Proper characterization of aminoacyl- and peptidyl-tRNA, key components of protein biosynthesis, is of crucial importance in the study of the multifaceted aspects of translation. Analysis of ribosome-associated aminoacyl- and peptidyl-tRNAs has often been approached via sucrose gradients followed by scintillation counting [1]. This method is suitable to gain information about the entire ribosomal complex but it fails to provide direct knowledge on the molecular size of aminoacyl- and peptidyl-tRNA. Previous attempts to perform direct analysis on peptidyl-tRNA were based on native gel electrophoresis [2]. On the preparative scale, aminoacyl- and peptidyl-tRNAs have been isolated by ion exchange [3,4], reverse phase, size exclusion or affinity chromatography. However, the latter approaches are not as convenient as gel electrophoresis and may require larger amount of samples.
Small scale gel electrophoresis is an effective tool for the analytical characterization of biomolecules. While the highly negatively charged RNA is typically detected via urea-containing gels, peptides and proteins require the presence of SDS, which serves as a denaturing agent able to coat the polypeptide surface with negative charges. The above strategies enable proteins or RNA to be both unfolded and coated with surface negative charges, allowing molecular discrimination to be based solely on molecular size. Electrophoretic analysis of peptidyl-tRNA, however, poses special challenges because this molecule embraces both polypeptide and RNA character. Furthermore, the ester bond linking the two moieties is easily hydrolysable. Here, we report a versatile method for the direct analysis of both aminoacyl- and peptidyl-tRNAs of variable chain length via 3-layer sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 1 in the presence of tris-acetate (TA) at pH 5.7. This gel system is particularly powerful in that (a) it is compatible with both the peptide and RNA components, (b) it minimizes hydrolysis of the ester bond linking the 3′ tRNA end to the C-terminal amino acid carboxyl group, and (c) it is suitable for the detection of a wide range of peptide sizes both in the presence and absence of covalently bound tRNA. Fluorescence detection is proposed here in conjunction with the TA gels [5].
Basic pH is known to favor hydrolysis of the ester bond connecting tRNA to its conjugate amino acid or polypeptide. Investigations of the pH-dependence of ester hydrolysis, from basic values down to pH 7 [6], indicates that the extent of hydrolysis is progressively reduced as pH decreases. The data in Figure 1 of this work confirm the above, and show that slightly acidic pH is even better than neutral pH at preventing hydrolysis. Conventional Laemmli tris-HCl [7] or tris-tricine (TT) [8] gels used for polypeptides and proteins typically employ components at relatively high pH (c.a. 8.5 - 8.8) for the resolving layer. In contrast, the approach presented here takes advantage of tris-acetate solutions at acidic pH (pH 5.7) in all three gel layers and the running buffer, to minimize the extent of in-gel hydrolysis.
Fig. 1.
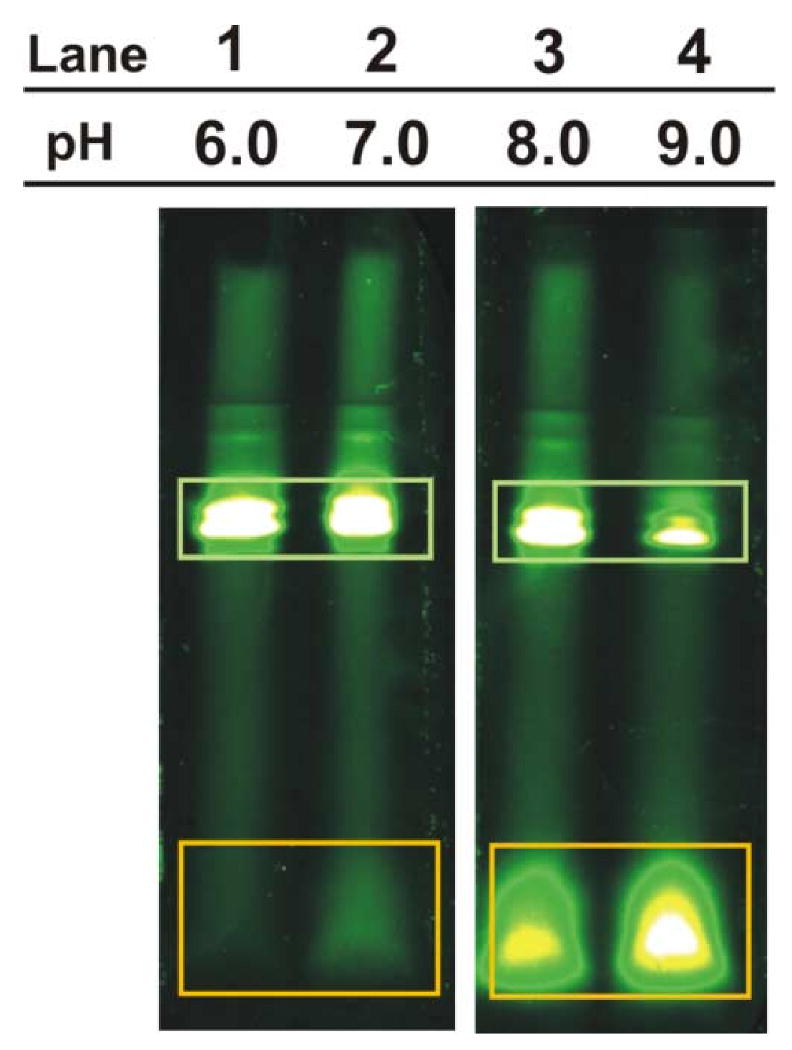
Analysis of BODIPYFL-Met-tRNA fMet incubated under different conditions by low pH TA SDS-PAGE. BODIPYFL-Met-tRNAfMet samples were hydrolyzed in 0.1 M tris buffers, adjusted to pH values ranging from 6. to 9 by addition of aqueous HCl, at 37 °C for 1.5 hrs. Bands corresponding to BODIPYFL-Met-tRNAfMet and its ester bond hydrolysis products are enclosed in green and orange rectangles, respectively.
Unlike low pH urea-PAGE gels, low pH SDS-PAGE gels have only occasionally been used before [9]. Typical applications of the latter type employ an unusually low pH (2-4) and suffer from a number of practical shortcomings, largely due to the nature of the gel composition. Namely, they employ catalysts such as ascorbic acid and iron sulfate, and require low temperatures (2-4 °C) for both gel polymerization and running [10]. Long polymerization times are typically involved. The gel system introduced here, in contrast, uses the common catalysts ammonium persulfate (APS) and tetramethylethylenediamine (TEMED) and it polymerizes promptly at room temperature. The minimization of ester bond hydrolysis and fast polymerization/run time render the three-layer TA SDS-PAGE gels convenient and effective for the analytical characterization of both aminoacyl- and peptidyl-tRNA.
The low pH SDS-based TA gels (10 cm total height) consist of a 5 cm resolving layer (14.1% T, 3.3% C, 6% glycerol), a 1 cm spacer layer (8.6% T, 3.3% C) and a 1 cm stacking layer (4.6% T, 3.3% C). Here, T and C denote acrylamide plus bis-acrylamide, and bis-acrylamide alone, respectively. All three layers are prepared as aqueous solutions in 1 M tris base and 4 mM SDS, The solution pH is then adjusted to 5.7 with acetic acid. All the tris base solutions that have been pH-adjusted with acetic acid are denoted here as tris-acetate. Polymerization was initiated by addition of 1, 1 and 3 mM APS and 5, 5, 15 mM TEMED to the resolving, spacer and stacking layers, respectively. Polymerization of each TA gel layer takes 10-15 minutes. The loading buffer consists of 84 mM tris-acetate (pH 5.7), 116 mM SDS and 28% glycerol. Samples were mixed with loading buffer and incubated at 100°C for three minutes before loading onto the gel. Gels were run at room temperature at constant voltage (100V) for 210 minutes in 3.5 mM SDS and 100 mM Tris-acetate (pH 5.7).
The reference TT gels (10 cm total height) consist of a 5 cm resolving layer (13.3% T, 3.3% C, 6% glycerol), a 1 cm spacer layer (8.1% T, 3.3% C) and 1 cm stacking layer (3.2% T, 3.3% C). The TT gels were prepared according to procedures by Schägger et al. [8]. Samples were mixed with loading buffer and incubated at 100°C for three minutes before loading. TT gels were run in 100 mM tris-tricine adjusted to pH 8.5 and 3.5 mM SDS at constant voltage (100V) for 100 minutes. Gel band detection was accomplished by fluoroimaging with a UC4×4 fluorescence scanner (Genomic Solutions, Ann Arbor, MI) using the blue 488 nm laser excitation source and a 512 nm bandpass filter.
The effect of aminoacyl-tRNA exposure to aqueous solutions of different pH (prior to SDS-PAGE), including slightly acidic conditions, was tested. This experiment was designed to probe (a) the effect of pH on ester hydrolysis, and (b) whether the low pH TA gel is suitable to properly evaluate the extent of ester bond hydrolysis occurring prior to gel analysis. BODIPYFL-Met-tRNAfMet, prepared as described [5], was incubated for 1.5 hrs at 37°C in 0.1 M tris at pH values ranging from 6 to 9. The solutions were then analyzed by TA SDS-PAGE. As shown in Figure 1, the bands corresponding to BODIPYFL-Met-tRNAfMet increase in intensity as the pH of the hydrolysis buffer decreases. Conversely, the bands corresponding to the fluorescence-labeled hydrolysis products become less intense at lower pH. This result shows that (a) the uncatalyzed hydrolysis of the ester joining amino acid and tRNA is pH-depedent, (b) the extent of ester hydrolysis decreases monotonically from pH 9 to 6 in 0.1 M tris at 37°C, (c) BODIPYFL-Met-tRNAfMet does not undergo ester hydrolysis at pH 6.0, and (d) the TA gel, which operates at pH close to 6, is highly effective at reporting aminoacyl-tRNA hydrolysis as it does not appear to cause any further in-gel reaction.
Fluorophore-labeled ribosome-bound model polypeptides derived from sperm whale apomyoglobin (apoMb) were also analyzed. Homogeneous populations of ribosome-bound polypeptide-tRNA conjugates bearing either 9 (ApoMb0-8) or 141 (ApoMb0-140) amino acids were generated by in vitro coupled transcription/translation in a home-made E. coli cell-free system prepared as described [11]. Briefly, plasmid DNA bearing an open reading frame gene sequence encoding the protein of interest was added to the cell-free mixture. An excess of antisense oligodeoxynucleotides complementary to specific regions of the target mRNA transcript [12] were then added. The antisense oligodeoxynucleotides hybridize to the newly transcribed mRNA at the desired ribosome stalling site. The resulting DNA:RNA hybrid acts as a substrate for the RNase H enzyme, which mediates mRNA cleavage. The translating ribosomes stall upon reaching the truncated 3′ mRNA end. Nascent polypeptides were cotranslationally labeled with the fluorescent probe BODIPY®FL, denoted here as BODIPYFL. BODIPYFL-sulfosuccinimidyl ester (Invitrogen, Carlsbad, CA) was reacted with Met-tRNAfMet to generate BODIPYFL-Met-tRNAfMet [5]. N-terminal methionines were then tagged by supplementing cell-free systems with BODIPYFL-Met-tRNAfMet [5].
The antibiotic puromycin binds the ribosomal A site and it undergoes transpeptidation with the nascent chain, causing peptide release from the tRNA [13]. Puromycin is active only in the presence of the ribosomal peptidyl-transferase center. Treatment with puromycin before gel sample preparation was used to assay for the presence of ribosome bound peptidyl-tRNA.
The low pH TA (pH 5.7) and TT (pH 8.5) SDS-PAGE profile for the peptidyl-tRNAs is shown in Figure 2. Bands corresponding to tRNA-linked and puromycin-treated nascent ApoMb0-8 and ApoMb0-140 are enclosed by green and orange rectangles, respectively. Puromycin causes a decrease in the molecular mass of the nascent chain due to loss of tRNA (Fig. 2A: lanes 2, 4, 6, 8). Spontaneously hydrolyzed peptidyl-tRNA gives rise to bands (lanes 1, 5, 7) running similarly to those due to the puromycin-released polypeptides. In general, ApoMb0-140-tRNA undergoes more spontaneous hydrolysis than ApoMb0-8-tRNA (Fig. 2A).
Fig. 2.
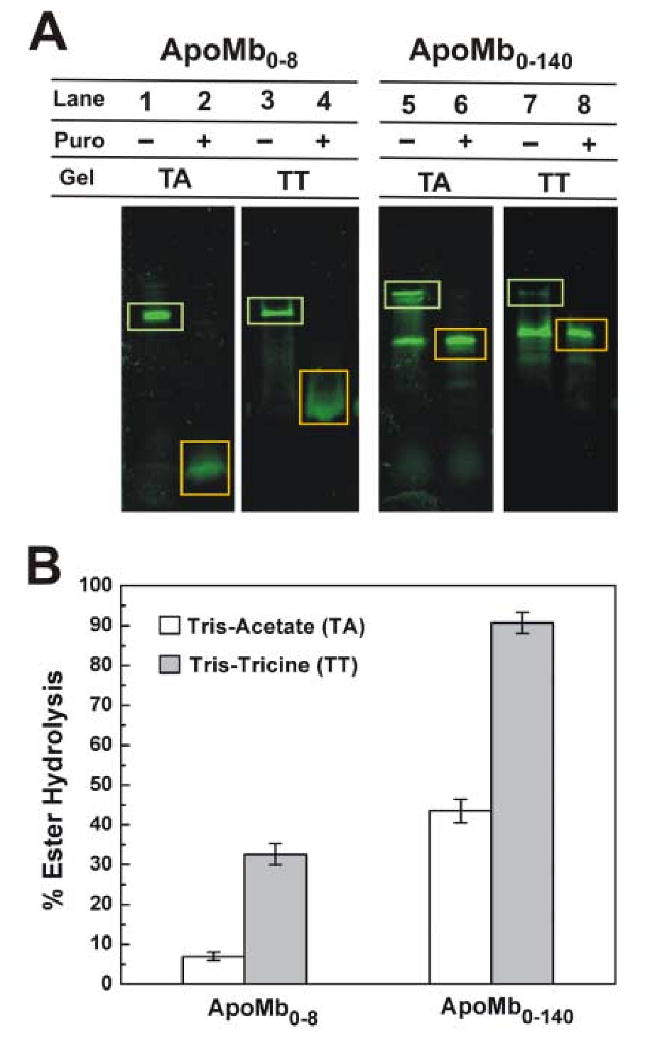
(A) Analysis of cell-free-generated peptidyl-tRNAs bearing N-terminal portions of the amino acid sequence of apoMb by low pH TA (Lanes 1, 2, 5 and 6) and reference pH TT (Lanes 3, 4, 7, and 8) SDS PAGE. All ribosome-associated nascent apoMb polypeptides were N-terminally labeled with BODIPYFL-Met-tRNAfMet. Bands corresponding to nascent polypeptides covalently linked to tRNA and tRNA-released by treatment with puromycin are enclosed in green and orange rectangles, respectively. (B) Diagram providing the quantitative extent of ester hydrolysis for peptidyl-tRNA run on the TA and TT gels in panel A (lanes 1, 3, 5, and 7). For each lane, the percent of ester hydrolysis was determined as the ratio of hydrolyzed product to total fluorescence intensity.
ApoMb0-8-tRNA gives rise to distinct bands by TA gel analysis (Fig. 2A, lane 1), showing that ester hydrolysis is minimal. However, some in-gel hydrolysis occurs in the TT gels, as evidenced by the smearing (Fig. 2A, lane 3). Quantitative analysis of band intensities by the software IMAGEJ [14], Fig. 2B) confirms that TT gels promote more ApoMb0-8-tRNA ester hydrolysis than TA gels.
TT SDS-PAGE erroneously suggests that ApoMb0-140-tRNA is nearly completely hydrolyzed (Fig. 2A, lane 7). However, only partial hydrolysis has occurred prior to gel analysis, as shown by TA SDS-PAGE (Fig. 2A, lane 5). This concept is confirmed by the block diagram of Figure 2B.
In summary, conventional high pH TT-type SDS-PAGE is not ideal for the analysis of aminoacyl- and peptidyl-tRNA. Low pH TA SDS-PAGE, in contrast, offers a convenient and easy-to-implement alternative. The pH 5.7 TA gel system introduced here is effective at preserving ester bonds of aminoacyl- and peptidyl-tRNA over a wide range of target polypeptide chain lengths. TA gels are preferable to the Laemmli-type [7] and TT gels [8], which typically operate at pH 8.5-8.8, for the analysis of aminoacyl and peptidyl-tRNA.
The success and simplicity of TA gels coupled with fluorescence detection provide exciting opportunities for studies on peptidyl- and aminoacyl-tRNA. For instance, the ability to reliably access populations of ribosome-bound peptidyl-tRNAs (by discriminating them from the corresponding hydrolytic products) will be instrumental in protein biosynthesis and cotranslational folding investigations. The gel system reported here is unique in that it utilizes a uniformly low pH environment for all layers, it polymerizes efficiently at room temperature, and it is suitable for the analysis of both the nucleic acid and amino acid components of aminoacyl- and peptidyl-tRNA.
Acknowledgments
We thank Gisela Kramer (Department of Chemistry and Biochemistry, University of Texas-Austin) for helpful discussions. We are grateful to Jamie P. Ellis for the gift of BODIPYFL-Met-tRNA fMet. This research was supported by the NIH (grant R21GM071012). M.K.I. and R.N.K were awarded fellowships by the UW-Madison Honors Summer Sophomore Research Apprenticeship Program and the NSF (REU program), respectively. J.J.H. is the recipient of a Taiwan Merit Scholarship (TMS-094-1-B-004).
Footnotes
Abbreviations used: SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TA, tris-acetate; TT, tris-tricine; APS, ammonium persulfate; TEMED, tetramethylethylenediamine; T, percentage of monomer (acrylamide plus bis-acrylamide); C, percentage of crosslinker (bis-acrylamide); apoMb, apomyoglobin.
Category: Peptides, Amino Acids, and Amino Acid Derivatives.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Voigt J, Nagel K. Regulation of elongation factor g GTPase activity by the ribosomal state. J Biol Chem. 1993;268:100–106. [PubMed] [Google Scholar]
- 2.Plapp FV, Tilzer L, Hayes LC, Chiga M. A rapid method for isolation of peptidyl-tRNA from liver polysomes. Anal Biochem. 1975;64:170–176. doi: 10.1016/0003-2697(75)90418-2. [DOI] [PubMed] [Google Scholar]
- 3.Kiely ML, McKnight GS, Schimke RT. Studies on attachment of carbohydrate to ovalbumin nascent chains in hen oviduct. J Biol Chem. 1976;251:5490–5495. [PubMed] [Google Scholar]
- 4.Kirk TZ, Evans JS, Veis A. Biosynthesis of type I procollagen. J Biol Chem. 1987;262:5540–5545. [PubMed] [Google Scholar]
- 5.Gite S, Mamaev S, Olejnik J, Rothschild K. Ultrasensitive fluorescence-based detection of nascent proteins in gels. Anal Biochem. 2000;279:218–225. doi: 10.1006/abio.1999.4472. [DOI] [PubMed] [Google Scholar]
- 6.Bresler S, Gravjevskaja R, Kirilov S, Saminski E. Stability of peptidyl-tRNA-The intermediate of protein synthesis. Biochim Biophys Acta. 1968;155:465–475. doi: 10.1016/0005-2787(68)90192-5. [DOI] [PubMed] [Google Scholar]
- 7.Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Schägger H, Vonjagow G. Tricine sodium dodecyl-sulfate polyacrylamide-gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 9.Lichtner R, Wolf HU. Dodecyl sulfate polyacrylamide-gel electrophoresis at low pH values and low-temperatures. Biochem J. 1979;181:759–761. doi: 10.1042/bj1810759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa FJR, Lima L, Pacheco ABF, Oliveira CLP, Torriani I, Almeida DF, Foguel D, Silva JL, Mohana-Borges R. Tetramerization of the lexA repressor in solution: Implications for gene regulation of the E-coli SOS system at acidic pH. J Mol Biol. 2006;359:1059–1074. doi: 10.1016/j.jmb.2006.03.069. [DOI] [PubMed] [Google Scholar]
- 11.Bakke CK, Jungbauer LM, Cavagnero S. In vitro expression and characterization of native apomyoglobin under low molecular crowding conditions. Protein Expr Purif. 2006;45:381–392. doi: 10.1016/j.pep.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Kramer G, Kudlicki W, Hardesty B. Cell-free coupled transcription-translation systems from Escherichia Coli. In: Hames BD, editor. Protein Expression. Oxford University Press; 1999. pp. 201–223. [Google Scholar]
- 13.Ramachandiran V, Willms C, Kramer G, Hardesty B. Fluorophores at the N terminus of nascent chloramphenicol acetyltransferase peptides affect translation and movement through the ribosome. J Biol Chem. 2000;275:1781–1786. doi: 10.1074/jbc.275.3.1781. [DOI] [PubMed] [Google Scholar]
- 14.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]


