± 1.8 (911) | 31.5 ± 2.5 (147) | 46.7 ± 50.3 (14) | 39.3 ± 14.6 (130) | 20.1 ± 3.1 (310) | 30.1 ± 11.0 (108) |
| Layer 5 | 16.8 ± 2.8 (258) | 24.4 ± 0.8 (75) | 6.9 ± 6.4 (32) | 25.6 ± 9.8 (95) | 25.9 ± 8.5 (353) | 25.4 ± 1.7 (139) |
| Layer 6 | 19.5 ± 5.3 (248) | 41.7 ± 11.8 (99) | 21.4 ± 18.5 (51) | 24.8 ± 14.3 (159) | 23.0 ± 4.9 (347) | 20.4 ± 6.2 (247) |
| Percentage of EGFP+ cells per layer that are marker+ | ||||||
| Mean % ± SD (n=3-4); Numbers in parenthesis are total number of EGFP+ cells counted; NA, not applicable. | ||||||
| I12b-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| Layer 1 | NA (256) | 8.6 ± 2.8 (140) | 0.4 ± 0.6 (363) | 7.3 ± 3.2 (249) | NA (207) | NA (313) |
| Layer 2/3 | 19.2 ± 1.1 (1082) | 17.1 ± 4.4 (714) | 1.7 ± 1.5 (1327) | 14.6 ± 7.3 (1508) | 18.4 ± 5.3 (847) | 15.1 ± 6.0 (975) |
| Layer 4 | 2.5 ± 0.9 (1546) | 16.1 ± 1.7 (568) | 0.6 ± 0.6 (841) | 12.9 ± 1.4 (910) | 39.8 ± 4.3 (744) | 17.6 ± 3.0 (812) |
| Layer 5 | 19.3 ± 2.3 (951) | 5.8 ± 1.4 (571) | 1.8 ± 0.8 (1207) | 8.3 ± 2.5 (1117) | 32.2 ± 11 (570) | 20.2 ± 6.3 (906) |
| Layer 6 | 23.5 ± 5.9 (837) | 4.9 ± 2.6 (689) | 4.2 ± 1.0 (1010) | 13.7 ± 4.3 (1154) | 22.1 ± 3.2 (982) | 20.6 ± 6.6 (1327) |
| URE2-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| Layer 1 | NA (89) | 16.8 ± 3.5 (67) | 0.0 ± 0.0 (98) | 4.0 ± 6.8 (129) | NA (89) | NA (116) |
| Layer 2/3 | 7.6 ± 2.4 (508) | 19.7 ± 3.4 (646) | 0.3 ± 0.2 (586) | 15.5 ± 4.2 (538) | 7.5 ± 0.9 (405) | 9.0 ± 4.5 (411) |
| Layer 4 | 1.3 ± 2.3 (709) | 13.6 ± 1.0 (384) | 0.8 ± 0.9 (334) | 13.8 ± 3.6 (369) | 20.5 ± 1.1 (304) | 7.9 ± 3.2 (411) |
| Layer 5 | 9.8 ± 2.1 (442) | 7.1 ± 1.7 (240) | 0.4 ± 0.3 (527) | 5.2 ± 1.0 (465) | 28.1 ± 9.5 (326) | 9.4 ± 2.3 (375) |
| Layer 6 | 12.7 ± 4.9 (379) | 7.2 ± 0.6 (138) | 2.3 ± 2.3 (302) | 9.4 ± 2.9 (420) | 21.0 ± 6.1 (380) | 13.4 ± 2.6 (375) |
CR, nNOS, NPY, PV, and SS were co-expressed in both I12b-Cre and URE2-Cre lineage cells; however less co-expression was detected for URE2-Cre labeled cells consistent with the ∼2.3-fold fewer interneurons labeled by URE2-Cre (Table 1). Nevertheless, in some instances, especially for CR, SOM and PV, the labeling ratio was substantially different than 2.3-fold suggesting different activities of I12b-Cre and URE2-Cre in these interneuron subtypes. For CR in layers 1 and 6, the I12b-Cre/URE2-Cre labeling ratio was closer to 1.0; for instance, in layer 1, the ratio was ∼1.1 (Table 1). On the other hand, for PV, SOM, CB, and nNOS, in most layers, the ratio was ∼3-4; for example, for PV in layer 1, the ratio was ∼4, and was ∼4.2 for SOM in layer 6. The labeling ratio of NPY did not show a trend away from 2.3 except in the deep layers where the ratio was biased towards I12b-Cre.
We next quantified the percentage of EGFP+ cells that co-express each GABAergic marker as an indication of the proportion of EGFP+ cells per cortical layer that are represented by each GABAergic subtype (Table 1). This analysis showed a similar trend in which URE2-Cre preferentially correlated with CR (in layers 1, 2/3, 5 and 6) but not with PV (in layers 2/3 and 4) or SOM (in all layers). Interestingly, many of the EGFP+ cells in layer 1 (∼80%) were not identified using the GABAergic markers in this study; we do not have an explanation for this. In sum, I12b-Cre labels the vast majority of cortical local circuit neurons and URE2-Cre labels ∼2.3-fold fewer, which in particular layers, are biased towards expressing CR and not expressing PV, SOM, CB and nNOS.
Since EGFP marks cells and their progeny in which CRE was expressed at any time point in their life, it does not directly indicate when I12b or URE2 enhancer elements are active in Cre;Z/EG mice. To determine when I12b or URE2 are active during neurogenesis, we analyzed embryonic and adult expression of CRE and EGFP in I12b-Cre;Z/EG and URE2-Cre;Z/EG animals. CRE expression in URE2-Cre mice was below the level of detection (data not shown). At E12.5 and E15.5 in I12b-Cre;Z/EG, CRE was co-expressed in the majority of EGFP+ migrating immature neurons (Fig. 9A-9E). Thus, I12b activity is maintained during tangential migration. Within the ganglionic eminences, CRE-only expressing cells were detected in the SVZ, while most cells in the MZ co-expressed EGFP and CRE. This pattern indicates that CRE expression precedes reporter expression, immature interneurons rapidly migrate from their site of origin, and I12b remains active during migration. In the adult olfactory bulb, CRE was expressed by migrating neuroblasts in the SVZ, granule cell neurons, and periglomerular neurons (Fig. 9F). In the adult somatosensory cortex, interneurons continued to express CRE, however the presence of EGFP+/CRE- cells indicates that I12b activity is downregulated in a subset of adult interneurons (Fig. 9G). In contrast, most striatal neurons express only EGFP, indicating that I12b is no longer active in this adult population. Finally, we detect CRE+ cells within the neurogenic niche of SVZ cells that line the lateral ventricles and give rise to olfactory bulb interneurons (Fig. 9H).
Hippocampus Labeling
PV, CR, and CB expression labels largely non-overlapping subpopulations of GABAergic neurons in the adult rodent hippocampus (Freund and Buzsaki, 1996; Jinno and Kosaka, 2006). PV labels basket and axoaxonic interneurons, CB is expressed in both interneurons and glutamatergic granule cells in the DG and some CA1 pyramidal neurons, and CR+ neurons can be divided into spiny and aspiny local circuit neurons, which innervate the dendrites of principal cells and other GABAergic interneurons, respectively (Danglot et al., 2006).
To determine the GABAergic neuron subtypes labeled by I12b-Cre and URE2-Cre within the adult hippocampus, we compared the expression of PV, CR, CB, nNOS, NPY, PV, and SOM to EGFP expression in I12b-Cre;Z/EG and URE2-Cre;Z/EG mice (Fig. 10 and Table 2). We quantified three regions of the hippocampus: DG, CA3 and CA1/2 (see Fig. 10A and Fig. 5D). Similar to the co-expression analysis in the somatosensory cortex, a higher percentage of the GABAergic markers nNOS, NPY, PV, and SOM were co-labeled with EGFP in I12b-Cre;Z/EG animals (78-93%) compared to URE2-Cre;Z/EG mice (17-41%) consistent with the ∼1.6-fold fewer interneurons labeled by URE2-Cre (Table 2). However, in some instances, especially for PV, SOM and nNOS, the labeling ratio was substantially different than 1.6-fold suggesting different activities of I12b-Cre and URE2-Cre in these interneurons subtypes. For PV, SOM, and nNOS, the ratio was ∼3-5.5 in most regions. For example, the ratio was ∼5.5 for PV in DG, and ∼3.2 for SOM in CA3.
TABLE 2. Characterization of Labeled Cells in Adult Hippocampus.
Ratio of number of I12b-lineage to URE2-lineage cells in the hippocampus: ∼1.6; ratios significantly different are highlighted
| Percentage of marker+ cells per region that are EGFP+ | ||||||
|---|---|---|---|---|---|---|
| Mean % ± SD (n=3-4); Numbers in parenthesis are total number of marker+ cells counted. | ||||||
| I12b-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| DG | 2.1 ± 1.1 (1118) | 17.3 ± 7.1 (210) | 86.2 ± 4.4 (198) | 85.2 ± 0.7 (169) | 92.7 ± 7.1 (45) | 81.9 ± 14.3 (174) |
| CA3 | 74.3 ± 9.0 (161) | 31.9 ± 12.2 (98) | 81.9 ± 11.9 (164) | 80.1 ± 14.3 (182) | 78.3 ± 7.0 (216) | 83.3 ± 10.3 (272) |
| CA1/2 | 24.7 ± 4.7 (861) | 47.9 ± 12.9 (191) | 81.9 ± 10.3 (329) | 85.3 ± 12.8 (347) | 84.3 ± 6.8 (193) | 86.9 ± 11.8 (278) |
| URE2-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| DG | 1.7 ±1.3 (1408) | 12.8 ±3.2 (308) | 22.5 ± 3.5 (119) | 37.0 ± 15.5 (233) | 16.9 ± 3.9 (51) | 26.6 ± 9.3 (149) |
| CA3 | 25.6 ± 4.7 (202) | 24.9 ± 2.5 (185) | 30.7 ± 6.2 (144) | 40.5 ± 18.0 (257) | 22.3 ±7.3 (210) | 22.4 ± 11.0 (162) |
| CA1/2 | 13.1 ± 5.4 (1011) | 28.4 ± 2.4 (322) | 23.5 ± 5.9 (272) | 37.8 ± 16.2 (408) | 17.6 ± 10.6 (240) | 29.6 ± 7.0 (239) |
| Percentage of EGFP+ cells per region that are marker+ | ||||||
| Mean % ± SD (n=3-4); Numbers in parenthesis are total number of EGFP+ cells counted. | ||||||
| I12b-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| DG | 6.2 ± 0.7 (383) | 9.3 ± 1.7 (392) | 37.2 ± 11.5 (459) | 31.1 ± 0.6 (463) | 10.8 ± 1.2 (386) | 31.2 ± 3.8 (456) |
| CA3 | 19.6 ± 0.5 (609) | 4.5 ± 0.7 (691) | 18.8 ± 5.8 (716) | 22.9 ± 0.6 (635) | 28.8 ± 5.9 (587) | 27.7 ± 4.9 (817) |
| CA1/2 | 20.4 ± 4.4 (1043) | 11.9 ± 1.8 (765) | 25.4 ± 7.0 (1060) | 23.5 ± 1.8 (1260) | 16.0 ± 6.3 (1015) | 20.1 ± 3.4 (1200) |
| URE2-Cre; Z/EG | ||||||
| Calbindin | Calretinin | nNOS | NPY | Parvalbumin | Somatostatin | |
| DG | 10.9 ± 7.7 (221) | 18.4 ± 6.6 (213) | 13.6 ± 0.7 (196) | 26.7 ± 8.6 (323) | 3.8 ± 1.0 (229) | 11.9 ± 7.4 (333) |
| CA3 | 16.3 ± 9.0 (317) | 13.0 ± 0.8 (354) | 13.2 ± 6.8 (336) | 24.9 ± 12.3 (418) | 12.4 ± 6.6 (378) | 7.1 ± 1.2 (507) |
| CA1/2 | 17.7 ± 1.7 (750) | 13.8 ± 3.1 (664) | 10.5 ± 1.4 (607) | 19.2 ± 8.0 (802) | 5.4 ± 0.9 (781) | 8.8 ± 2.7 (797) |
We next quantified the percentage of EGFP+ cells within each region of the hippocampus that co-expressed the interneuron markers as an indication of the subtype preferences within the i12b-Cre and URE2-Cre lineages (Table 2). This analysis showed a similar trend with URE2-Cre labeled cells preferentially correlated with CR but not with PV and SOM. The percentage of EGFP+ cells that co-expressed CR in the DG and CA3 was the only interneuron marker that was greater from the URE2-Cre lineage.
Post-mitotic EGFP+ neurons destined for the olfactory bulb, hippocampus, and cortex are generated with different time courses in I12b-Cre;Z/EG and URE2-Cre;Z/EG mice
Peak neurogenesis of cortical and hippocampal GABAergic interneurons occurs within the ganglionic eminences between E12 and E15 in the mouse (Danglot et al., 2006; Flames and Marin, 2005; Rymar and Sadikot, 2007). Olfactory bulb interneurons are generated by at least E12.5, and neurogenesis continues after birth (Batista-Brito et al., 2008; Tucker et al., 2006). We analyzed the birthdate of EGFP+ neurons by intraperitoneal administration of the thymidine analogs CldU or IdU to pregnant I12b-Cre;Z/EG or URE2-Cre;Z/EG dams at 12.5, 13.5, 14.5, or 15.5 days gestation. Newborn pups (P0) were collected and quantified for co-expression of EGFP and either CldU or IdU (hereafter referred as XdU). When delivered in equimolar concentrations, CldU and IdU label equivalent populations of cells during the S phase of DNA replication (Vega and Peterson, 2005). At P0, EGFP+ cells in I12b-Cre;Z/EG and URE2-Cre;Z/EG are detected in the granule cell layer, mitral cell layer, external plexiform layer (EPL) and the glomerular layer (GL) of the olfactory bulb (Fig. 4 and 11A). In the olfactory bulb at P0, administration of XdU at E12.5 strongly labeled many cells in the GCL and the MCL, but few cells in the GL. Interestingly, XdU injection at E13.5 generated XdU+ cells in the GL layer, suggesting that many glomerular cells are first generated around E13 in the mouse. XdU administered at E14.5 and E15.5 labeled cells within the GCL, MCL, and GL. The percentage of olfactory bulb EGFP+ cells that were becoming post-mitotic between E12.5 and E15.5 was relatively constant in I12b-Cre;Z/EG mice (∼20%), while differentiation increased from ∼10% at E12.5 to ∼20% at E15.5 in URE2-Cre;Z/EG animals (Fig. 11D). Significantly more EGFP+ cells were born at E12.5 and E14.5 in I12b-Cre versus URE2-Cre lineages.
Figure 11. Birthdate analysis of EGFP+ neurons in the olfactory bulb, hippocampus and cortex of I12b-Cre;Z/EG and URE2-Cre;Z/EG mice.
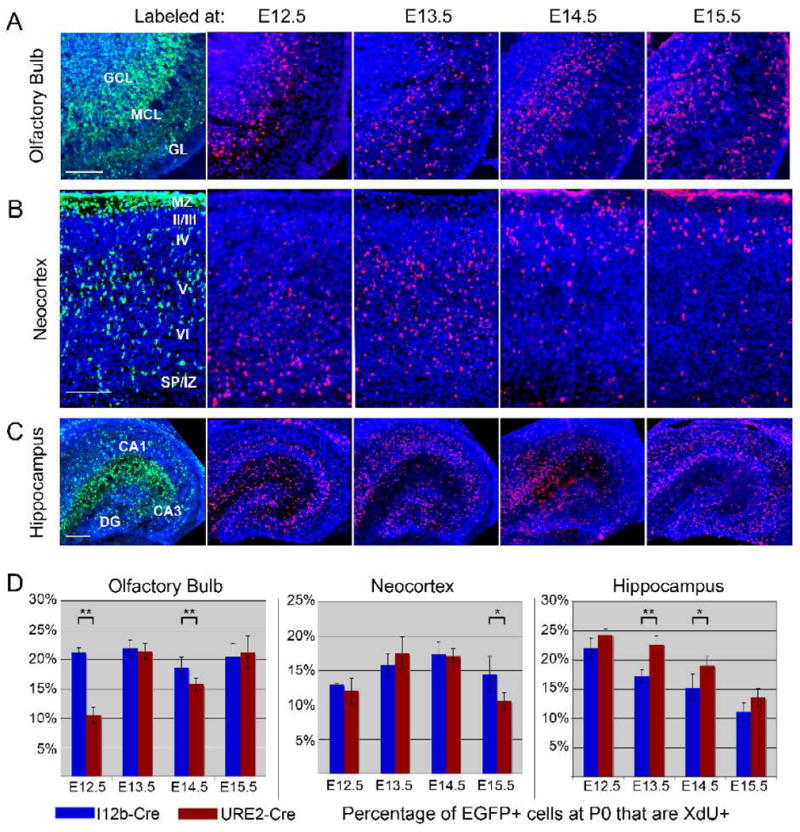
(A-C) Pregnant I12b-Cre;Z/EG and URE2-Cre;Z/EG dams were treated with thymidine analogs CldU or IdU (collectively labeled as XdU) at gestational day 12.5, 13.5, 14.5, or 15.5. Pups were collected at birth and coronal sections through the olfactory bulb (A), neocortex (B) and hippocampus (C) were double labeled for EGFP (green) and XdU (red). A representative image of EGFP expression is shown in the first column and XdU-labeling of each treatment timepoint shown in the right four columns.
(D) Quantification of the percentage of EGFP+ cells that co-expressed XdU at P0 in the olfactory bulb (left graph), neocortex (middle graph), and hippocampus (right graph). At least 3 coronal sections from 3-4 animals were analyzed. Each bar represents the mean and error bars are the SEM. * p < 0.04; ** p < 0.008.
Scale bar in A, B = 100 μm; C = 200 μm.
Within the neocortex, projection neurons are generated in an inside-out pattern with earlier born neurons populating deeper layers and later-born neurons residing in more superficial layers. In contrast, the determinants of GABAergic interneuron position within the cortex depend upon birthdate and GABAergic interneuron subtype. For example, while PV+ interneurons follow an inside-out gradient with their birthdates isochronous to cortical projection neurons, CR+ interneurons populate the cortex with an outside-in gradient and are born heterochronically with respect to nearby projection neurons (Rymar and Sadikot, 2007). At PO, while EGFP+ cells from I12b-Cre;Z/EG and URE2-Cre;Z/EG are detected in all neocortical layers, most cells are found in the marginal zone (MZ), layer V, and near the subplate (SP) (Fig. 4 and 11B). A large fraction of EGFP+ cells are likely still migrating to their final laminar position as the distribution of EGFP+ cells in the PO and adult somatosensory cortex is different (Fig. 5C). Interneuron subtype identity appears to be largely specified before they start their tangential migration (Xu et al., 2005); therefore, we analyzed the percentage of EGFP+ cells that are XdU positive in the neocortex as an indication of the birthdate of neocortical GABAergic interneurons generated in I12b-Cre;Z/EG and URE2-Cre;Z/EG mice. XdU labeling showed an inside-out pattern, with E12.5-labeled cells near the SP and E15.5-labeled cells near the MZ and layer II/III (Fig. 11B). Birthdate analysis showed that both I12b-Cre and URE2-Cre labeled cortical cells showed a peak in neurogenesis around E14 (Fig. 11D). The percentage of EGFP+ cells that were born between E12.5 and E14.5 was similar between I12b-Cre and URE2-Cre, while at E15.5 there were significantly more I12b-Cre lineage cells born. Both Cre lines showed a trend towards reduced neurogenesis by E15.5, although our data indicate that the rate of this decrease might be slower in the I12b-Cre lineage.
The production of CA1-3 and DG interneurons is though to occur in the CGE and MGE between E12 and E15, about 2-3 days before the production of glutamatergic pyramidal cells (Danglot et al., 2006). To determine if the birthdate of hippocampal neurons within I12b-Cre and URE2-Cre lineage cells differed, we examined the co-expression of EGFP and XdU in I12b-Cre;Z/EG and URE2-Cre;Z/EG hippocampus at birth (Fig. 11C, 11D). EGFP+ cells from both I12b-Cre and URE2-Cre were present in the DG, CA3 and CA1 regions. The highest concentration of EGFP+ cells was present within the SVZ and the inner MZ (future stratum radiatum), consistent with previous analysis of GAD67-GFP mice (Manent et al., 2006). Birthdate analysis between E12.5 and E14.5 revealed XdU+ cells were localized to all regions of the newborn hippocampus including the inner MZ (Fig. 11C). However, few XdU+ cells born at E15.5 were detected in the inner MZ, suggesting that most interneurons were already generated by this time. In support of this idea, co-expression of EGFP and XdU at birth for both I12b-Cre;Z/EG and URE2-Cre;Z/EG animals was highest for neurons that became postmitotic at E12.5 (∼22%) and progressively decreased over time (∼12% at E15.5 treatment) (Fig. 11D). Significantly more hippocampal EGFP+ cells from the URE2-Cre lineage were postmitotic at E13.5 and E14.5 compared to the I12b-Cre lineage, indicating that a greater proportion of URE2-Cre lineage cells are born at those time points.
DLX2 directly binds I12b and URE2 in vivo and DLX1&2 are necessary for I12b and URE2 activity
Both I12b and URE2 contain putative DLX DNA binding sites: TAATT/AATTA (Fig. 12A). To test whether DLX transcription factors bound to I12b or URE2, we assayed for direct binding to I12b and URE2 by DLX2 in vivo using a chromatin immunoprecipitation (ChIP) assay. E15.5 ganglionic eminences from wildtype mice were treated with formaldehyde to cross-link protein-DNA complexes. Soluble nucleoprotein complexes were immunoprecipitated using anti-DLX2 antibodies. We then used PCR to amplify the precipitated chromatin for potential DLX binding regions in the I12b and URE2 elements (Fig. 12A-B). The Dlx5/6i enhancer element contains a DLX binding motif and has been shown to bind DLX2 in vivo (Zhou et al., 2004). As a positive control, we confirmed that Dlx5/6i was detected in our ChIP analysis (Fig. 12B). Using the same conditions and chromatin preparations, we assayed whether DLX2 binds directly to I12b or URE2. Within I12b, an ultra-conserved (>95% identity between all vertebrate species) potential DLX binding motif was chosen for analysis. Three ultra-conserved regions of URE2 contain putative DLX binding sites. We chose to assay one of these regions (URE2a) because of its strong homology to the DLX2 binding domain in Dlx5/6i (Fig. 12A). By ChIP analysis, we detected that both I12b and URE2 were directly bound to DLX2 in vivo. Control ChIP assays were negative (no antibody or with anti-DLX2 antibodies followed by PCR to irrelevant genomic loci, such as the Opsin promoter region or an enhancer region upstream of the progesterone receptor [ERC9a]).
Figure 12. DLX2 binds directly to I12b and URE2 in vivo and DLX1&2 function is necessary for I12b and URE2 activity.
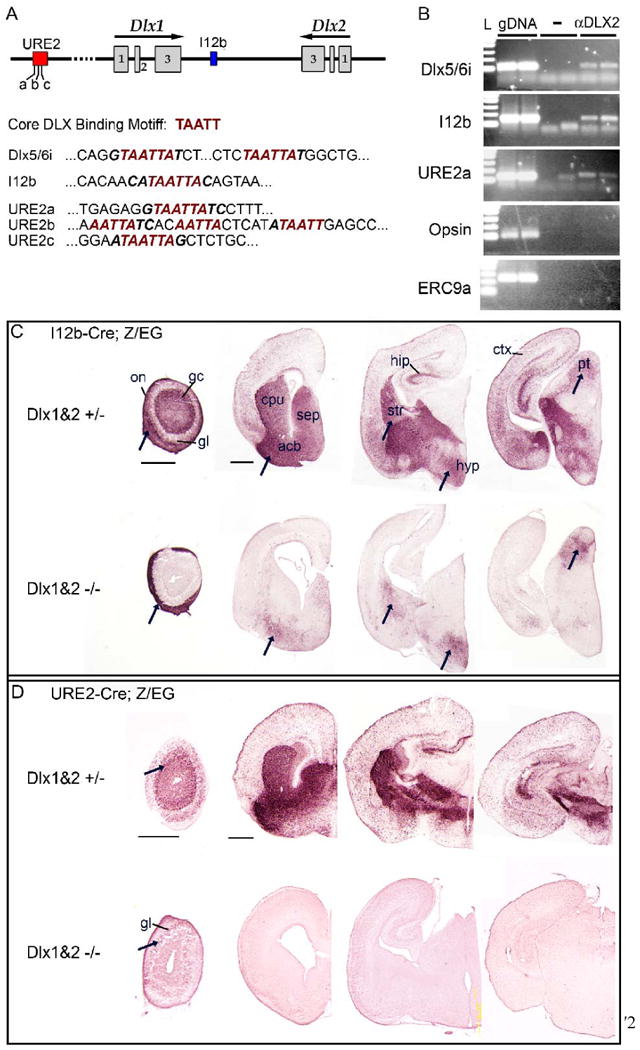
(A) Schematic of the Dlx1 and Dlx2 genomic locus showing the location of I12b and URE2 regulatory elements; the three putative DLX binding sites within URE2 are labeled (a-c). The core binding motif for DLX transcription factors is predicted to be TAATT. Dlx5/6i is a regulatory element in the intergenic region between Dlx5 and Dlx6 known to bind DLX1 and DLX2 in vivo. The DLX binding motif is present in Dlx5/6i, I12b, and URE2 enhancer elements (red italics).
(B) Chromatin immunoprecipitation (ChIP) assays from E15.5 ganglion eminences using DLX2-specific antisera (aDLX2) shows that DLX2 binds directly to Dlx5/6i (top panel) and also to I12b (second row) and URE2 (third row) in vivo. Control assays without antibody or with DLX2 antisera followed by PCR to irrelevant genomic loci (Opsin promoter region or ERC9a, an enhancer region upstream of the progesterone receptor), were negative. L, 100 bp DNA ladder; gDNA, genomic DNA; -, no antibody.
(C) I12b activity was greatly diminished in Dlx1&2 mutants (Dlx1&2-/-) with some notable exceptions (arrows). I12b-Cre;Z/EG animals were bred with Dlx1&2 mutant mice to generate Dlx1&2 null mutants that were transgenic for I12b-Cre and Z/EG. EGFP expression was visualized in coronal brain sections from control (top row) or Dlx1&2-/- mutants (bottom row) at P0.
(D) URE2 activity is nearly abolished in Dlx1&2 mutants. URE2-Cre;Z/EG animals were bred with Dlx1&2 mutant mice to generate Dlx1&2 null mutants that were transgenic for URE2-Cre and the EGFP reporter. EGFP expression was visualized in coronal forebrain sections from control (top row) or DLX1&2-/- mutants (bottom row) at P0. Expression was lost throughout the brain except in a few cells within the glomerular layer of the olfactory bulb (arrows).
acb, accumbens; cpu, caudate-putamen; ctx, cortex; gc, granule cell layer; gl, glomerular layer; hip, hippocampus; hyp, hypothalamus; on, olfactory nerve layer; pt, pretectal nucleus; sep, septum.
Scale bars = 500 μm.
We next determined whether DLX proteins are required for I12b and URE2 activity in vivo. Because DLX1 and DLX2 are functionally redundant during embryonic development (Anderson et al., 1997a; Anderson et al., 1997b; Panganiban and Rubenstein, 2002), we analyzed I12b and URE2 activity in mice that were null for Dlx1 and Dlx2 (Dlx1&2-/-). We generated Dlx1&2-/- and Dlx1&2+/- (as controls) mice that were transgenic for I12b-Cre;Z/EG and URE2-Cre;Z/EG. Since Dlx1&2 mutants die at birth, we assayed I12b or URE2 activity using immunohistochemistry for EGFP at P0.
In the absence of Dlx1&2, both I12b and URE2 activity was greatly reduced (Fig. 12C, 12D). In Dlx1&2-/-, EGFP expression in I12b-Cre;Z/EG olfactory bulb was lost in the granule cell and glomerular layers, but was present in the olfactory sensory neuron axons, consistent with the known functions of Dlx1&2 in the generation of olfactory bulb interneurons but not for the development of olfactory neurons (Long et al., 2007). EGFP expression was greatly diminished in the cortex, hippocampus, septum, and rostral striatum, but maintained at lower levels in the caudal striatum, hypothalamus, and pretectal nucleus (the latter is a site not known to express Dlx1 and Dlx2). Interestingly, these regions maintain Dlx5 or Dlx6 expression in Dlx1&2-/-, suggesting that Dlx5 and/or Dlx6 are maintaining I12b expression in these regions (Anderson et al., 1997b; Long et al., 2007). We also note that some of these regions are domains that show lower URE2-Cre activity: septum, ventral striatum, pallidum, and anterior hypothalamus (Fig. 4 and 12).
In the absence of Dlx1&2, EGFP expression in URE2-Cre;Z/EG mice was almost completely abolished, except for a weak signal within the glomerular cell layer of the olfactory bulb. Thus, URE2 activity appears to be completely dependent upon Dlx1&2 function.
Discussion
DLX proteins maintain their own expression in the embryonic forebrain
Forebrain GABAergic neuron production relies upon several mechanisms, including Shh-signaling and the Gsh1/Gsh2 and Nkx2.1 transcription factors to delineate progenitor identity (Sussel et al., 1999; Toresson and Campbell, 2001; Xu et al., 2005; Yun et al., 2003) and Mash1 and Dlx transcription factors to direct GABAergic neuron differentiation (Casarosa et al., 1999; Long et al., 2007; Long et al., 2008; Stühmer et al., 2002a; Yun et al., 2002; Yun et al., 2003). The molecular mechanisms that promote GABAergic neuron migration, subtype specification, and survival are under active investigation, but it is likely that continuous DLX expression is necessary for all of these processes. During GABAergic differentiation a sequential cascade of Dlx expression is induced and maintained; Dlx2 followed by Dlx1, Dlx5 and Dlx6 (Liu et al., 1997). Dlx genes operate singly or in combination with each other to regulate distinct functions. Dlx1&2-/- mutants have reduced GAD expression, a block in tangential migration, abnormal neurite morphogenesis and decreased neuronal survival (Anderson et al., 1997a; Anderson et al., 1997b; Cobos et al., 2007; Long et al., 2007; Long et al., 2008); Dlx1-/-; Dlx2+/- mutants exhibit defects in laminar position of neonatal cortical interneurons (Cobos et al., 2007); Dlx1-/- mutants have selective postnatal loss of a subset of cortical interneurons (Cobos et al., 2005); Dlx5-/- mutants exhibit a global defect in the differentiation of olfactory bulb interneurons (Levi et al., 2003; Long et al., 2003; Perera et al., 2004).
We show that I12b and URE2 drive Cre-recombinase transgene expression in a pattern closely resembling Dlx1 and Dlx2 expression in forebrain regions that generate GABAergic neurons, and that nearly all Cre-lineage cells are GABAergic neurons. Our data provide evidence that the DLX proteins regulate Dlx1 and Dlx2 telencephalic expression using the I12b and URE2 regulatory elements. URE2 activity appears to depend entirely on Dlx1&2 function, whereas a subset of regions maintain I12b activity in the Dlx1&2-/- mutants (Fig. 12). These Dlx1&2-independent regions maintain Dlx5&6 expression in Dlx1&2-/- mutants (Anderson et al., 1997b; Long et al., 2008; Zerucha et al., 2000), suggesting that Dlx5 and/or Dlx6 might activate I12b in these regions.
We find that DLX2 binds directly to I12b and URE2 (Fig. 12B), and that the activity of these elements depends on Dlx1&2 function (Fig. 12C and 12D). Thus, once DLX1 and/or DLX2 become expressed in the VZ we predict that they maintain their expression by binding to I12b and/or URE2, and thereby direct the progenitors towards a GABAergic neuron fate. Lack of I12b-Cre and URE2-Cre activity within the VZ suggests the existence of other regulatory elements that control Dlx1&2 expression in VZ progenitors. This is based in part on the observation that Dlx1&2-/- mutants continue to express low levels of Dlx1 and Dlx2 RNA in the subpallial progenitor zones (Anderson et al., 1997b; Long et al., 2008; Zerucha et al., 2000).
DLX1 and DLX2 expression is initiated in a subset of cells in the VZ (Eisenstat et al., 1999; Yun et al., 2002), whereas I12b and URE2 activity is first detected in the SVZ (Fig. 2 and 7). Fate mapping with I12b-Cre and URE2-Cre only labels neuronal descendents (Fig. 6); thus, the neuron/glial fate switch must occur in the VZ or early SVZ (SVZ1). In support of this idea, transgenic lines such as Nkx2.1-Cre, Gsh2-Cre, Mash1-Cre, or Olig2-Cre label both oligodendrocytes and neurons because of early expression of Cre within multipotent VZ progenitors (Fogarty et al., 2007; Kessaris et al., 2006; Kim et al., 2008; Miyoshi et al., 2007; Xu et al., 2008). As a consequence, I12b-Cre and URE2-Cre transgenic mice will be extremely useful for further lineage-analysis or conditional knock-out studies of forebrain GABAergic neurons.
Temporal regulation in the generation of GABAergic interneurons
GABAergic interneurons generated within the ganglionic eminences populate all regions of the forebrain, including the olfactory bulb, cortex, and hippocampus. Here we provide further evidence using lineage-tracing with floxed EGFP reporter mice and birth-date analysis with thymidine analogs that GABAergic neurons destined for the olfactory bulb, cortex, or hippocampus are born with different time-courses. Analysis of i12b-Cre;Z/EG animals at birth, in which nearly all cortical, hippocampal, and olfactory bulb interneurons are labeled, reveals that olfactory bulb interneurons are born at a steady rate, while the birthdates of cortical interneurons peak around E14 and hippocampal interneurons birthdates steadily decline during embryogenesis. In the case of cortical GABAergic interneurons, both birth date and progenitor zone location are important determinants of GABAergic subtype (Flames et al., 2007; Fogarty et al., 2007; Rymar and Sadikot, 2007; Xu et al., 2008).
Our birth-dating analysis suggests that the progenitor populations destined for the hippocampus or cortex are undergoing differentiation at different rates, with a larger population of hippocampal interneuron progenitors becoming post-mitotic before cortical interneuron progenitors. The influence of these different kinetics upon distribution of GABAergic subtypes to these two regions remains to be explored; nevertheless, our lineage-tracing analysis of I12b-Cre;Z/EG animals suggests that the distribution of GABAergic interneuron subtypes differ between the hippocampus and cortex. For example, the number of nNOS+ interneurons is ∼6 times greater in the hippocampus compared to the somatosensory cortex (Table 1 and Table 2).
The production of GABAergic interneuron subtypes that reside in the rodent olfactory bulb has recently been shown to be under temporal control. Using the I12b enhancer to drive Cre-ER (a tamoxifen-inducible form of Cre-recombinase) expression in transgenic mice, Batista-Brito et al. observed that at least seven subtypes of olfactory bulb interneurons are generated in the I12b-lineage in a unique temporal pattern (Batista-Brito et al., 2008). For example, the majority of tyrosine-hydroxylase and CB expressing interneurons and Blanes cells are produced embryonically, while CR+ and PV+ interneurons are generated mostly after birth. Batista-Brito et al. further suggest that temporally distinct neurogenic niches give rise to olfactory bulb interneuron diversity. While our analysis of I12b-Cre lineage cells did not include the olfactory bulb, our data indicates that I12b-Cre and URE2-Cre mark a diverse set of hipppocampal and cortical interneurons (Table 1 and Table 2). Thus, in light of the current studies of the olfactory bulb and prior analysis of the cortex (Flames and Marin, 2005; Fogarty et al., 2007) and hippocampus (Danglot et al., 2006), we propose that subtypes of I12b-lineage interneurons that reside in the hippocampus and neocortex are generated from restricted spatial and age-dependant neurogenic niches. Indeed, the neurogenic niche labeled by DLX2 and I12b changes over time, as localization of DLX2 and I12b-Cre expression contracts from the progenitor zones of the ganglionic eminences embryonically to the SVZ that lines the lateral ventricles postnatally (Fig. 7 and Fig. 9H).
Lastly, it is currently unknown how progenitors born at similar times and in similar locations within the MGE or LGE/CGE choose to become either cortical or hippocampal interneurons. Two possible mechanisms are: 1) distinct and separable progenitor populations exist that are pre-determined to become either hippocampal or cortical interneurons (but not both); or 2) a single progenitor can become a hippocampal or cortical interneuron either stochastically or under the influence of intrinsic (i.e. transcription factor) or extrinsic (i.e. cell-cell signaling) factors. Lineage tracing of retrovirus-labeled progenitors (McCarthy et al., 2001) within i12b-Cre;Z/EG MGE could help address these two possibilities: the first possibility is likely correct if an EGFP+ clone gives rise to only hippocampal (or cortical) interneurons.
I12b-Cre and URE2-Cre activity in cortical and hippocampal GABAergic interneuron subtypes
Both I12b-Cre and URE2-Cre were expressed in all cortical and hippocampal GABAergic interneuron subtypes examined in this study, although URE2-Cre was consistently expressed in a smaller population of each subtype (Table 1 and Table 2). Within the MGE and its descendants, URE2-Cre was active in fewer cells than I12b-Cre between E12.5 and P0. This could explain why there were 2.4-fold fewer adult cortical interneurons labeled by URE2-Cre versus I12b-Cre. Interestingly, the ratio of co-expression of CR, SOM and PV with EGFP in the cortex of adult I12b-Cre;Z/EG versus URE2-Cre;Z/EG mice varied from this ∼2.4-fold difference, suggesting that I12b and URE2 have different activities in these GABAergic interneuron populations. SOM and PV expressing interneurons arise from the MGE, while CR+ cortical interneurons arise from the CGE and MGE (Butt et al., 2005; Flames et al., 2007; Fogarty et al., 2007; Xu, 2004). Thus the lower levels of URE2 activity in the MGE and its derivatives such as the globus pallidus, compared to I12b, could explain the decreased labeling of adult SOM and PV interneurons. Furthermore, this is consistent with the low labeling of adult SOM+ interneurons by URE2-LacZ (Ghanem et al., 2007).
URE2 activity is reduced compared to I12b in the CGE at E12.5 (Fig. 2); however, this difference is less apparent at later developmental stages. The CGE has been defined using its anatomical features, yet recent gene expression analysis suggests that it may not contain progenitor pools that are intrinsically different than those found in the LGE or MGE (Flames et al., 2007). The CR-producing progenitors within the CGE appear to be extensions of the caudal pole of the LGE, which has comparable URE2-Cre and I12b-Cre activity. Thus, similar numbers of CR+ interneurons that arise from the URE2-Cre and I12b-Cre lineage might reflect an equivalent level of I12b and URE2 activity within the CR-producing progenitor population within the caudal LGE. Furthermore, URE2 activity may begin or increase after CGE cells migrate; if so, URE2 activity will be detected in CGE descendents. The same phenomenon may account for URE2-Cre labeling of MGE-derived interneurons.
Analysis of I12b-lacZ and URE2-lacZ transgenic mice suggested that I12b and URE2 were differentially active in cortical SOM (90% versus 10%, I12b versus URE2), CB (∼100% versus ∼15%, I12b versus URE2), and nNOS (∼20% versus ∼100%, I12b versus URE2) expressing interneurons (Ghanem et al., 2007). These striking differences were not present in cortical interneurons labeled in I12b-Cre and URE2-Cre mice. A likely explanation for this discrepancy is the difference between lacZ-labeling and Cre-mediated labeling of the interneuron subtypes. In lacZ transgenic mice, β-gal will be expressed when the regulatory element is active, and the β-gal signal will diminish as the protein is degraded in cells that no longer express the transgene. In contrast, EGFP reporter expression will be maintained indefinitely within cells and their progeny that expressed CRE at any time in Cre-transgenic mice. Thus, our analysis of adult cortical interneuron subtypes includes populations of interneurons that have at any point of their development expressed CRE. It is possible that URE2 or I12b might be transiently active in embryonic neuronal progenitors, immature migrating interneurons, or adult interneurons. In support of this idea, CRE is transiently expressed within a subpopulation of cortical EGFP+ interneurons in adult I12b-Cre;Z/EG mice (Fig. 9G).
Our detailed characterization of I12b-Cre and URE2-Cre activity during embryogenesis, and quantification of labeled cortical and hippocampal interneuron subtypes, lays the groundwork for future use of these mice in lineage analyses or conditional knock-out studies. These mice are ideally suited for specifically studying the function of genes that regulate the development and function of forebrain GABAergic neurons without altering the properties of glial cells.
Experimental Methods
Animals
All experimental procedures were approved by the Committees on Animal Health and Care at the University of California San Francisco (UCSF). Mouse colonies were maintained at UCSF in accordance with National Institutes of Health and UCSF guidelines. I12b-Cre and URE2-Cre mice were generated by oocyte injection of linearized I12b-Cre or URE2-Cre plasmids, respectively. Genomic DNA collected from the tails of transgenic mice were screened by PCR for Cre. BamHI-digested genomic DNA from Cre+ animals were then screened by Southern blot analysis using radioactively-labeled transgene-specific probes to determine transgene copy number and the number of integration sites (Supplemental Fig. S1). Transgenic mice that contained single integration sites were crossed to Z/EG reporter mice (Novak et al., 2000) and analyzed for EGFP fluorescence in the forebrain. Four I12b-Cre lines and four URE2-Cre lines produced similar expression patterns for each transgene at birth; two lines for each transgene was analyzed further by immunohistochemistry for EGFP on P0 coronal forebrain sections at and gave nearly identical patterns and levels of EGFP expression. One I12b-Cre line and one URE2-Cre line was chosen for additional analysis presented in this study. During the course of our analysis, it was observed that female I12b-Cre and URE2-Cre mice often produced ectopically-labeled EGFP+ mice when crossed to male Z/EG animals. Thereafter, I12b-Cre and URE2-Cre males were mated to Z/EG females to generate I12b-Cre;Z/EG animals with ectopic expression noted in ∼1% of mice. Mice with ectopic EGFP+ expression were easily identifiable and not used for analysis. Mutant mice lacking Dlx1&2 were generated previously in our laboratory as described (Qiu et al., 1997). I12b-Cre;Z/EG;Dlx1&2-/- mice were generated by mating I12b-Cre;Dlx1&2+/- males to Z/EG;Dlx1&2+/- females. A similar scheme was used to generate URE2-Cre;Z/EG;Dlx1&2-/- animals.
Histology
Pregnant females were anesthetized with isofluorene before euthanasia by cervical dislocation. E10.5, E12.5, or E15.5 pups were extracted from the uterus and the brains dissected (for E15.5) and fixed with 4% paraformaldehyde (PFA) in phosphate-buffered solution (PBS 0.1 M, pH 7.4). Newborn mice were anesthetized on ice for 1 minute before sacrifice and dissected brains were fixed with PFA in PBS overnight. Adult mice (P60-P100) were deeply anesthetized with Avertin (Sigma; 0.2 ml/10 g body weight) and underwent intracardiac perfusion with PBS followed by 4% PFA in PBS. The brains were removed and post-fixed overnight in the 4% PFA. After fixing, all brains were cryoprotected by immersion in 15% and 30% sucrose, frozen in OCT (Tissue-Tek) on dry ice, and stored at -80°C. Embryonic and P0 brain sections were cut at a thickness of 20 mm on a cryostat and mounted on Fisher Superfrost/Plus slides. For double immunohistochemistry, adult brains were cut at 40 mm on the cryostat and processed as floating sections.
Immunohistochemistry
Sections were blocked for 1 hour at room temperature with blocking solution (5% goat serum, 1% bovine serum albumin, and 0.03% Triton X-100). Primary antibodies were incubated at 4°C overnight in blocking solution and secondary antibodies for 1 hour at room temperature or overnight at 4°C in blocking solution. For fluorescent imaging, sections were counterstained with DAPI, cover-slipped using Fluoromount-G (Southern Biotech), and analyzed using an Olympus AX700 or Nikon 6D microscope. For non-fluorescent immunohistochemistry, ABC Kit (Vector Laboratories) was used to form avidin-biotin complexed to horseradish peroxidase and visualized using VIP (Vector Laboratories) color reaction. Sections were dehydrated overnight, washed in xylene, and cover-slipped using Permount (Fisher Scientific). Primary antibodies and concentration used: guinea-pig anti-DLX2, 1:2000 (gift from K. Yoshikawa), rabbit anti-OLIG2, 1:20000 (gift from C. Stiles), rabbit anti-GFP, 1:2000 (Chemicon), chicken anti-GFP, 1:1000 (Chemicon), guinea pig anti-SOX10, 1:1000 (gift of M. Wegner), rabbit anti-OLIG1, 1:10000 (gift from C. Stiles), mouse anti-CRE, 1:800 (Chemicon), goat anti-ChAT, 1:250 (Chemicon, AB144P), rabbit anti-GFAP, 1:1000 (Chemicon, Abcam), rat anti-somatostatin, 1:150 (Chemicon, MAB354), rabbit anti-calretinin, 1:2500 (Chemicon, AB5054) or 1:2000 (Swant); rabbit anti-NPY, 1:5000 (Sigma, N9528) or 1:2000 (Immunostar); mouse anti-parvalbumin, 1:2000 (Sigma, P3088); rabbit anti-calbindin, 1:4000 (Swant, Switzerland); rabbit anti-neuronal nitric oxide synthase (nNOS), 1:1000 (Zymed). Secondary antibodies: (all Alexa Fluors used at 1:500, Molecular Probes): goat anti-chicken 488, goat anti-rabbit 594/488/350, goat anti-rat 594/488, goat anti-mouse 594/488, goat anti-guinea pig 594/488.
X-gal staining
E10.5 Cre;R26R-lacZ embryos were fixed in fresh 2% PFA/0.2% glutaraldehyde in PBS (without Mg2 or Ca2+) for 1 hr at 4°C, then rinsed three times, 30 minutes each, in PBS at room temperature. Embryos were transferred to X-gal buffer overnight at room temperature, washed 3 times for 30 minutes each, and then post-fixed overnight at 4°C in 4% PFA. After fixation, embryos were cleared in methyl salicylate. Embryos were washed with distilled water two times, 30 minutes each at room temperature, dehydrated by incubation for 30 minutes in 70% then 95% then 100% EtOH, and then transferred to 100% methyl salicylate at room temperature for about 10 minutes. Embryos were photographed in 100% methyl salicylate. X-gal buffer: 1 × PBS (w/o MgCl2 or Ca2+), 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal.
Thymidine-analog birth-dating
Equimolar solutions of the thymidine analogs were injected into pregnant dams. For IdU at 23 mg/ml and CldU at 17 mg/ml, injections at 2.5 ml/kg resulted in an equimolar final effective delivery of 57.5 mg/kg IdU or 42.5 mg/kg CldU. The solutions were administered by intraperitoneal injection. Mouse anti-BrdU (Becton Dickinson #347580; clone B44) at 1:500 was used to detect IdU, and rat anti-BrdU (Accurate #OBT-0030; clone BU1/75) at 1:250 was used to detect CldU. No cross reaction was detected using these antisera at these dilutions and both antibodies gave no signal in non-injected controls (data not shown) (Vega and Peterson, 2005). For immuno-detection, sections were treated with 2N HCl for 10 min at 37°C before antibody incubation.
Statistical analysis
GABAergic interneuron subtype analysis
40 μm coronal sections from adult I12b-Cre;Z/EG or URE2-Cre;Z/EG animals were double-immunolabled for EGFP and the interneuron marker. At least 3 serial sections of the hippocampus or somatosensory cortex from 3 different animals were photographed using a Olympus AX700 or Nikon 6D fluorescent microscope. Images were imported into Photoshop CS3 (Adobe Systems Incorporated) and six or three layers were created for cortical or hippocampal sections, respectively. EGFP only, marker-only, and double-positive cells were overlayed manually by color-coded dots in the new layers. The number of labeled cells (dots) was calculated using the Record Measurements tool of Photoshop CS3 and imported into Excel 2004 (Microsoft Corporation). The percentage of labeled cells per region per section was calculated for each brain, and the final mean percentage and SEM where calculated across all brains.
Birthdate analysis
Three to four 20 μm sections that contained the olfactory bulb, neocortex, or hippocampus from three P0 I12b-Cre;Z/EG and URE2-Cre;Z/EG animals that had received thymidine-analog injections were analyzed. Sections were photographed using a Olympus AX700 or Nikon 6D fluorescent microscope. Images were imported into Photoshop CS3 and two layers were created for each section. EGFP+ only and double-positive cells were manually marked by color-coded dots in the new layers. The number of labeled cells (dots) was calculated using the Record Measurements tool of Photoshop CS3 and imported into Excel 2004. The percentage of labeled cells per section was calculated for each brain, and the final mean percentage and SEM where calculated across all brains. Statistical analysis was performed in Excel using the two-tailed ttest function. p values < 0.05 were considered significant.
ChIP analysis
Chromatin Prep
Medial and lateral ganglionic eminences were dissected from E15.5 wild type mice and transferred to 1 mL of 1% PFA. The tissue was mechanically dissociated, incubated for 15 min, and quenched with 100 μL of 2.5M glycine. After centrifugation, tissue was resuspended in 1 mL lysis buffer and rotated for 10 minutes at 4°C. The tissue was centrifuged, resuspended in 1 mL Wash Buffer 1, incubated for 10 min at 4°C, mixed by rotation for 10 min at 4°C, centrifuged, and resuspended in 4 mL Wash Buffer 2. Samples were sonicated until DNA fragments of 100-1000 bp were produced. Sarcosyl was added to chromatin samples to a final concentration of 0.5% and incubated at RT for 10 min. Cesium Chloride (CeCl) was added to a final concentration of 0.568 g/mL and samples and centrifuged at 40,000 rpm for 48 hrs. Fractions obtained from the CeCl gradient were run on a gel; chromatin rich fractions were isolated and pooled. Pooled fractions were dialyzed in Dialysis Buffer overnight.
Lysis Buffer
0.25% Triton-X, 0.5% NP40, 1 mM EDTA, 10 mM Tris pH 8, 1mM EGTA, 1 mM PMSF and proteinase inhibitors; Wash Buffer 1: 0.2 M NaCl, 1 mM EDTA, 10 mM Tris pH 8, 1 mM EGTA, 1 mM PMSF and proteinase inhibitors; Wash Buffer 2: 1 mM EDTA, 10 mM Tris pH 8, 1 mM EGTA, 1 mM PMSF and proteinase inhibitors; Dialysis Buffer: 10mM Tris pH 8, 5% Glycerol, 1 mM PMSF.
Immunoprecipitations (IP)
10 μg of chromatin per IP were resuspended to 500 μL in RIPA buffer and pre-cleared for 2 hrs with 20 μL Protein A DynaBeads. The Protein A beads were removed, 1 μL of guinea pig anti-DLX2 antibody (Gift of Dr. Yoshikawa) was added. For (–) antibody control, no antibody was added at this step. Samples were incubate overnight at 4°C. Separately, Protein A DynaBeads (10 μL per IP) were left to block overnight at 4°C in blocking solution. Blocked beads were then resuspended in RIPA buffer at 500 μL Buffer/ 10 μL Beads/ IP. Chromatin samples were rocked with 500 μL blocked beads in RIPA (10 μL beads total) at 4°C for 3 hours. Protein A beads/antibody/chromatin complexes were washed at 4°C seven times by pulldown and resuspension in Wash RIPA Buffer. Chromatin was eluted at 37°C in 50 μL elution buffer with shaking for 30 minutes, the beads were pulled down, and 50 μL of supernatent transferred to a new tube. Chromatin was reverse crosslinked by addition of 150 μL of TE, 2.5 μL proteinase K, incubation at 55°C for 3 hrs and 65°C overnight. DNA was isolated by phenol/chloroform extraction and ethanol precipitation. 2X RIPA Buffer: 2% Triton-X, 1% Na deoxycholate, 280 mM NaCl; Wash RIPA: 1% Triton, 0.5% Na Deoxycholate, 500 mM NaCl, 0.001% SDS; Blocking Solution: 200 μL BSA, 200 μL E.coli tRNA 10 mg/ml, 100 μL chicken core hisones 1mg/ml.
PCR primers
URE2a (143 bp product): 5′-GAT TCT TAT TCA GGC ATT CTG TGG-3′, 5′-GCA AAT GTG CCT CTG TAG TTA GC-3′; Dlx5/6i (127 bp product): 5′-AGT GCC ATC CAA TTT GAA GCA GAC-3′, 5′-:GCT GTA ATC AGC GGG CTA CAT GA-3′; i12b (155 bp product): 5′-CGG GCC CAT CAA ACA CAA CAT AA-3′, 5′-GCA GCA AAT TTG GCT TTC AGT GC-3′; Opsin promoter (100 bp product): 5′-CTT CCA CAA GCC CTA CTG CAA-3′, 5′-TTC CAT GTT TCT GAG TCC AAT CC-3′; ECR9a (242 bp product) 5′-ACA GTA ATT TGA GAT CCC TCT GC-3′, 5′-GAA TTG CAC CAA CAC AAA CAT TTG-3′. Each experiment was performed at least two times on two different chromatin preparations with similar results.
Acknowledgments
UCSF Comprehensive Cancer Center Transgenic Core performed the oocyte injections and generated the I12b-Cre and URE2-Cre transgenic mice. Louis Reichardt, Ben Cheyette and the Nikon Imaging Center at UCSF provided access to microscopes used in this study. We would like to thank M. Wegner for the SOX10 antibody, K. Yoshikawa for DLX2 antibody, and C. Stiles for OLIG1 and OLIG2 antibodies. We thank N. Ghanem for providing data prior to publication. G.B.P. made the I12b-Cre and URE2-Cre transgenes, characterized the different lines of I12b-Cre and URE2-Cre transgenic mice, maintained the mouse colonies, designed and implemented the experiments, photographed and quantified brain sections, performed statistical analysis on the data, wrote the manuscript, and prepared all tables and figures. M.A.P. prepared, sectioned, immunolabeled and photographed brain tissue. E.S. genotyped and maintained transgenic mice, sectioned, immunolabeled and photographed tissue, and quantified the birth-date data. G.L.M. performed the ChIP experiments. M.E. provided plasmids containing I12b and URE2 DNA, feedback on transgene construction, and comments on the manuscript. J.L.R. provided mentorship on experimental design, analysis, and manuscript preparation. G.B.P. was supported by the California Institute of Regenerative Medicine's Postdoctoral Fellowship; M.A.P. was funded in part by NICHD HD-07162H; M.E. was supported by CIHR grant #MOP-14460; JLR was supported by Nina Ireland, NIMH RO1 MH49428 and K05 MH065670.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Close J, Machold R, Fishell G. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28:3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Cobos I. Cellular Patterns of Transcription Factor Expression in Developing Cortical Interneurons. Cerebral Cortex. 2006;16:i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cossart R, Bernard C, Benari Y. Multiple facets of GABAergic neurons and synapses: multiple fates of GABA signalling in epilepsies. Trends in Neurosciences. 2005;28:108–115. doi: 10.1016/j.tins.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Danglot L, Triller A, Marty S. The development of hippocampal interneurons in rodents. Hippocampus. 2006;16:1032–1060. doi: 10.1002/hipo.20225. [DOI] [PubMed] [Google Scholar]
- DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Jones EG. High-Resolution Light and Electron Microscopic Immunocytochemistry of Colocalized GABA and Calbindin D-28k in Somata and Double Bouquet Cell Axons of Monkey Somatosensory Cortex. Eur J Neurosci. 1992;4:46–60. doi: 10.1111/j.1460-9568.1992.tb00108.x. [DOI] [PubMed] [Google Scholar]
- Demeulemeester H, Arckens L, Vandesande F, Orban GA, Heizmann CW, Pochet R. Calcium binding proteins and neuropeptides as molecular markers of GABAergic interneurons in the cat visual cortex. Exp Brain Res. 1991;84:538–544. doi: 10.1007/BF00230966. [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Flames N, Marin O. Developmental Mechanisms Underlying the Generation of Cortical Interneuron Diversity. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman D, Rubenstein J, Puelles L, Marin O. Delineation of Multiple Subpallial Progenitor Domains by the Combinatorial Expression of Transcriptional Codes. Journal of Neuroscience. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Furusho M, Ono K, Takebayashi H, Masahira N, Kagawa T, Ikeda K, Ikenaka K. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293:348–357. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Gaveriaux-Ruff C, Kieffer BL. Conditional gene targeting in the mouse nervous system: Insights into brain function and diseases. Pharmacol Ther. 2007;113:619–634. doi: 10.1016/j.pharmthera.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2008;1:11. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- He W, Ingraham C, Rising L, Goderie S, Temple S. Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci. 2001;21:8854–8862. doi: 10.1523/JNEUROSCI.21-22-08854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Li X, McEvilly R, Rosenfeld M, Lufkin T, Rubenstein JL. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw- and upper jaw-specific genetic programs. Development. 2008 doi: 10.1242/dev.019778. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S, Kosaka T. Cellular architecture of the mouse hippocampus: A quantitative aspect of chemically defined GABAergic neurons with stereology. Neuroscience Research. 2006;56:229–245. doi: 10.1016/j.neures.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Kalanithi PS, Zheng W, Kataoka Y, DiFiglia M, Grantz H, Saper CB, Schwartz ML, Leckman JF, Vaccarino FM. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc Natl Acad Sci U S A. 2005;102:13307–13312. doi: 10.1073/pnas.0502624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katarova Z, Sekerková G, Prodan S, Mugnaini E, Szabó G. Domain-restricted expression of two glutamic acid decarboxylase genes in midgestation mouse embryos. J Comp Neurol. 2000;424:607–627. doi: 10.1002/1096-9861(20000904)424:4<607::aid-cne4>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Battiste J, Nakagawa Y, Johnson JE. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci. 2008 doi: 10.1016/j.mcn.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Petryniak M, Long J, Ekker M, Obata K, Yanagawa Y, Rubenstein J, Alvarez-Buylla A. A Subpopulation of Olfactory Bulb GABAergic Interneurons Is Derived from Emx1- and Dlx5/6-Expressing Progenitors. Journal of Neuroscience. 2007;27:6878–6891. doi: 10.1523/JNEUROSCI.0254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Lufkin T. Dlx homeobox gene control of mammalian limb and craniofacial development. Am J Med Genet A. 2006;140:1366–1374. doi: 10.1002/ajmg.a.31252. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kawaguchi Y. Three classes of GABAergic interneurons in neocortex and neostriatum. Jpn J Physiol. 1994;44 2:S145–148. [PubMed] [Google Scholar]
- Levi G, Puche AC, Mantero S, Barbieri O, Trombino S, Paleari L, Egeo A, Merlo GR. The Dlx5 homeodomain gene is essential for olfactory development and connectivity in the mouse. Mol Cell Neurosci. 2003;22:530–543. doi: 10.1016/s1044-7431(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Lewis D, Hashimoto T, Volk D. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Garel S, Depew MJ, Tobet S, Rubenstein JL. DLX5 regulates development of peripheral and central components of the olfactory system. J Neurosci. 2003;23:568–578. doi: 10.1523/JNEUROSCI.23-02-00568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan CH, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 Transcription Factors Control Striatal Patterning and Differentiation Through Parallel and Overlapping Pathways. J Comp Neurol. 2008 doi: 10.1002/cne.21854. Accepted manucript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manent JB, Jorquera I, Ben-Ari Y, Aniksztejn L, Represa A. Glutamate acting on AMPA but not NMDA receptors modulates the migration of hippocampal interneurons. J Neurosci. 2006;26:5901–5909. doi: 10.1523/JNEUROSCI.1033-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Marshall CA, Goldman JE. Subpallial dlx2-expressing cells give rise to astrocytes and oligodendrocytes in the cerebral cortex and white matter. J Neurosci. 2002;22:9821–9830. doi: 10.1523/JNEUROSCI.22-22-09821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M, Turnbull DH, Walsh CA, Fishell G. Telencephalic neural progenitors appear to be restricted to regional and glial fates before the onset of neurogenesis. J Neurosci. 2001;21:6772–6781. doi: 10.1523/JNEUROSCI.21-17-06772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin C, Baudoin J, Rakic S, Parnavelas J. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur J Neurosci. 2006;23:894–900. doi: 10.1111/j.1460-9568.2006.04630.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monory K, Massa F, Egertova M, Eder M, Blaudzun H, Westenbroek R, Kelsch W, Jacob W, Marsch R, Ekker M, Long J, Rubenstein JL, Goebbels S, Nave KA, During M, Klugmann M, Wolfel B, Dodt HU, Zieglgansberger W, Wotjak CT, Mackie K, Elphick MR, Marsicano G, Lutz B. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron. 2006;51:455–466. doi: 10.1016/j.neuron.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Perera M, Merlo GR, Verardo S, Paleari L, Corte G, Levi G. Defective neuronogenesis in the absence of Dlx5. Mol Cell Neurosci. 2004;25:153–161. doi: 10.1016/j.mcn.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pow DV. Immunocytochemical detection of amino acid neurotransmitters in paraformaldehyde-fixed tissues. Methods Mol Biol. 1997;72:103–123. doi: 10.1385/0-89603-394-5:103. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Rubenstein J, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymar V, Sadikot A. Laminar fate of cortical GABAergic interneurons is dependent on both birthdate and phenotype. J Comp Neurol. 2007;501:369–380. doi: 10.1002/cne.21250. [DOI] [PubMed] [Google Scholar]
- Rymar V, Sasseville R, Luk K, Sadikot A. Neurogenesis and stereological morphometry of calretinin-immunoreactive GABAergic interneurons of the neostriatum. J Comp Neurol. 2004;469:325–339. doi: 10.1002/cne.11008. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stühmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002a;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002b;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Koos T, Wilson CJ. GABAergic microcircuits in the neostriatum. Trends Neurosci. 2004;27:662–669. doi: 10.1016/j.tins.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Toresson H, Campbell K. A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development. 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- Tucker ES, Polleux F, LaMantia AS. Position and time specify the migration of a pioneering population of olfactory bulb interneurons. Dev Biol. 2006;297:387–401. doi: 10.1016/j.ydbio.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Vega CJ, Peterson DA. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nat Methods. 2005;2:167–169. doi: 10.1038/nmeth741. [DOI] [PubMed] [Google Scholar]
- Wonders C, Anderson S. Cortical Interneurons and Their Origins. The Neuroscientist. 2005;11:199–205. doi: 10.1177/1073858404270968. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Taylor L, Welagen J, Mbata IC, Xiang JZ, Anderson SA. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314:127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q. Origins of Cortical Interneuron Subtypes. Journal of Neuroscience. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–4998. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Yun K, Garel S, Fischman S, Rubenstein JL. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol. 2003;461:151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- Yung SY, Gokhan S, Jurcsak J, Molero AE, Abrajano JJ, Mehler MF. Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species. Proc Natl Acad Sci U S A. 2002;99:16273–16278. doi: 10.1073/pnas.232586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, Eisenstat DD. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]


