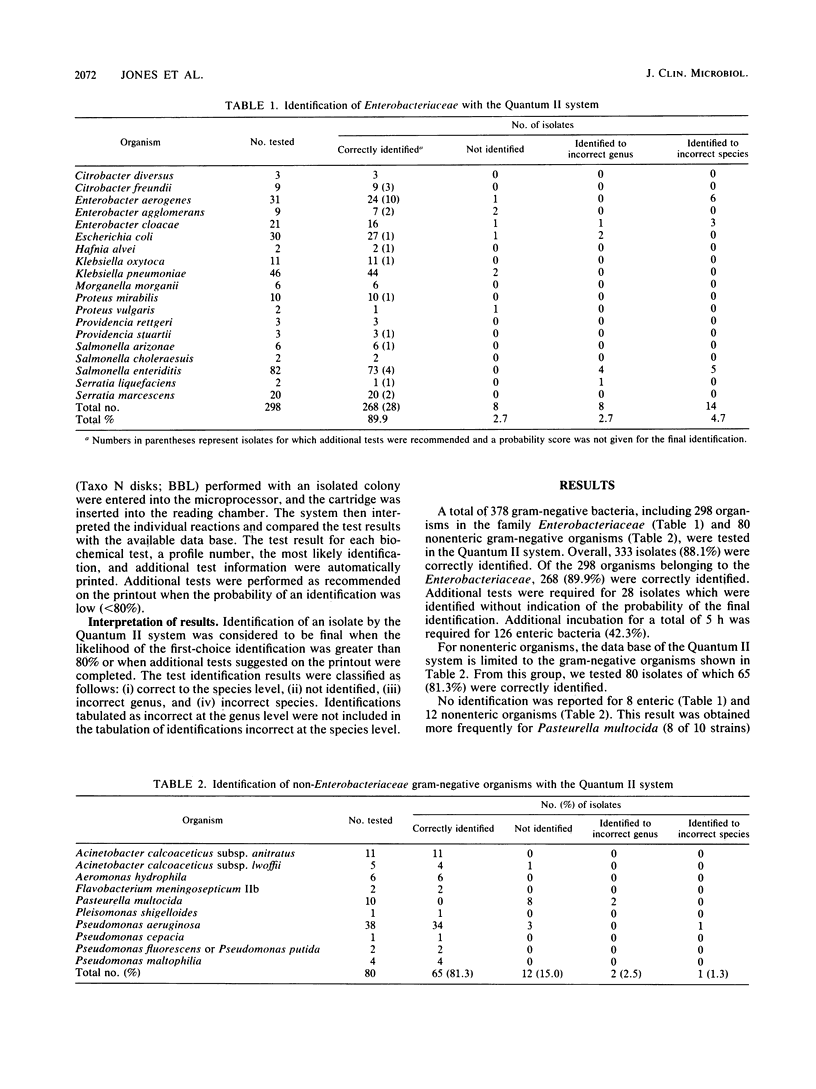
Abstract
The ability of a rapid, semiautomated bacterial identification system, the Quantum II microbiology system (Abbott Laboratories, Irving, Tex.), to accurately identify gram-negative bacteria from veterinary sources was evaluated. A total of 378 isolates were tested, including 298 organisms in the family Enterobacteriaceae and strains representing Acinetobacter sp., Aeromonas sp., Flavobacterium sp., Pasteurella multocida, Plesiomonas sp., and Pseudomonas spp. Of these isolates, 333 (88.1%) were correctly identified, 20 (5.3%) were not identified, 10 (2.6%) were incorrectly identified at the genus level, and 15 (4.0%) were incorrectly identified at the species level. The Quantum II system correctly identified 268 (89.9%) of the isolates of Enterobacteriaceae and 65 (81.3%) of the nonenteric isolates. P. multocida was not identified correctly, and some nonenteric gram-negative bacteria of clinical significance in veterinary medicine are not included in the data base. The Quantum II system provided an accurate identification system for isolates of Enterobacteriaceae but had limited usefulness for the identification of other gram-negative bacteria of clinical significance in veterinary medicine.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adney W. S., Jones R. L. Evaluation of the RapID-ANA system for identification of anaerobic bacteria of veterinary origin. J Clin Microbiol. 1985 Dec;22(6):980–983. doi: 10.1128/jcm.22.6.980-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. T., Swanson E. C. Use of the API 20E system to identify non-Enterobacteriaceae from veterinary medical sources. Am J Vet Res. 1981 Jul;42(7):1269–1273. [PubMed] [Google Scholar]
- Devenish J. A., Barnum D. A. Evaluation of the API 20E system for the identification of gram-negative nonfermenters from animal origin. Can J Comp Med. 1982 Jan;46(1):80–84. [PMC free article] [PubMed] [Google Scholar]
- Ewing W. H. The nomenclature of Salmonella, its usage, and definitions for the three species. Can J Microbiol. 1972 Nov;18(11):1629–1637. doi: 10.1139/m72-252. [DOI] [PubMed] [Google Scholar]
- Fierer J., Fleming W. Distinctive biochemical features of Salmonella dublin isolated in California. J Clin Microbiol. 1983 Mar;17(3):552–554. doi: 10.1128/jcm.17.3.552-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P. R., Gauthier A., Niles A. Evaluation of the Quantum II and Rapid E identification systems. J Clin Microbiol. 1984 Sep;20(3):509–514. doi: 10.1128/jcm.20.3.509-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Bale M. J., Schulte K. R., Koontz F. P. Comparison of the Quantum II Bacterial Identification System and the AutoMicrobic System for the identification of gram-negative bacilli. J Clin Microbiol. 1986 Jan;23(1):1–5. doi: 10.1128/jcm.23.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M., Henrichsen C., Smith M. A collaborative evaluation of a rapid, semi-automated identification system for gram-negative bacilli: the Quantum II BID. J Med Microbiol. 1986 Mar;21(2):145–150. doi: 10.1099/00222615-21-2-145. [DOI] [PubMed] [Google Scholar]
- Swanson E. C., Collins M. T. Use of the API 20E system to identify veterinary Enterobacteriaceae. J Clin Microbiol. 1980 Jul;12(1):10–14. doi: 10.1128/jcm.12.1.10-14.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester M. K., Washington J. A., 2nd Evaluation of the Quantum II Microbiology System for bacterial identification. J Clin Microbiol. 1984 Dec;20(6):1196–1197. doi: 10.1128/jcm.20.6.1196-1197.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J. R. Bacteriological, biochemical and virulence studies on Salmonella dublin from abortion and enteric disease in cattle and sheep. Vet Rec. 1972 Feb 26;90(9):236–240. doi: 10.1136/vr.90.9.236. [DOI] [PubMed] [Google Scholar]


