SUMMARY
even-skipped (eve) has been proposed to set up parasegment borders at the anterior edge of each of its seven stripes by providing a sharp expression boundary, where engrailed is activated on one side and wingless on the other. By expressing bell-shaped early eve stripes without the sharp boundary provided by narrow, late stripes, we find that the early gradient is sufficient for generating stable parasegment borders. Based on several lines of evidence, we propose that the anterior portion of each early stripe has morphogenic activity, repressing different target genes at different concentrations. These distinct repression thresholds serve to both limit and subdivide a narrow zone of paired expression. Within this zone, single cell rows express either engrailed, where runt and sloppy-paired are repressed, or wingless, where they are not. While the early eve gradient is sufficient to establish parasegmental borders without refined, late expression, late eve expression has a role in augmenting this boundary to provide for strong, continuous stripes of engrailed expression. In addition, we show that the early eve gradient is sufficient, at its posterior edge, for subdividing the ftz domain into engrailed expressing and non-expressing cells.
Keywords: segmentation, morphogen, Drosophila, homeodomain, transcriptional repression, even-skipped
INTRODUCTION
Segmentation in the Drosophila embryo begins in a syncytial blastoderm with the actions of three groups of genes, most of which encode transcription factors (for reviews, see Akam, 1987; Ingham, 1988; Ingham and Martinez-Arias, 1992). First, maternal effect gene products establish the basic anteroposterior coordinate in the form of gradients. These gradients are then used by zygotic gap genes to establish their region-specific expression. The maternal effect and gap gene products, in turn, activate periodic expression of pair-rule genes, usually in seven stripes in the blastoderm embryo. Pair-rule genes themselves are organized hierarchically into at least two groups. The primary pair-rule genes, hairy, runt, even-skipped (eve), and possibly paired (prd), establish periodic stripe expression and regulate secondary pair-rule genes. Following cellularization of the blastoderm, the periodic cues of these pair-rule genes are used to establish expression of segment-polarity genes, usually in 14 stripes, leading to the formation of 14 parasegments. The products of the segment polarity genes have a much less restricted range of function. In addition to transcription factors, they include diffusible factors, membrane proteins, and members of intracellular signaling cascades (Ingham and Martinez-Arias, 1992). Previous studies of the expression patterns of pair-rule genes and the consequences of alterations in these patterns have been crucial to the elucidation of the segmentation cascade. However, the expression patterns of many pair-rule genes, like those of most other segmentation genes, undergo changes with time, thereby altering their relative phase relationships. This renders a static analysis imprecise. Therefore it would be useful to create an embryo in which only part of the normal temporal pattern of a pair-rule gene is expressed, so that one can examine how different temporal and spatial aspects of expression relate to function. This is the primary motivation of the work presented here.
Two pair-rule genes, eve and fushi tarazu (ftz) play critical roles in defining parasegments, as reflected in their expression patterns. eve expression starts as three broad bands that cover most of the trunk region. At precellular blastoderm, these resolve into seven stripes that center on the odd-numbered parasegmental primordia (Harding et al., 1986; Macdonald et al., 1986; Frasch et al., 1987). In a roughly complementary pattern, ftz stripes start broadly and come to coincide with the seven even-numbered parasegmental primordia (Carroll and Scott, 1985; Hiromi et al., 1985; Lawrence et al., 1987). After cellularization, both patterns undergo ‘refinement’, in which stripes narrow from about four cells to about two cells by loss of expression from the posterior, with a concomitant increase of expression in the anterior-most cells, thereby sharpening the anterior borders. During this period, the segment polarity genes engrailed (en) and wingless (wg) begin to appear in the anterior-most and the posterior-most row of cells, respectively, in each parasegmental primordium (Lawrence et al., 1987; Ingham et al., 1988). In particular, the sharpened borders of eve and ftz stripes coincide, cell by cell, with the anterior borders of en stripes, thus demarcating parasegmental boundaries (Lawrence et al., 1987). Consistent with the regulatory relationships suggested by this coincidence, even-numbered en stripes are missing in ftz mutants (Howard and Ingham, 1986; DiNardo and O’Farrell, 1987), and even-numbered parasegments are not formed (Wakimoto et al., 1984; Martinez-Arias and Lawrence, 1985). Similarly, odd-numbered en stripes and odd-numbered parasegments are not formed in eve hypomorphic mutants (Nüsslein-Volhard et al., 1985; Frasch et al., 1988). However, eve null mutants lack both even- and odd-numbered en stripes (Harding et al., 1986; Macdonald et al., 1986), and no segmentation occurs in the trunk region (Nüsslein-Volhard et al., 1985). Thus, eve is required for expression of all en stripes, but low levels of activity suffice for even-numbered stripes. It is not known what aspects of the eve pattern are important for this regulation: eve could act as a local morphogen for downstream genes; if so, the overall level and shape of the expression pattern may be the most important qualities (Lawrence, 1987; Lawrence and Johnston, 1989). Alternatively, eve may act in combination with other genes to provide distinct signals in different parts of each stripe. In this case, the extent of expression at different times relative to that of other pair-rule genes may be more important (DiNardo and O’Farrell, 1987; Ingham et al., 1988). Therefore, it is of considerable interest to relate eve’s changing expression pattern to its role in activating en and wg.
Transgenic studies showed that a 6.4 kb fragment from the upstream region of eve is sufficient to drive expression of stripes 2, 3, and 7 (Goto et al., 1989; Harding et al., 1989). Further dissection of the fragment uncovered two classes of regulatory elements: stripe-specific ‘early elements’ which have been shown to translate the non-periodic cues of gap genes, stripe by stripe, into the periodic pattern of eve, and an element which governs expression of all late, narrowed stripes (Goto et al., 1989; Harding et al., 1989; Small et al., 1991; Jiang et al., 1991). This latter element has been shown to respond to early eve expression, providing autoregulatory feedback. By removing this autoregulatory contribution, we have recently shown that the late element is in fact responsive to other pair-rule genes in the absence of feedback (M.F. and T.G., unpublished data). Thus, it carries out refinement of the eve pattern in response to the primary pair-rule genes (Goto et al., 1989). In this work, we used transgenes to create embryos that express only unrefined early stripes, or both unrefined early stripes and refined late stripes. This allowed us to study the changing roles of eve expression before and after refinement. Our results show that early and late eve expression have distinct roles in regulating downstream genes. Notably, only early expression is required for the activation of both even- and odd-numbered en stripes, while late eve stripes strengthen expression of the odd-numbered en stripes. A model is proposed for how early eve expression, acting as a bell-shaped, concentration-dependent morphogenetic field, biased by combinatorial interactions with other primary pair-rule genes, subdivides each of its domains into multiple functional subregions. These findings add to the previous discoveries of embryolength morphogenetic gradients, composed of maternal gene products, and the more localized gradients of gap gene products, and show that Drosophila uses the same principle again to further refine pattern information to the single cell level.
MATERIALS AND METHODS
Drosophila strains
eve transgenes based on pCaSpeR3 (Thummel et al., 1988) were constructed as follow; E+L-eve carries an eve genomic DNA fragment from -6.3 kb (NdeI restriction site) to +1.85 kb (MluI site) relative to the transcription start site, which includes the late element, early stripe 2, 3, and 7 elements, the protein coding sequence, and the polyadenylation site. E-eve contains genomic DNA from -4.95 kb (KpnI) to +1.85 kb (MluI site). L-eve has a fragment from -6.3 kb to -4.95 kb combined with a fragment from -0.27 kb (SfiI) to +1.85 kb. These fragments were inserted into pCaSpeR3 with the direction of transcription opposite to that of the white gene. The RVΔSac construct is an E+L-eve construct with the 5′ deletion endpoint at -5.65 kb (EcoRV site) containing an internal deletion from -5.50 kb, a site downstream of the MAS (Jiang et al., 1991), to -5.01 kb (SacI site). A construct having the same 5′ endpoint but no internal deletion is fully active as a late element (Goto et al., 1989).
Flies of the genotype w;Df(eve)/CyO;+/+ were used as host for injection of pCaSpeR-eve constructs. Transgenic flies were crossed with a marked balancer strain (y w;Sco/CyO P[hb-lacZ];D/TM3) and screened for Df(eve)/CyO P[hb-lacZ], so that Df(eve)/Df(eve) embryos were identifiable by staining for β-galactosidase.
mRNA and protein localization
In situ hybridization to whole mount embryos using digoxigenin-(DGG) labeled probes was performed as described (Tautz and Pfeifle, 1989). DGG-labeled anti-sense mRNA probes were used. Those probes were visualized using alkaline phosphatase-conjugated anti-DGG antibody (Boehringer Mannheim). In double-staining, in situ hybridization with an RNA probe was followed by antibody staining, with either anti-Engrailed (Patel et al., 1989; kindly provided by Steve DiNardo) or anti-Ftz (kindly provided by Dianne Mattson and Ian Duncan) monoclonal antibodies (Kellerman et al., 1990). Biotinylated secondary antibodies were detected using avidin-conjugated peroxidase (VectorLabs). Embryos were mounted in Fluoromount.
Cuticle preparations
Cuticles were prepared as described (Wieschaus and NüssleinVolhard, 1986). Embryos were collected on grape juice plates, aged for 18 to 24 hours at 25°C, dechorionated with 50% bleach, and devitellinized with methanol/heptane. After incubation at 60°C for 40 minutes in PBS, the embryos were mounted in Hoyer’s medium/lactic acid (1:1) and incubated at 60°C until cleared.
RESULTS
Localized rescue patterns
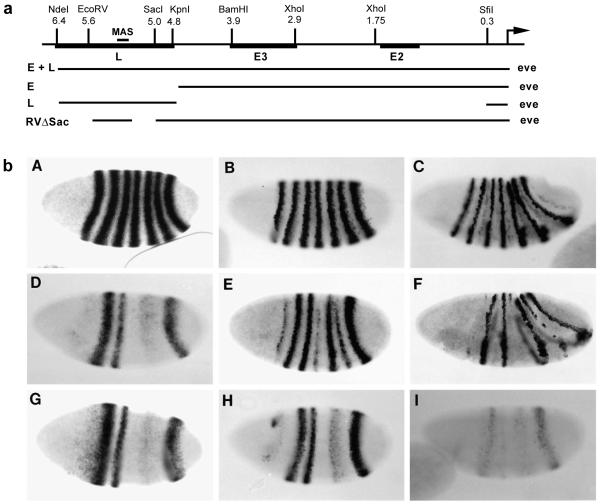
Previous studies showed that a 4.8 kb fragment from the upstream region of eve contains stripe-specific ‘early elements’ (‘Es’) sufficient to drive the expression of broad, unrefined early stripes 2, 3, and 7 during blastoderm. In contrast, a 6.4 kb fragment expresses not only these early stripes but also all seven refined, late stripes in gastrulating embryos (Goto et al., 1989; Harding et al., 1989). This difference in cis-regulatory activity was attributed to the presence in the 6.4 kb fragment of a ‘late element’, or ‘L’, which can direct late stripe expression of a reporter gene in wild-type embryos. Since these early and late stripes are different in both the timing of their appearance and their shape, they may have distinct functions in vivo. We investigated this possibility by examining the phenotypic consequences of expressing Eve (the eve protein) driven by either the 4.8 kb fragment (E-eve) or the 6.4 kb fragment (E+L-eve) in the absence of endogenous eve. This was conveniently carried out by transforming an eve deficiency mutant, Df(eve). The construction of these two transgenes, E-eve and E+L-eve, is diagrammed in Fig. 1a. Since these transgenes reproduce only part of the normal eve pattern in an eve deficiency host, we will refer to this approach as ‘localized rescue’.
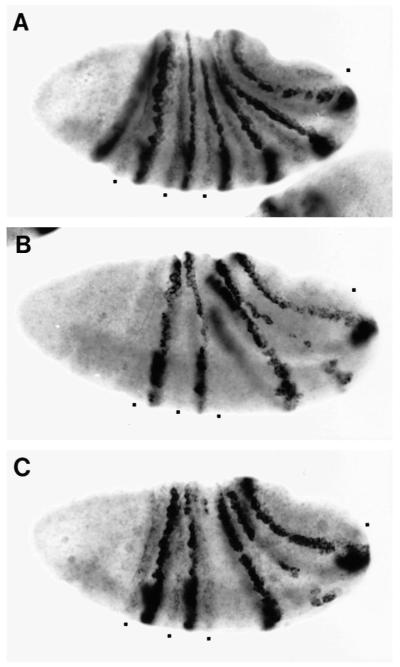
Fig. 1.
Localized rescue of an eve deficiency mutation. (a) Map of the 5′ upstream region of the eve gene and sequences used in eve transgenes. L is the late element, and E2 and E3 refer to stripe-specific, early elements. MAS is the minimal autoregulatory sequence defined by Jiang et al. (1991). E+L-eve and E-eve were used, in a Df(eve) background, to generate EL embryos and E embryos, respectively. (b) Expression of eve in wild-type and locally rescued embryos. Whole mount in situ hybridization using a digoxigenin-labeled eve probe was carried out to wild-type embryos (A,B,C), EL embryos (D,E,F), or E embryos (G,H,I). (A,D,G) Early cellular blastoderm stage; (B,E,H) late cellular blastoderm; (C,F,I) gastrulation. At the early stage (D,G), E and EL embryos express early stripes 2, 3, and 7, and a broad, weak stripe in the region of normal stripes 5 and 6. Later, EL embryos initiate expression of all seven late stripes (E), but, in the absence of early stripes 1 and 4, late stripes 1 and 4 are not maintained (F). Early stripe 5/6 does lead to rescue of a somewhat broad, late stripe 5 and, frequently, a weak, late stripe 6.
Shown in Fig. 1b are the eve mRNA patterns at three developmental stages in wild-type embryos (WT; A,B,C) and in Df(eve) embryos locally rescued with either E+L-eve (EL embryos; D,E,F) or E-eve (E embryos; G,H,I). The patterns of the two transgenes at pre-cellular blastoderm, consisting of three early stripes, are essentially identical to each other (D,G), and are also similar to the β-galactosidase (β-gal) pattern previously observed with the equivalent lacZ construct (Goto et al., 1989; Harding et al., 1989). However, in addition to the fact that they have only three stripes, these patterns differ from the wild-type eve pattern in two respects. First, stripe 2 is considerably stronger than stripe 3, which appears slightly narrower than in wild-type embryos, and stripe 2 has an anterior trailing edge that is somewhat more intense than that seen in either the endogenous eve pattern or the β-gal pattern (D,G). Both the disparity of stripe intensities and the anterior trailing disappear gradually during blastoderm, although stripe 3 remains somewhat weaker than stripe 2. Second, an aberrant, weak ‘stripe’ is observed that covers the early stripe 5 and 6 domains. It is not clear whether this ‘stripe 5/6’ reflects normal regulation of stripe formation in this part of the embryo, but it has significant bearing on the regulation of downstream genes in this region by the transgenes, as described below.
After cellularization, stripes in the wild-type embryo become narrow, with their anterior borders sharply demarcated (B). The patterns in E and EL embryos have diverged markedly by this stage. In E embryos, stripes have begun to fade and become somewhat narrower (H), perhaps due to uniform loss of expression. In EL embryos, stripes 2, 3, and 7 undergo refinement after cellularization, and then fade as the germband extends; that is, they behave normally (E,F). The other late stripes are also initiated, but in the absence of early stripes, late stripes 1 and 4 fade rapidly. However, late stripes 5 and 6 persist: stripe 5 is strong and somewhat broader than normal; stripe 6 is much weaker and ‘patchy’. The behavior of these stripes is likely to be linked to the earlier expression of stripe 5/6. Their locations are more posterior than normal, which may reflect the fact that the level of expression of stripe 5/6 is considerably weaker than normal (see below and Discussion). In summary, the expression pattern of E+L-eve is more or less normal in the domains of stripes 2, 3, and 7, while the regions of stripes 1 and 4 remain eve deficient, and the domains of stripes 5 and 6 are aberrant due to the expression of an early but weak stripe 5/6.
Localized rescue of en and wg expression by E-eve and E+L-eve
The E and EL embryos provide an opportunity to dissect differences in early and late eve function. Since a major part of eve’s role in segmentation is in the initiation of en expression, we examined en expression patterns in the transformants (Fig. 2). We were particularly interested in testing the idea that broad, early stripes are involved in activating (even-numbered) en stripes in neighboring ftz domains, while narrow, late stripes activate (odd-numbered) en stripes in their own domains (Goto et al., 1989).
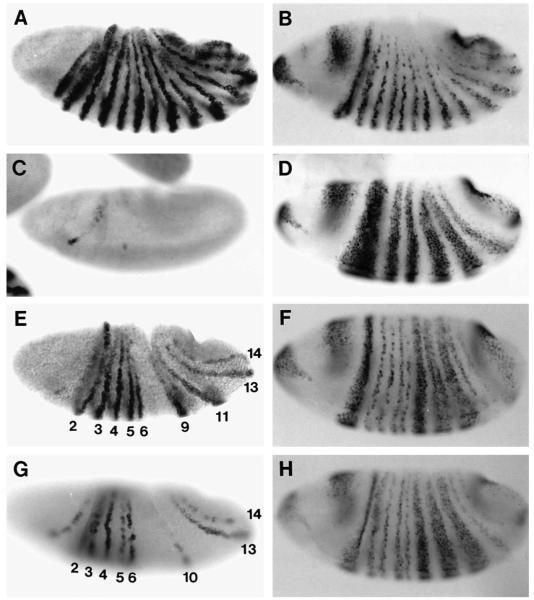
Fig. 2.
en and wg expression in locally rescued embryos. Gastrulation stage embryos were hybridized with en probe (A,C,E,G) or wg probe (B,D,F,H). (A,B) wild type. (C,D) Df(eve). (E,F) EL embryo. (G,H) E embryo. Expression of early eve stripes 2, 3, 5/6, and 7 is sufficient to activate en stripes 2 (partially hidden in this embryo), 3, 4, 5, 6, 13, 14, and occasionally 10 (G); however, odd-numbered stripes are relatively weak and incomplete. en stripe 5 is slightly posteriorly shifted, and stripe 6 may be slightly anteriorly shifted (see text). Normal wg expression is rescued in the regions of early eve stripes 2, 3, and 7, but not that of 5/6 (H). Addition of late expression restores normal expression of both en and wg more completely (E,F). Broad, derepressed wg stripes are ‘split’ where eve late stripes 5 and 6 are expressed (F).
In EL embryos, en stripes 2, 3, 4, 5, 6, 9, 11, 13, and 14 are expressed (Fig. 2E), although stripe 11 is often weak and incomplete. The identities of these stripes have been confirmed by En/wg double staining (data not shown). The appearance of most of these stripes can be ascribed to expression of specific eve stripes: en stripes 3, 5, and 13 are in the domains of eve stripes 2, 3, and 7, while en stripes 4, 6, and 14 are in the ftz domains immediately posterior to these eve domains. These correlations suggest that stripes 2, 3, and 7 expressed by E+L-eve function normally in initiating en expression. (en stripe 5 is slightly shifted posteriorly, and stripe 6 is weaker than in wild type and may also be slightly shifted anteriorly. These features correlate with early stripe 3 being somewhat weaker and/or narrower than in wild type; see Discussion.) en stripes 9 and 11 are in the domains of eve late stripes 5 and 6 which, as noted above, are apparently induced by early stripe 5/6; these en stripes also appear to be shifted posteriorly (the anteriorly adjacent wg stripes are expanded, Fig. 2F). Eve early stripe 5/6 does not rescue en stripes 10 and 12, which would be in the ftz domains just posterior to early eve stripes 5 and 6, although in some embryos weak stripe-like expression is visible just posterior to stripe 9, which may correspond to a weak stripe 10. The expression of en stripe 2 is unexpected (see Discussion). Finally, en stripes 1, 7, and 8 are completely absent, correlating with the complete absence of eve early stripes 1 and 4.
In E embryos, en stripes 2, 3, 4, 5, 6, 13, and 14 are rescued by the early eve expression (Fig. 2G); in addition, as in EL embryos, a weak stripe 10 is often present. This pattern differs from that of EL in two ways. First, en stripes 9 and 11 are absent. This correlates with the absence of eve late stripes 5 and 6. Second, odd-numbered en stripes (particularly 3 and 5) are relatively weak and incomplete. Some rescue of odd-numbered en stripes is perhaps not surprising due to the temporal and spatial overlap of early and late expression, and the incompleteness of the rescue demonstrates the importance of late eve in establishing normal levels of en. Strikingly, however, this en expression is properly restricted to the anterior portion of the normal early stripes, showing that this aspect of regulation does not require the sharp, refined late expression that has been shown to coincide with these en stripes (see below and Discussion). In contrast, the normal expression of even-numbered en stripes in immediately posterior ftz domains indicates that early eve stripes are sufficient for this function. In summary, expression of an early eve stripe alone completely rescues the even-numbered en stripe in the immediately posterior ftz domain and rescues a weak version of the embedded odd-numbered en stripe, but late expression is additionally required for normal levels of the latter.
wg is another gene regulated by eve and ftz that is essential for segmentation. The previous observation that wg is ectopically expressed throughout the normal eve domain in eve mutants shows that Eve is a repressor of wg (Ingham et al., 1988; Fig. 2D). That this effect is direct is supported by the finding that wg is rapidly repressed following ectopic Eve expression (Manoukian and Krause, 1992). We found that in both E and EL embryos, wg expression is normal in the domains of eve stripes 2, 3, and 7, but remains derepressed in the domains of eve stripes 4-6 (Fig. 2F,H), as it is in Df(eve) mutants (Fig. 2D). Thus, early eve stripes are sufficient for the normal restriction of wg expression within eve domains. The failure of stripe 5/6 to repress wg in E embryos is likely to be due to its low level of expression. In contrast, late eve stripes 5 and 6 in EL embryos, which are expressed at higher levels, do repress wg, resulting in narrow stripes of cells devoid of wg expression within broad wg stripes in this region (F). These observations are consistent with Eve acting as a concentration-dependent morphogen (see Discussion).
Finally, we have investigated whether late eve expression alone can rescue odd-numbered en stripes in the absence of early expression. Embryos transformed with L-eve initiate expression of weak late stripes which are not sharply defined at the anterior border (data not shown). These stripes fade prematurely during gastrulation without rescuing en expression. Since this failure to rescue en may be due to a low level of Eve expression from the transgene, we also tested transgenes containing either a dimer or a trimer of L, which drive late eve expression at higher levels (see Jiang et al., 1991). For most cells, the outcome with an L multimer is the same as with a single L, although in a small number of cells there is enhancement and maintenance of eve expression and initiation of en to varying levels. These exceptional cells notwithstanding, the results clearly show that eve late stripes initiated without early stripes cannot properly organize the eve domain. The regulation of the late element in the absence of early eve expression will be described further elsewhere (M.F. and T.G., unpublished data).
Cuticle patterns confirm the roles of early and late eve expression
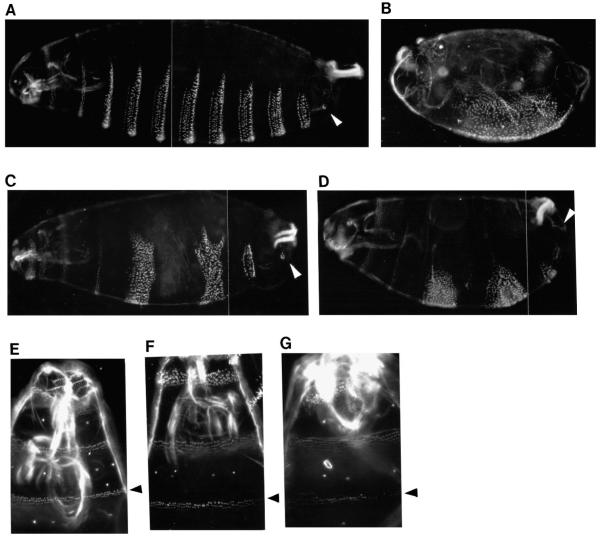
Although en stripes 3 and 5 in the E embryo are weak, their expression recovers to a certain degree in time (data not shown). This recovery is likely a consequence of the intercellular communication known to exist between en- and wg-expressing cells, which is required to maintain their expression (Ingham and Martinez-Arias, 1992). A consequence of this recovery is that the cuticle patterns of E and EL embryos (Fig. 3) are more similar to each other than might be expected from their initial en patterns (see Fig. 2E and G). Nonetheless, there are differences between the two that suggest the functional importance of late eve stripes. For example the T3 denticle band, which derives from the domain of eve stripe 3, is completely recovered in EL embryos but is incomplete in E embryos (Fig. 3F,G). In addition, the portion of the A8 segment that derives from the domain of eve stripe 7 is normal in EL embryos, whereas in E embryos it is missing several structures, including the caudal sensory cone (arrowhead in Fig. 3C,D). Interestingly, in both E and EL embryos an abnormally wide band of naked cuticle is formed in the middle trunk region. This may be a consequence of the presence of stripe 5/6, which results in prolonged expression of wg in this region relative to that in Df(eve), presumably by an indirect mechanism. This aspect of the phenotype is dosage sensitive and is not seen in most embryos containing only one copy of the transgene (data not shown). Overall, the cuticle patterns confirm the effectiveness of early eve expression both in establishing stable parasegmental borders and in organizing intervening odd-numbered parasegments. However, late expression is additionally required for complete functional rescue.
Fig. 3.
Late eve expression assists early eve in rescuing the cuticle pattern. (A,E) wild type. (B) Df(eve). (C,F) EL larva. (D,G) E larva. The caudal sensory cone (csc) in A8 (white arrowhead) is present in EL embryos but is missing in E embryos, indicating the importance of late eve expression, despite the fact that the en pattern in late E embryos recovers significantly and resembles that in EL embryos. (E,F,G) The head/thoracic region, with the black arrowhead indicating the T3 denticle band, which derives from the region of eve stripe 3. Rescue of the T3 band in EL embryos (F) is more complete than in E embryos (G).
odd-skipped mediates the regulation of even-numbered en stripes by eve
Although genetic experiments have shown that eve plays a role in activating both even- and odd-numbered en stripes, they do not tell us whether the interaction is direct. In fact, evidence suggests that it is not. In studying the consequences of ectopic eve expression from a heat-shock promoter, Manoukian and Krause (1992) concluded that the activation of en expression by eve is likely to be an indirect effect. A possible mediator of the activation of ftz-dependent (even-numbered) en stripes by eve is odd-skipped (odd), since even-numbered en stripes, which fail to form in eve hypomorphic mutants, ‘re-appear’ in eve-/odd-double mutants (DiNardo and O’Farrell, 1987). Primary odd stripes overlap the posterior portion of ftz stripes. Thus en, which is expressed in the anterior part of ftz stripes, may be activated by ftz and repressed by odd (Coulter et al., 1990). Furthermore, Manoukian and Krause (1992) concluded, from their studies on ectopic Eve expression, that Eve represses odd and ftz directly, but odd is sensitive to lower levels of Eve than is ftz. Based on this observation, they postulated that the role of eve in regulating en in the ftz domain is to repress odd in the anterior row of ftz-expressing cells. We tested this model by double staining wild-type, Df(eve), and E embryos with probes for odd mRNA and Ftz protein (Fig. 4). Staining of wild-type embryos shows that the Ftz and odd stripes are completely coincident at the precellular blastoderm stage, but odd expression disappears from the anterior-most cells of Ftz stripes during gastrulation (A), as was shown previously (Manoukian and Krause, 1992). Moreover, this ‘clearing’ begins in anterior stripes and progresses toward the posterior in a manner similar to the progressive appearance of en stripes (Manoukian and Krause, 1992). In contrast, in Df(eve) embryos, ftz stripes and odd stripes are completely coincident throughout gastrulation (B). In E embryos, the anterior-most cells of ftz stripes 1, 2, 3, and 7 are devoid of odd expression, correlating with the appearance of en in these locations, whereas stripes 4, 5, and 6 of ftz and odd overlap even at late gastrulation (C). Double staining for En protein and odd mRNA shows that odd message is indeed absent from en expressing cells (D). These results strongly support the eve/odd double negative model of en activation discussed above, and further show that early eve expression alone is sufficient for this effect in the ftz domain.
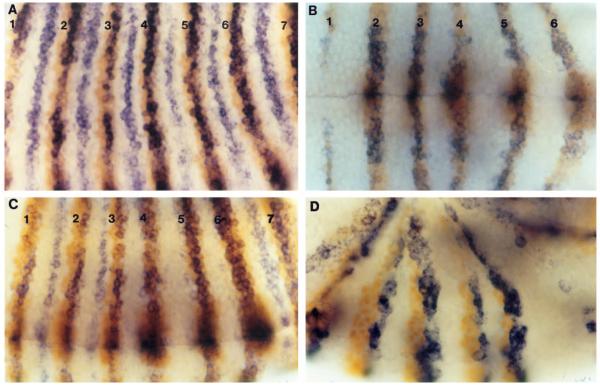
Fig. 4.
Early eve stripes mediate the clearing of odd expression from the anterior-most cells of posteriorly adjacent Ftz stripes. Embryos were hybridized with odd probe (blue), followed by staining with a Ftz monoclonal antibody (orange, A,B,C; Ftz stripes are numbered) or an en monoclonal antibody (orange, D). (A) Wild type; odd mRNA is cleared from the anterior-most cells of Ftz stripes. (B) Df(eve); odd expression is coincident with Ftz expression; both Ftz stripe 1 and odd stripe 1 are weaker than in wild type. (C) E embryo; odd expression is cleared from the anterior cells of Ftz stripes 1, 2, 3, and 7. (D) E embryo; even-numbered en stripes (2, 4, and 6 are shown) are induced just anterior to odd stripes (where Ftz is expressed). (odd is also expressed, more weakly, just posterior to odd-numbered en stripes.)
eve minor stripes are not required for en regulation in the ftz domain
The results thus far leave an unanswered question as to the source of Eve in the ftz domain. As suggested previously (Manoukian and Krause, 1992), a logical candidate has been seven eve minor stripes, which appear during gastrulation and alternate with the strong late stripes, apparently coinciding with late, narrowed ftz stripes (Harding et al., 1986; Macdonald et al., 1986; see Fig. 5A). According to this idea, the level of Eve in these minor stripes, which is very low, is sufficient to repress odd but not sufficient to repress ftz, effectively dividing the ftz stripes into odd expressing and non-expressing cells (Manoukian and Krause, 1992). However, some of our observations are not consistent with this model. For example, the clearing of odd expression from the anterior of ftz stripes 1, 2, and 3 begins during the cellular blastoderm stage, prior to the appearance of the minor stripes (not shown). To clarify the matter further, we examined E and EL embryos for expression of minor stripes (Fig. 5). Gastrulating E embryos do not express minor stripes at detectable levels. However, during germ-band elongation, they do express seven faint stripes near where minor stripes are expected (data not shown). It is not clear whether these late appearing stripes are belated minor stripes. In fact, there is a cis-acting element downstream of the eve transcription unit that can drive reporter gene expression in seven late ftz-like stripes (Charles Sackerson and T.G., unpublished data), and the E-eve construct may contain part of this cis-acting element. However, irrespective of how these stripes relate to normal eve expression, they cannot be the source of odd repression, since these stripes still appear in the regions that lack early eve stripes, and yet they do not rescue the corresponding en stripes (8 and 12). In contrast to E embryos, EL embryos do detectably express some minor stripes, although at lower levels than in wild type (this expression is not clearly reproduced in Fig. 5B). We find that expression of minor stripes is boosted by certain deletions within L in the locations of rescued even-numbered en stripes, suggesting that L is at least partly responsible for the minor stripes; one such embryo (a transformant of RVΔSac in Fig. 1) is shown in Fig. 5C (compare C with the EL embryo shown in B). These observations indicate that E embryos, lacking L, do not have proper minor stripe expression, yet there is rescue of even-numbered en stripes. Therefore, we conclude that the minor stripes are not required for the regulatory effects of eve in the ftz domain, and that it is probably the posterior trailing edge of early stripes that mediate this aspect of eve function.
Fig. 5.
eve minor stripes are derepressed by a deletion within the late element (L). Gastrulating embryos were hybridized with an eve probe. (A) wild type; minor stripe expression is centered between eve major stripes. Minor stripes in the domains of ftz stripes 1, 2, 3, and 7 are marked with square dots. (B) EL embryo; weak minor stripes are just detectable in the domains of ftz stripes 1, 2, 3, and 7 (square dot; more clearly visible under the microscope). Embryos in A and B were from the same staining population. (C) RVΔSac-eve; the transgene contains a small deletion within a shortened, but fully active L (see Fig. 1A). Minor stripes apparently corresponding to those seen in EL embryos (C) are strongly enhanced, indicating that L plays a major role in driving minor stripe expression.
Other pair-rule genes mediate early eve regulation of en
Odd-numbered en stripes fail to appear in loss-of-function mutants of two pair-rule genes, eve and prd, and the posterior of ‘early’ prd stripes overlap the anterior of early eve stripes where en is activated (DiNardo and O’Farrell, 1987; Ingham et al., 1988). From these observations, it was suggested that the combination of eve and prd may directly activate odd-numbered en stripes. For the simplest version of this model to be correct, both Eve and Paired must be transcriptional activators. While Paired does act as a transcriptional activator in cell culture (Han et al., 1989), a growing body of evidence suggests that Eve is a transcriptional repressor (Jaynes and O’Farrell, 1988; Han et al., 1989; Biggin and Tjian, 1989; Johnson and Krasnow, 1992; Manoukian and Krause, 1992; Han and Manley, 1993). If this is its molecular activity in activating odd-numbered en stripes, we might be able to identify intermediates among the known pair-rule genes. Therefore, we examined the expression patterns of all known pair-rule genes (i.e. hairy, runt, prd, odd-paired and sloppy-paired (slp), in addition to ftz and odd) in E and EL embryos. We found that for each of these genes, altered expression patterns are similar in E and EL embryos during blastoderm and early gastrulation (data not shown). This finding suggests that late eve expression does not strongly affect the patterning of other pair-rule genes during the period critical to the activation of en, just as it is not crucial to the proper localization of odd-numbered en stripes. However, late eve does enhance expression of these en stripes. Intermediates thus may share a border of expression with late eve. Therefore, we double-stained EL embryos for expression of eve and each of three genes, prd, runt, and slp, which previous work had shown might be involved in en regulation (see below).
As shown in Fig. 6A, early prd stripes 2 and 3 are sharply defined, while stripes 4-6, in the absence of strong early eve stripes 4-6, are less clearly defined, due to ectopic prd expression throughout the region. This is consistent with the previous observation that eve maintains prd repression between early prd stripes (Gutjahr et al., 1993). (Stripe 1 is regulated differently from the rest (Gutjahr et al., 1993) while the appearance and refinement of prd stripe 7 is delayed relative to the others.) In addition, the relative sharpness of prd stripes 2 and 3 also indicates that peak concentrations of early eve restrict the posterior borders of prd stripes. That prd is not sensitive to lower concentrations of eve is seen in the inability of the weak stripe 5/6 to repress prd (Fig. 6A). In embryos after cellularization (B,C), prd apparently becomes resistant to repression by eve, as stripes 2 and 3 continue to overlap with the high concentrations of Eve in the anterior portions of narrowing eve stripes, while prd stripes 4-6 remain broad despite strong eve late expression in the region. Thus, early eve appears to restrict the posterior borders of early prd stripes, and this border is maintained as eve stripes are refined.
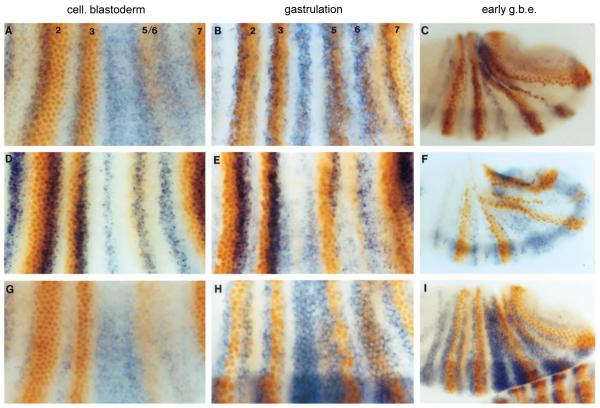
Fig. 6.
Eve represses prd, runt, and slp, regulators of late eve and en. EL embryos were stained for Eve protein (orange) and either prd, runt, or slp RNA (blue) at three developmental stages. (A,B,C) prd and Eve. Eve stripes are numbered. At cellular blastoderm (A), prd stripes 2 and 3 overlap the anterior portions of early Eve stripes (this is less clear with prd stripe 7, which develops later than the others). Early prd expression is posteriorly restricted by Eve, as suggested by the ectopic prd expression observed in the region of stripes 4-6, which also shows that the level of Eve in early stripe 5/6 is not sufficient to repress prd. prd and Eve continue to overlap at gastrulation (B) and early germ band extension (C), indicating that late prd expression is insensitive to Eve repression. (D,E,F) runt and Eve (same developmental stages as A,B,C above). Early runt stripes overlap the posterior portion of early eve stripes and do not expand in the Eve-deficient region (D), suggesting that they are not sensitive to Eve repression. However, late runt expression, which appears at the anterior of eve stripes, seems to be sensitive to repression by Eve (see text), and possibly vice versa, as their patterns show mutual exclusivity with sharp boundaries, particularly at the anterior border of late eve (E,F; see also Manoukian and Krause 1993). (G,H,I) slp and Eve (same stages as above). slp expression, appearing later than eve, is ectopically expressed in the Eve-deficient regions (G), suggesting that it is repressed by early Eve. This apparent sensitivity continues to later stages (H,I).
runt has been shown to be a potent and apparently direct repressor of expression of both late eve and odd-numbered en stripes (Frasch and Levine, 1987; Goto et al., 1989; Manoukian and Krause, 1993). Conversely, ectopic eve can repress runt, perhaps directly (Manoukian and Krause, 1992). The seven early runt stripes, which do not appear to be sensitive to eve repression, overlap the posterior half of early eve stripes (Fig. 6D), consistent with a contribution from runt to the localization of late eve stripes and odd-numbered en stripes. After cellularization, the runt pattern evolves into 14 stripes that eventually cover all the trunk region, except where late eve (and odd-numbered en) and ftz (and even-numbered en) are activated (E,F). An antagonistic relationship between late runt and late eve is suggested by the mutual exclusivity of their expression, even in the region of late eve stripes 5 and 6 in EL embryos, which are not entirely normal, as described earlier. In contrast to its effect on late runt, early eve expression appears to reinforce early runt expression posterior to prd (Fig. 6D,E; Klingler and Gergen, 1993). The sensitivity of late eve and late (14-stripe) runt to repression by Runt and Eve, respectively, may generate the observed mutually exclusive patterns, with early expression providing an initial spatial bias, and late expression sharpening the boundaries. By repressing runt in cells that normally express en, late eve may thus boost the level of odd-numbered en stripes relative to that seen in E embryos.
Previous studies with both slp mutants and ectopic Slp expression indicated that slp is a repressor of eve and en, and may be involved in restricting the anterior borders of late eve and odd-numbered en stripes (Cadigan et al., 1994a,b). slp expression starts relatively late, and the initial slp pattern is complementary to that of eve, as shown in Fig. 6G, suggesting that slp is negatively regulated by eve. In E embryos at this stage as well, while slp expression is normal in the stripe 2, 3, and 7 region, it is derepressed in the stripe 4 region (data not shown), showing that bell-shaped early stripes are responsible for this effect. It appears that slp is more sensitive to repression by eve than is prd, since the low level of Eve in stripe 5/6 is sufficient to repress slp, but not prd. This is also consistent with the fact that slp stripes are normally localized further from the center of early eve stripes than are the corresponding prd stripes. This apparent sensitivity to repression by eve continues as the slp pattern evolves into 14 stripes, as slp is absent in cells expressing high levels of eve (Fig. 6H,I). (Cells in the region of eve stripe 1 do express both eve and slp (not shown), but regulation of several genes in this region appears to be different from that in the rest of the trunk.) Similar to the crossregulation by late eve and runt, the mutual exclusivity of the eve and slp patterns is consistent with the notion that mutual repression by slp and late eve helps to determine the anterior borders of both late eve and odd-numbered en stripes.
DISCUSSION
The expression patterns of most pair-rule genes are seemingly in a state of flux. In the case of eve, the course of these changes can be divided into an early establishment phase and a later refinement phase based both on the shape of the stripes and on the cis-regulatory elements involved. The refinement process has been described as a progressive loss of expression from the posterior of early, broad stripes. This process, as we have shown here, can be effectively reproduced as a simple combination of two static patterns. Broad, early stripes are initiated in response to combinations of earlier acting genes, particularly the gap genes, acting on stripe-specific regulatory elements within the eve gene. These early stripes simply fade, probably uniformly, giving way to late expression driven by a separate regulatory element. This element responds both to early eve expression and to regulatory inputs from other pair-rule genes to give narrow, late expression in the anterior portion of early stripes. The temporal overlap of these discrete early and late stripes gives the observed, apparently progressive refinement. In this work we have also shown that Eve protein expression driven by a combination of early and late elements can functionally substitute for the endogenous eve gene during embryonic pattern formation. We used these separable regulatory elements to express Eve in either an early-only pattern (E embryos) or an early-plus-late pattern (EL embryos) in portions of otherwise eve deficient embryos. This allowed us to separately determine the contributions of the early and late eve patterns to the regulation of its target genes. We found that there are important functional distinctions between early and late expression, as discussed below. This approach provides a number of new insights into how the domains of pair-rule gene expression are divided into multiple functional subregions, providing for correct activation and restriction of expression of downstream genes. Our results suggest that the early eve domain is subdivided by interactions that depend on both concentration-dependent and combinatorial mechanisms.
Activation of en is an important function of eve in both even- and odd-numbered parasegments. We find that early expression alone is sufficient for normal activation of en in even-numbered parasegments. This has implications for the mechanism of eve action in the ftz domain, as discussed below. In contrast, we find that the odd-numbered en stripes, which form in the domain of late eve expression, require both early and late expression for complete activation. While this dual requirement is partially attributable to the fact that the early eve pattern serves to enhance and maintain late expression, there is activation of functional levels of en by early expression alone. Strikingly, this en expression is properly restricted to the anterior portion of the normal early stripes in E embryos. This, along with other results discussed below, suggests that the early bell-shaped eve stripes function as morphogenetic gradients over the distance of just a few cell diameters, regulating downstream genes in a concentration-dependent manner to specify the fates of individual cell rows.
Activation of even-numbered en stripes
Activation of even-numbered en stripes in the ftz domain is apparently attributable to the low level of eve expression at the posterior edges of early stripes. A previously proposed model of en activation suggested that en stripes are initiated in nuclei that express ftz but not odd, with eve being responsible for the ‘clearing’ of odd from the anterior-most cells of each ftz stripe (Manoukian and Krause, 1992). Our data confirm these features of the model, showing that odd disappears from the anterior row of ftz-expressing cells immediately posterior to each early eve stripe, where en is activated. However, there is a discrepancy between our data and the previous proposal that the minor stripes of eve that appear relatively late in the anterior cells of ftz stripes might be the source of eve function there (Manoukian and Krause, 1992). Our data point to early expression as the primary source of eve function in the ftz domain. Since early stripes are bell-shaped, the source of Eve in the ftz domain is presumably the posterior trailing edge of the gradient, although the functional level of Eve there may be below the detection limits of conventional staining procedures. A similar conclusion has been drawn about functional levels of Hairy protein in apparent interstripe regions (Landelli and Ish-Horowicz, 1993).
In the model by Manoukian and Krause (1992) cited above, it was postulated that the differential sensitivity of odd and ftz to repression by eve was sufficient to subdivide the ftz domain into odd-expressing and non-expressing cells. Based on the observation that several other potential target genes of eve also show differential sensitivities to ectopic Eve induction, these researchers further concluded that Eve acts as a concentration-dependent morphogen. Our examination of pair-rule gene expression in E and EL embryos shows that late stripes contribute relatively little to the regulation of downstream pair-rule genes by early stripes. Thus, the morphogenetic activity of Eve arises from the bell-shaped early stripes, rather than from the saw-toothed gradient that results from the sum of early and late expression. However, our data also call for caution in invoking a morphogenetic activity for Eve in the ftz domain. For example, the eve expression within ftz stripe 5 of E embryos, part of eve stripe 5/6, is apparently uniform, and the level is higher than that normally found within the ftz domain (as well as more persistent). Nonetheless, there is some clearing of odd in the anterior portion of ftz stripe 5, and this leads to a variable, weak rescue of en stripe 10. This suggests that something other than a difference of early eve concentration contributes to the distinction between anterior and posterior portions of ftz stripes; this additional regulation may involve a combinatorial mechanism. This notion is also suggested by the observation that en stripe 2 is rescued by E-eve within the anterior-most cells of ftz stripe 1 (Fig. 2E,G), apparently by the abnormally extended anterior edge of eve stripe 2, which increases in expression from anterior to posterior throughout ftz stripe 1 (Fig. 1D,G). However, it should be pointed out that this en stripe is partially eve independent (a semblance of this stripe still forms in eve null mutants), and thus may be regulated differently from other en stripes. Thus, while differential sensitivity of ftz and odd to a gradient of eve may contribute to odd clearing and en activation in the ftz domain, additional mechanisms are apparently also involved.
Activation of odd-numbered en stripes by eve
Odd-numbered en stripes in E embryos, which express only early eve stripes, are weak but properly placed. The proper localization of these stripes without late eve expression supports the notion that the bell-shaped gradient of early eve provides cues that these en stripes normally respond to. eve probably acts indirectly in activating en, since Eve is a repressor in cultured cells and in in vitro transcription assays (Han et al., 1989; Biggin and Tjian, 1989; Johnson and Krasnow, 1992; Han and Manley, 1993), and abnormal en activation by ectopically expressed Eve is delayed (Manoukian and Krause, 1992). It is therefore likely that this regulation involves secondary factors that are normally under the control of early eve stripes. One of these genes is prd. It encodes a transcriptional activator (Prd), based on cell culture assays (Han et al., 1989), and is required for the activation of odd-numbered en stripes. The posterior of ‘early’ prd stripes and the anterior of early eve stripes overlap where en is activated, and these en stripes fail to appear in loss-of-function mutants of either gene (DiNardo and O’Farrell, 1987; Ingham et al., 1988). It was suggested therefore that the combination of eve and prd specifies odd-numbered en stripes. Consistent with this model, ectopic expression of Prd causes posterior expansion of these en stripes (Morrissey et al., 1991). Thus, in addition to activating these stripes, Prd is apparently involved in determining the posterior borders of odd-numbered en stripes. However, rather than simply acting combinatorially with prd, early eve regulates the posterior border of prd expression, as eve mutations cause ectopic activation of prd in this region (Fig. 6A; Gutjahr et al., 1993). Thus, this determinant of the posterior border of odd-numbered en stripes is itself restricted by high concentrations of early eve.
The restriction of the anterior borders of odd-numbered en stripes may be provided, at least in part, by secondary (late) runt stripes. These stripes appear just prior to en activation and abut the en anterior borders, occupying the anterior portion of late prd stripes (Fig. 6; Manoukian and Krause, 1993; Klingler and Gergen, 1993). Eve is probably a direct repressor of runt (Manoukian and Krause, 1992), and we observe an expansion of late runt expression in the Df(eve) portion of locally rescued embryos (Fig. 6; see also Frasch and Levine, 1987). Thus, late runt expression is apparently repressed at a lower concentration of Eve than is prd, thereby allowing en activation in cells that express prd but not runt. However, runt is unlikely to be the only repressor at this border, because these en stripes do not obviously expand in runt mutants. Moreover, odd-numbered en stripes do not re-appear in eve/runt double mutants (our unpublished observation). It is likely that stripes of slp, which also abut the anterior of odd-numbered en stripes (Fig. 6; Grossniklaus et al., 1992), provide a redundant repression function here, since odd-numbered en stripes expand in slp mutants and are repressed by ectopic slp expression (Cadigan et al., 1994a,b). Our data show that slp is repressed by Eve in a concentration dependent manner, and by a lower concentration of Eve than that required to repress prd, since the level of Eve in the stripe 5/6 region of E embryos represses slp but not prd (Fig. 6). Thus, the anterior border of odd-numbered en stripes is abutted by two redundant repressors of these stripes (based on the fact that ectopic expression of either Runt or Slp causes en repression), both of which are repressed by early eve expression.
A model of en activation by a morphogenetic gradient of early eve expression
Previous models for activation of en invoked a combinatorial interaction between prd and late eve to explain the localization of odd-numbered en stripes. Our results show that the effects of eve are more complex. We have found that late eve expression is not required for proper localization of these en stripes. Moreover, both late eve and prd are regulated by early eve, and late eve can be activated by a low level of early eve that cannot repress prd. Moreover, two other pair-rule genes, slp and runt, that negatively regulate en and share stripe borders with en are also regulated by early eve. It was previously shown that ectopically expressed Eve represses both prd and runt, but that a higher concentration of Eve is required to repress prd (Manoukian and Krause, 1993); we have found that repression of prd requires the high concentration of eve present in the middle of early stripes. Further, we have found that slp is repressed by early eve in a concentration dependent manner, and that slp is repressed at a lower concentration of eve than is prd. Based on these observations, we propose that eve acts as a concentration-dependent morphogen in its activation of odd-numbered en stripes, as illustrated in Fig. 7.
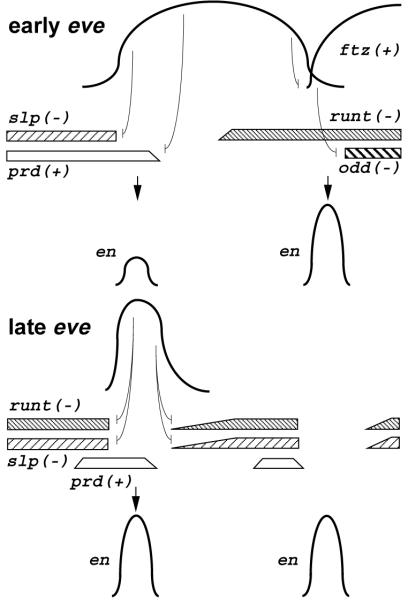
Fig. 7.
A model of en activation by the Eve morphogen. Target genes are repressed by different concentrations of Eve in early stripes, leading to specific patterning of en activators (prd and ftz) and repressors (slp, runt, and odd). Combinations of these gene activities lead to activation of both odd-numbered en stripes and late eve stripes at the anterior edge of early eve stripes, and even-numbered en stripes at the posterior edge of early eve stripes. Late eve expression reinforces the repression of en repressors, allowing strong expression of odd-numbered en stripes. See the text for details.
In this model, high levels of early eve repress prd, restricting its posterior border, while lower levels allow prd expression. In eve mutants, prd is ectopically expressed, but en is not activated. This is accounted for in the model by the proposed negative regulators of the en anterior border, slp and runt, which are also ectopically activated in eve mutants. These repressors set the anterior border of odd-numbered en stripes by being repressed at lower levels of early eve than is prd. These two concentration thresholds for repression by Eve provide a narrow region where prd is activated, but not runt and slp, resulting in activation of en. en is activated only toward the anterior, and not the posterior, of the early eve stripe due to the expression of early runt in the posterior.
wg is also repressed by early eve, since its expression expands throughout the eve-deficient portion of E embryos. prd is required for wg activation adjacent to odd-numbered en stripes (Ingham and Hidalgo, 1993), and wg is very sensitive to repression by both ectopically expressed Eve (Manoukian and Krause, 1993) and En (Heemskerk et al., 1991). Activation of en in the posterior row of prd-expressing cells is therefore capable of restricting wg expression to the anterior portion of late prd stripes, adjacent to the odd-numbered en stripes. Consistent with this model for subdivision of the prd stripe in E embryos, weaker than normal odd-numbered en stripes induced in E embryos are flanked by slightly expanded wg stripes (Fig. 2H). Thus, multiple target genes are repressed at different concentrations of Eve at the anterior border of early stripes to provide for the two adjacent cell identities (wg- and en-expressing cells) that establish the parasegmental border.
There is a strong correlation between the locations of stable late eve stripes and odd-numbered en stripes in all known circumstances, suggesting that they respond to a very similar set of regulatory interactions. Thus, like en, the anterior border of late eve is probably determined by the combined repressive action of secondary runt and slp stripes, whose sharp posterior borders abut late eve stripes (Fig. 6). Consistently, eve, like odd-numbered en stripes, is sensitive to repression by ectopic expression of either Slp or Runt (Cadigan et al., 1994a). Furthermore, eve stripes expand anteriorly in slp mutants during germ-band elongation (Cadigan et al., 1994b), after runt has faded. The reinforcing effect of late eve expression on odd-numbered en stripes is most likely due to its preventing late runt and slp expression from encroaching into the posterior-most row of prd-expressing cells, where en is activated (Fig. 7). Late eve can also restrict wg to the anterior-most row of prd-expressing cells, adjacent to the incipient en stripes. Thus a presumptive parasegment border may initially result from mutual repression between eve on one side and runt and slp on the other. Interestingly, prd, which is completely repressed by high levels of early eve expression, is not repressed by the high levels of late eve. This may be the result of prd having distinct early and late regulatory programs, analogous to those of eve (Gutjahr et al., 1993).
It is interesting to note that a number of pair-rule genes, including eve, prd, runt, and slp, have both pair-rule expression patterns and, later, narrower expression patterns in every segmental primordium. This suggests an evolutionary relationship between molecular interactions that occur at the pair-rule and segment polarity stages of pattern refinement. However, assuming such an evolutionary relationship, our results suggest that the molecular interactions at these two stages of development have been modified during evolution, such that, as with the effects of eve on prd, a strong interaction at one stage may be completely absent at the other. These modifications are apparently important to pattern refinement, in this case allowing the early Eve gradient to first restrict and then subdivide a domain of prd expression to establish the parasegment border. Early prd expression appears to activate late prd, so that the posterior restriction of early prd by eve is reflected in the late prd pattern.
A satisfying aspect of the model that eve acts as a bell-shaped gradient morphogen is that it can explain the relative shift of late eve and ftz stripes that has been observed in eve hypomorphs. Earlier work described this as an anterior shift of ftz expression relative to that of eve (Frasch et al., 1988). However, one could also account for this data as a posterior shift of late eve expression (DiNardo and O’Farrell, 1987). The latter interpretation is supported by the fact that we see little or no ftz shift between the eve+ and eve-deficient portions of locally rescued embryos, while there is an apparent posterior shift of late eve expression in the stripe 5/6 region, where the early eve expression level is lower than normal. In eve hypomorphs, the level of eve activity is reduced, presumably resulting in a similar posterior shift of late eve stripes. Since late eve stripes of approximately normal width still form, the determinants of both their anterior and posterior borders apparently shift toward higher concentrations of early eve. Therefore, this coordinated shift in response to reduced eve activity further supports the model that, in addition to acting combinatorially with other pair-rule gene products, Eve behaves as a concentration-dependent morphogen.
Acknowledgments
We thank Zhifang Li, Anita John, and Yoshiko Ishikawa for technical assistance, and Dr Charles Sackerson for encouragement. We are grateful to Drs Kenneth Cadigan, Douglas Coulter, Steve DiNardo, Ian Duncan, Peter Gergen and Dianne Mattson for providing us with antibodies and probes.
REFERENCES
- Akam M. The molecular basis for metameric pattern formation in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Biggin MD, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Cadigan K, Grossniklaus U, Gehring W. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994a;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Cadigan K, Grossniklaus U, Gehring W. Functional redundancy: The respective roles of the two sloppy paired genes in Drosophila segmentation. Proc. Nat. Acad. Sci. USA. 1994b;91:6324–6328. doi: 10.1073/pnas.91.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieshaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990;8:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, O’Farrell P. Establishment and refinement of segmental patterning in Drosophila embryo: Spatial control of engrailed expression by pair-rule genes. Genes Dev. 1987;1:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Frasch M, Warrior R, Tugwood J, Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 1988;2:1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- Goto T, MacDonald P, Maniatis T. Early and late patterns of even-skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1035–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Frei E, Noll M. Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair-rule genes. Development. 1993;117:609–623. doi: 10.1242/dev.117.2.609. [DOI] [PubMed] [Google Scholar]
- Han K, Levine M, Manley J. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Han K, Manley J. Transcriptional repression by the Drosophila Even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M. Auto-regulatory and gap response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–10. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y, Kuroiwa A, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez-Arias A. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even-skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Martinez-Arias A. Boundaries and fields in early embryos. Cell. 1992;68:221–235. doi: 10.1016/0092-8674(92)90467-q. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Hidalgo A. Regulation of wingless transcription in the Drosophila embryo. Development. 1993;117:283–289. doi: 10.1242/dev.117.1.283. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: Combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Johnson BF, Krasnow MA. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- Kellerman KA, Mattson DM, Duncan I. Mutations affecting the stability of the fushi tarazu protein of Drosophila. Genes Dev. 1990;4:1936–1950. doi: 10.1101/gad.4.11.1936. [DOI] [PubMed] [Google Scholar]
- Klingler M, Gergen JP. Regulation of runt transcription by Drosophila segmentation genes. Mech. Dev. 1993;43:3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- Landelli M, Ish-Horowicz D. Drosophila hairy pair-rule gene regulates embryonic patterning outside its apparent stripe domains. Development. 1993;118:255–266. doi: 10.1242/dev.118.1.255. [DOI] [PubMed] [Google Scholar]
- Lawrence PA. Pair-rule genes: Do they paint stripes or draw lines? Cell. 1987;51:879–880. doi: 10.1016/0092-8674(87)90573-3. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P. Pattern formation in the Drosophila embryo: Allocation of cells to parasegments by even-skipped and fushi tarazu. Development. 1989;105:761–767. doi: 10.1242/dev.105.4.761. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham PW, Struhl G. Isolation, structure, and expression of even-skipped: A second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause H. Concentration dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause H. Control of segmental asymmetry in Drosophila embryos. Development. 1993;118:785–796. doi: 10.1242/dev.118.3.785. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A, Lawrence PA. Parasegments and compartments in the Drosophila embryo. Nature. 1985;313:639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Morrissey D, Askew D, Raj L, Weir M. Functional dissection of the paired segmentation gene in Drosophila embryos. Genes Dev. 1991;5:1684–1696. doi: 10.1101/gad.5.9.1684. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Kluding H, Jurgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harbor Symp. Quant. Biol. 1985;50:145–154. doi: 10.1101/sqb.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element mediated transformation and tisssue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Turner FR, Kaufman TC. Defects in embryogenesis in mutants associated with the Antennapedia gene complex of Drosophila melanogaster. Dev. Biol. 1984;102:147–172. doi: 10.1016/0012-1606(84)90182-9. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila, A Practical Approach. IRL Press; Oxford, UK: 1986. pp. 199–228. [Google Scholar]