Abstract
The signaling programs that enforce negative selection in early transitional (T1) B cells in response to B cell receptor (BCR) engagement remain poorly defined. We carried out a comprehensive comparison of BCR signaling in T1 vs. follicular mature (FM) splenic B cells. T1, in contrast to FM B cells, failed to express key NF-κB target genes in response to BCR engagement; and exhibited a striking defect in assembly of an active transcriptional complex at the promoter of the survival and proliferative genes, A1 and c-Myc. Surprisingly, and contrary to previous models, classical PKC and IKK activation, NF-κB nuclear translocation and DNA binding were intact in T1 B cells. Further, despite a marked reduction in NFAT1 expression, differential NFAT or AP-1 activation cannot explain this transcriptional defect. Our combined findings demonstrate that T1 B cells are programmed for signal- and stage-specific ‘nuclear non-responsiveness’ upon encounter with self-antigens.
INTRODUCTION
B cell development begins within the bone marrow, starting from common lymphoid progenitor cells. Development proceeds through discrete stages that ultimately lead to the generation of immature B cells expressing a functional B cell receptor (BCR) on the cell surface. Immature B cells subsequently emigrate to the periphery where progression through transitional developmental stages within the spleen leads to the formation of naïve mature B cells capable of activation and differentiation into antibody-secreting cells upon encounter with cognate antigen (1). During this developmental sequence, tolerance mechanisms progressively shape the BCR repertoire and are critical to ensuring a self-tolerant B cell pool (2). In the BM, encounter with self-antigen can lead to one of three outcomes: deletion by apoptosis, induction of anergy or receptor editing and re-expression of a new BCR. Deletion primarily occurs when receptor editing fails to produce a self-tolerant BCR (3). While the precise events that govern the decision to undergo receptor editing vs. anergy remain incompletely defined, high avidity antigens appear to preferentially promote editing or deletion, while weaker interactions more commonly induce anergy (2).
B cell tolerance mechanisms are less well-characterized in the periphery. However, detailed analysis of BCR specificity in human peripheral B cells has demonstrated a ~50% reduction in self-reactivity in mature vs. newly formed B cells indicating that tolerance mechanisms are also active at this stage (4). In addition, deletion of self-reactive B cells occurs in vivo when self-antigen is specifically expressed in the periphery (5). In contrast to BM B cells, however, transitional B cells in the periphery lose the capacity to receptor edit and instead undergo deletion upon encounter with high avidity self-antigens in vivo (5, 6). Based on the abundance of autoimmune diseases associated with antibodies that recognize multimeric, high-avidity, self-antigens such as DNA and RNA, it is critical to better define the signaling events that regulate the generation of naïve mature B cells capable of recognizing such antigens.
Deletion-mediated tolerance to self-antigen can be mimicked in vitro by stimulation of early transitional B cells with a BCR crosslinking antibody that recognizes surface IgM (sIgM). Mature B cells enter cell cycle and proliferate in response to stimulation with anti-IgM. In contrast, early transitional B cells rapidly die by apoptosis and fail to proliferate following an identical stimulus (7–10). Several lines of evidence indicate that this response in early transitional B cells is not secondary to a global defect in BCR signaling. Indeed, tonic BCR signals are required for cell survival even at the transitional stage (11) and, contrary to the response of anergic cells, transitional cells exhibit robust phosphotyrosine (pY) signals and calcium flux in response to BCR engagement (12–14). These data and other findings have supported the idea that developmentally-programmed differences in BCR signal transduction function to specifically modulate peripheral tolerance.
Correlated with the strong apoptotic response and lack of proliferation, early transitional B cells fail to upregulate key NF-κB target genes that express protein products required for B cell survival and division (7, 10). This observation, in conjunction with an apparent inability to produce inositol-1,4,5-trisphosphate (InsP3) in response to BCR engagement and other indirect evidence, has led to the hypothesis that early transitional B cells fail to activate proximal signals that trigger PKCβ-mediated NF-κB activation (15, 16). This model, however, has remained incompletely tested. In addition, it has remained unclear whether deficient NF-κB target gene expression is due primarily to abortive signaling caused by the strong apoptotic response, or instead, represents a unique difference in the downstream signaling capacity of the BCR.
In the current study, we show that even in near absence of the apoptotic response, early transitional B cells fail to transcribe genes necessary for survival and progression through cell cycle. This BCR-driven transcriptional defect is not due to altered proximal NF-κB signaling, NF-κB nuclear translocation or DNA binding. In addition, altered activation of other key transcription factors, including NFAT and AP-1, is unlikely to account for this deficit. However, despite intact proximal signals and transcription factor activation, T1 B cells fail to assemble an active transcriptional complex at the A1 and c-Myc gene promoters in response to BCR engagement. Taken together, these data support a model wherein developmentally-controlled, nuclear events limit BCR-specific transcriptional activity and thereby control the fate of autoreactive transitional B cells.
MATERIALS AND METHODS
Mice
Balb/c, MD4 Hel-Ig Tg and Bim KO mice were obtained from The Jackson Laboratories. NFAT1 KO mice were kindly provided by Anjana Rao. All animals were maintained in the SPF animal facility at Seattle Children’s Research Institute (Seattle, WA) and handled according to IACUC approved protocols. Mice were used at 5 to 8 weeks of age.
Antibodies and reagents
Antibodies specific for: PLCγ2, ERK, pERK1/2, JNK1, p65, c-Rel, IKKα/β, IκBα, HDAC1, RNA Polymerase II, NFAT1 and NFAT2 were purchased from Santa Cruz Biotechnology; AKT, p-AKT, p-JNK1/2, p-PKD, p38, p-p38, p-p65 S536, p-IKKα/β, p-IκBα, and p-Ser PKC substrates from Cell Signaling Technologies; pY, from Millipore (Upstate); CD21, B220, anti-active caspase 3 from BD Biosciences; CD24 from BioLegend; and Cy5 labeled non stimulatory Fab fragment and unlabeled crosslinking F(ab)2 anti-IgM from Jackson ImmunoResearch. CpG was purchased from Alexis and Indo-1 AM was purchased from Invitrogen.
Cell sorting
Splenic B cells were initially isolated by depletion of CD43+ cells using magnetic beads conjugated with anti-CD43 Abs according to the manufacturer’s instructions (Miltenyi Biotech). CD43− cells were then surface stained with fluorescently labeled Abs recognizing CD21, CD24 and B220 or IgM Fab fragment and sorted using a FACS Aria cell sorter with Diva software (BD Biosciences). Post-sort purities were consistently >95%.
Cell culture, apoptosis and proliferation assays
Sorted cells were cultured in RPMI 1640 with 10% FCS, 4µM L-glutamine, 50µM 2-ME, 10mM HEPES and antibiotics at 37°C at 1×106 cells/ml with or without 10µg/ml anti-IgM or 500nM CpG. Proliferation assays were carried out for 48 h with addition of 1µCi 3H Thymidine for the final 8 h before harvesting and analyzing using a scintillation counter. For the CFSE assay, cells were labeled with 0.1µM CFSE for 10 minutes at 37°C followed by extensive washing. After 48 h in culture, 7AAD was added to distinguish live/dead cells and dilution of CFSE was analyzed on a FACS Calibur flow cytometer (BD Biosciences). To detect active caspase-3 levels, cells were cultured for the indicated time then fixed, permeabilized and stained with a fluorescently labeled antibody specific to the active form of caspase-3 according the manufacturers instructions.
Calcium flux
CD43 depleted splenic cells were loaded with 7µg/ml indo-1AM followed by surface staining with fluorescently labeled antibodies recognizing CD21 and CD24. Intracellular calcium levels were measured based on the ratio between indo-violet and indo-blue on an LSRII flow cytometer (BD Bioscences) with a warming jacket to maintain cells at 37°C. Cells were resuspended in HBSS with 1.3mM Ca2+ with or without the addition of 2mM EGTA. After 30 seconds of acquisition to determine the baseline, 10µg/ml anti-IgM was added. Results were subsequently analyzed using the kinetics module in FlowJo software (Treestar Inc).
Western blotting
Sorted cells were stimulated at 1×107/ml with 10µg/ml anti-IgM or left unstimulated. 1–2 ×106 cells were lysed in 1% NP40, 50mM Tris pH 7.4, 150mM NaCl, 2mM EDTA, and 10% glycerol and lysates run on an 8–10% acrylamide gel. Blots were incubated with primary antibodies overnight, then HRP-conjugated secondary antibodies for 1 hr followed by detection by ECL. For cytoplasmic and nuclear factions, 3×106 cells were lysed in hypotonic buffer (10mM Hepes, 10mM KCl, 0.1mM EDTA, 0.1mM EGTA, 1mM DTT, 0.2% NP40) for 10 minutes, and the supernatant was collected after spinning for 30 sec at 14,000rpm. The pellet was washed 1× with the hypotonic buffer, followed by incubation with frequent vortexing for 45 minutes in a high salt buffer (20mM Hepes, 0.4M NaCl, 1mM EDTA, 1mM EGTA, 1mM DTT). All lysis buffers were supplemented with 1µg/ml Aprotinin, 10µg/ml Leupeptin, 80mM AEBSF and 2µM Na3VO4.
Real-time PCR and statistical analysis
3×105 sorted cells were stimulated at 1×106/ml with 10µg/ml anti-IgM or 500nM CpG, pelleted and frozen at −80°C. RNA isolation and detection of transcript levels was performed as previously described (8). Primers used were: c-Myc fp -5’ TCTCCACTCACCAGCACAACTACG, c-Myc rp −5’ ATCTGCTTCAGGACCCT; A1 fp -5’ CCTGGCTGAGCACTACCTTCA, A1 rp-5’ CTGCATGCTTGGCTTGGA; Bcl-xL fp -5’ – CTGGGACACTTTTGTGGATCTCT, Bcl-xL rp -5’ GAAGCGCTCCTGGCCTTT; NFAT2 fp-5’ CCCACAGCACACTCTGCCTTG, NFAT2 rp-5’ CCAGAGGACAGGAAGTATCCCG NFAT1; fp- 5’ TTGGGAGATGGAAGCTACGGTG, NFAT1 rp- 5’ TGGTTCATACTCATCGCTGGGC; NFAT4 fp- 5’ TTTCCGCCTGGTGGTGCAAC, NFAT4 rp- 5’ GGCTTGGGAAACAGAAATCTGG. Statistical significance was determined using a two-tailed t test of independent sample means.
EMSA
A double-stranded oligonucleotide spanning the κB binding region of the Il2R promoter 5’ CAACGGCAGGGGAATTCCCCTCTCCTT, AP-1 binding region of the metallothionine promoter 5’ TCGAGTGACTCAGCGCGTCGA, or NFAT binding region of the A1 promotor 5’ TGCTCTGAAACATTTTCCTCCTTCACAGT was end labeled with γ32P (34, 43, 44). Cells were incubated at 2–5×106/ml with or without anti-IgM. Nuclear extracts were isolated from stimulated cells as for western blotting. Binding reactions containing 2µg nuclear protein, 1µg poly(dIdC) and 50,000 cpm probe were incubated 20 minutes at room temperature (NF-κB), 15 minutes at 4°C (AP-1) or 1 hour at 4°C (NFAT) and run on a 4.5% acrylamide gel in 0.5X TBE. For supershift analysis, 1µg antibody was added to binding reactions for 1hr at 4°C before adding radiolabeled probe.
ChIP assay
ChIP assays with nuclear extracts prepared from cells sorted from WT Balb/c mice were performed as described previously with some modifications (45). Cells were incubated at 3×106/ml with 10µg/ml anti-IgM or 500nM CpG and fixed with 1% Formaldehyde for 10 min at RT. Chromatin from lysed cells was sheared by sonication, diluted 3 times in dilution buffer (50mM Tris pH 8.0, 0.5% NP40, 0.5mM EDTA, 0.2M NaCl), precleared for 1hr at 4°C with salmon sperm saturated Protein A beads followed by overnight incubation with 1µg antibody. Beads were added to immunoprecipitates for 2 h and washed 2x with high salt buffer I (20mM Tris pH 8.0, 0.1% SDS, 1% NP40, 2mM EDTA, 500mM NaCl), 2x with high salt buffer II (250mM LiCl, 1% Tris, 1% deoxycholate, 1mM EDTA, 10mM Tris pH 8.0) and 2x with TE. After extraction with TE+1%SDS, immune crosslinking was reverted at 65°C for 4–6 h and DNA was ethanol precipitated overnight. DNA was resuspended in TE, digested with proteinase K, and purified with QIAquick columns (Qiagen). PCR (30 cycles) was then performed on 1/6th of the DNA obtained using primers spanning a 300–400bp region near the κB and NFAT binding sites of the A1 promoter: fp - 5’ CCTCTTCTCGGTTCTGTGGGTTTC, rp - 5’ AGAGGCCCTTCAAAGTCACTCTGG; Bcl-xL promoter: fp – 5’ CCTTATCTTGGCTTTGGATCCTGG, rp-5’ TTCACTGCTGCCATGGGAATCAC; or start site of the c-Myc gene: fp – 5’CGCACCAACCAGAGCTGGATAAC, rp-5’ AGCGACCGCAACATAGGATGGAG.
RESULTS
T1 B cells exhibit a BCR signal-dependent transcriptional defect
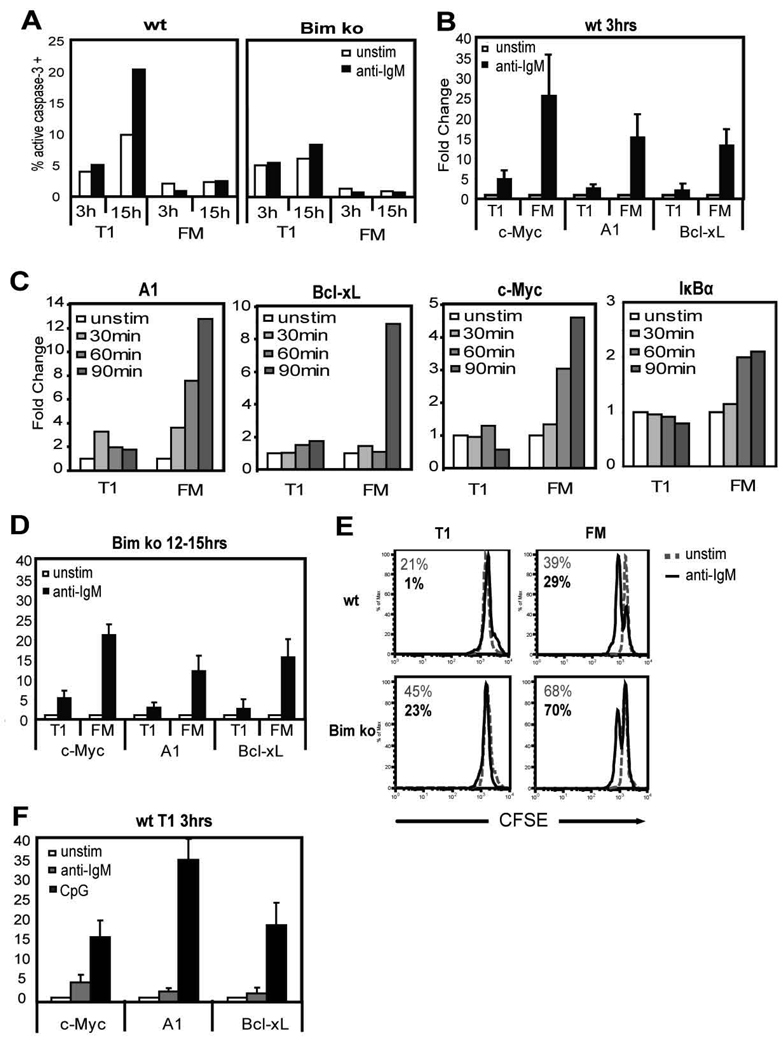
Previous studies have reported that early transitional murine B cells, in contrast to mature B cells, fail to upregulate expression of proteins required for cell cycle progression and survival including: c-Myc, Cdk2, A1, Bcl-xL and Cyclin E upon BCR engagement (7, 10). However, in these earlier studies, protein expression was measured at relatively late time points (12–24 h) after anti-IgM stimulation when the majority of T1 B cells have undergone apoptosis (7–10). To ensure that this lack of protein expression was not due simply to cell death, we measured the transcriptional expression of a subset of these genes three hours after BCR stimulation, a time point at which anti-IgM induced apoptosis is not evident in T1 B cells (Fig. 1A, left panel, and data not shown). T1 (B220+, CD24hi, CD21lo) and FM (B220+, CD24int, CD21int) B cells from WT mice were isolated by cell sorting as described previously (8). Sort purity was typically greater than 95%. BCR engagement with anti-IgM led to a 15–25 fold increase in A1, Bcl-xL and c-Myc mRNA levels in FM B cells (Fig. 1B). In contrast, minimal transcriptional expression occurred in T1 B cells during this time period. A detailed kinetic analysis demonstrated that transcriptional expression of all three genes and an additional NF-κB target gene, IκBα was detectable as early as 60–90 min post BCR engagement in FM, but remained minimal in T1 B cells (Fig. 1C). IκBα transcript levels did not increase in T1 B cells at later time points, similar to c-Myc, Bcl-xL, and A1 (data not shown).
Figure 1.
Transcriptional responses of T1 vs. FM B cells to anti-IgM and CpG. A, Caspase 3 activity in WT vs. Bim KO B cells. T1 and FM B cells from WT (left panel) and Bim KO (right panel) mice were stimulated with or without anti-IgM for the indicated time. Shown is the percentage of cells within the live gate expressing the active form of caspase-3. B–D, Transcription of NF-κB target genes after stimulating with anti-IgM. B, WT cells were stimulated for 3 h or C, for 30, 60, 90 min. D, Bim KO cells were stimulated for 12–15 h. E, Proliferative response based on dilution of CFSE in sorted cells after culture with (black solid lines) or without (gray dashed lines) anti-IgM for 48hrs. Numbers represent the percent live cells based on 7AAD staining with (black) or without (gray) anti-IgM. F, Expression of NF-κB target genes in WT T1 B cells after stimulation for 3 h with anti-IgM or CpG. All gene expression data are shown as fold change in transcript levels with unstimulated cells set as 1.
To verify that these findings did not simply reflect a delay in the transcriptional activity of T1 B cells, we also measured transcription at later time points after BCR engagement using cells purified from Bim deficient (Bim KO) mice. The pro-apoptotic Bcl-2 family member, Bim, is critical for the apoptotic program in immature B and T cells (17). Apoptosis upon BCR engagement, as measured by upregulation of active caspase-3 was largely abrogated in Bim KO T1 B cells (Fig. 1A, right panel) and cell survival was nearly identical in Bim KO T1 vs. WT FM B cells 48 h after anti-IgM stimulation (Fig. 1E). However, even at 12–15 h post-receptor engagement, transcriptional activity was minimal in T1 compared to FM B cells (Fig. 1D). Curtailing anti-IgM induced apoptosis also had no effect on the proliferative capacity of T1 B cells (Fig. 1E). Thus, in addition to the relatively well characterized BCR-induced apoptotic program, T1 B cells exhibit a cell-intrinsic, stage-specific transcriptional defect that limits the expression of gene products previously shown to play critical roles in BCR-dependent survival and proliferation (18–20). In contrast, T1 B cells were able to upregulate each of these gene products to high levels upon stimulation with the TLR9 ligand CpG (Fig. 1F). These findings also correlated with the ability of T1 cells to survive and proliferate in response to this signal (data not shown). Together, these data demonstrate that T1 B cells exhibit a BCR-signal specific transcriptional defect.
BCR signaling in highly purified sIgM matched T1 vs. FM B cells
The near absent transcriptional response in T1 B cells strongly implies that this subset has differences in the activation of a key signal(s) downstream of BCR engagement. Consistent with this idea, a number of previous studies have reported altered BCR-proximal signals in immature vs. mature primary B cells (7, 9, 10, 12–14, 21–24). These studies, however, have also led to contradictory findings likely due to differences in cell source and purity, surface IgM (sIgM) expression profile, and cell activation methods.
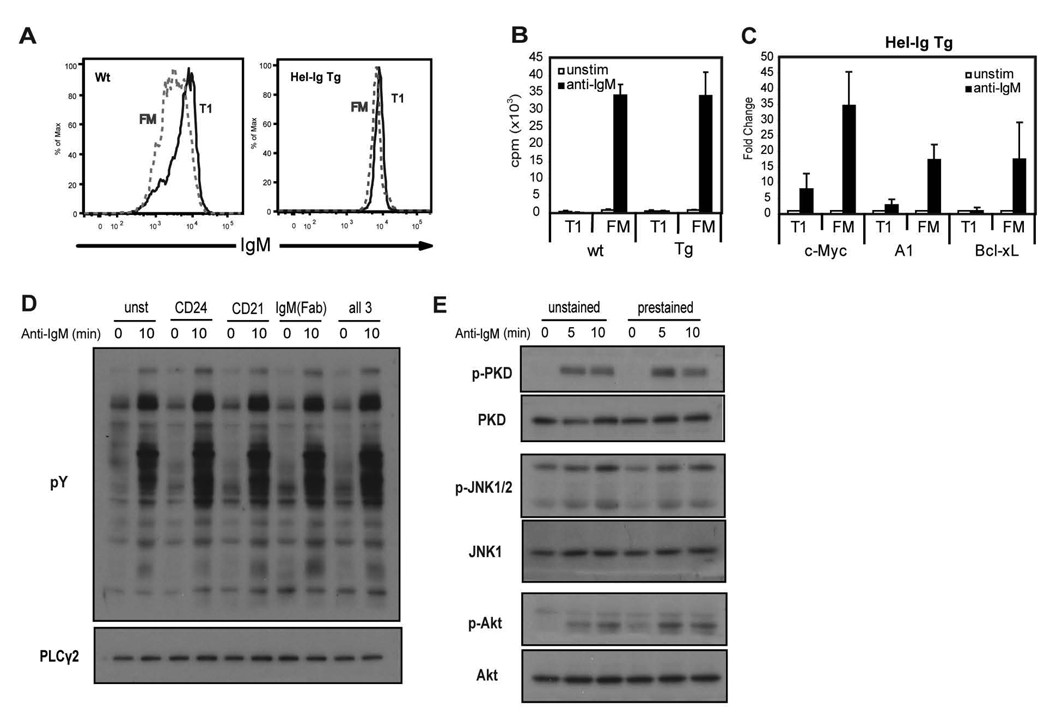
We directly compared BCR signaling using highly purified, functionally and phenotypically well-defined, T1 vs. FM B cells with matched sIgM levels. To achieve this goal, we sort purified CD24hiCD21lo T1 vs. CD24intCD21int FM B cells derived from MD4 Hel-Ig transgenic (Tg) mice (25) which express nearly identical sIgM levels (Fig. 2A). Recent work from our laboratory has extensively characterized the functional responses and developmental capacity of primary B cells purified according to these and related gating criteria (8). As shown in Fig. 2B–C (and data not shown), sorted Tg B cell subsets exhibited identical proliferative and transcriptional responses compared with the equivalent WT B cell subsets. Unless otherwise stated, all subsequent biochemical analyses were performed using purified Hel-Ig transgenic B cell subsets. Importantly, pre-staining with the cell surface antibodies used to isolate these subsets by cell sorting had no appreciable effect on downstream signaling as assessed by total pY levels, JNK, PKD or AKT phosphorylation (Fig. 2D–E).
Figure 2.
sIgM expression and anti-IgM responses of T1 and FM B cells from Hel-Ig Tg and WT mice and effect of cell surface antibodies on BCR staining. A, FACS analysis of surface IgM levels of T1 and FM B cells gated based on expression of cell surface markers CD24, CD21 from WT (left panel) and MD4 Hel-Ig Tg (right panel) B cells. B, Proliferative response following anti-IgM stimulation of sorted T1 and FM B cells from WT or Tg mice. C, Fold increase in transcript levels with anti-IgM stimulation (3 h) compared to unstimulated cells from purified WT or Hel-Ig Tg B cells subsets. Mean data with standard deviation (SD) from 5 experiments. D, Total B cells were stained with: anti-CD24, anti-CD21, anti-IgM Fab fragment, all three antibodies, or no antibody and then stimulated with anti-IgM F(ab)2 for 10 min. Whole cell lysates were blotted with anti-pY to measure activation and PLCγ2 to show equal loading. E, Total B cells were stained with anti-CD24, anti-CD21 and anti-IgM Fab fragment (prestained) or with no antibodies (unstained) and stimulated with anti-IgM as in D for the indicated time points. Whole cell lysates were probed with antibodies recognizing: p-JNK1/2, p-PKD and p-AKT. The blot was then stripped and reprobed with anti-JNK1, PKD and AKT antibodies as loading controls.
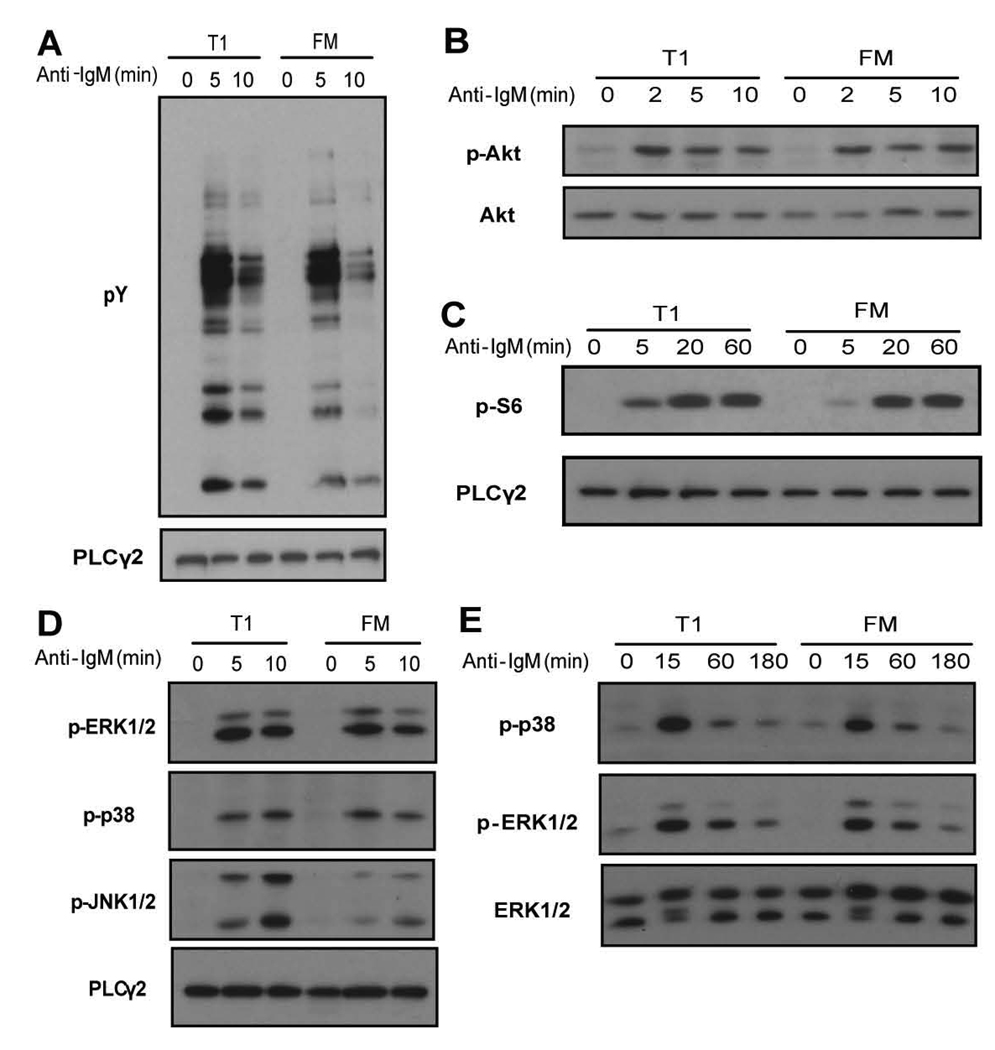
As an initial assessment of proximal tyrosine kinase activity, we compared total protein tyrosine phosphorylation (pY) in response to anti-IgM engagement. As shown in Fig. 3A, T1 B cells exhibited an equivalent or slightly increased pY signal compared to FM B cells. A critical signal initiated by BCR engagement is the production of phosphatidyl-inositol-1,4,5-trisphosphate (PIP3) via activation of phosphoinositide-3-kinase (PI3K). PIP3 is essential for membrane recruitment of PH-domain containing adapters and enzymes, and strongly promotes activation of Tec-family kinases and AKT. While direct measurement of PI3K activity was not possible due to limited cell numbers, we assessed trans-phosphorylation of AKT at S473 as a surrogate PIP3- dependent BCR signal (26, 27). T1 and FM B cells exhibited identical levels and kinetics of pAKT induction at early time points (Fig. 3B). In addition, based on equivalent activation of an AKT downstream target, S6, at later time points, sustained AKT and/or mTOR activity is also intact in T1 B cells (Fig. 3C).
Figure 3.
Activation status of proximal signaling molecules in sorted T1 and FM B cells upon anti-IgM stimulation. Whole cell lysates from stimulated and unstimulated cells were probed with antibodies that recognize: A, total pY B, phosphorylation of AKT at S473 C, phosphorylation of S6 D, phosphorylation of ERK1/2, p38 or JNK1/2 (short time points) or E, ERK1/2 and p38 (longer time points). Blots were stripped and reprobed with anti-PLCγ2, anti-AKT or anti-ERK1/2 to evaluate protein loading. Data shown are representative of at least 3 independent experiments.
When we measured activation of MAPK pathways, we found that site-specific phosphorylation of p38 and ERK was comparable in T1 and FM B cells even at late time points post-receptor engagement (Fig. 3D–E). In contrast, we reproducibly observed a significant increase in relative JNK phosphorylation in T1 B cells compared to FM B cells (Fig. 3D). The expression level of all three MAPK family members was identical between T1 and FM B cells (data not shown).
Activation of PLCγ2 and DAG signaling is intact in T1 B cells
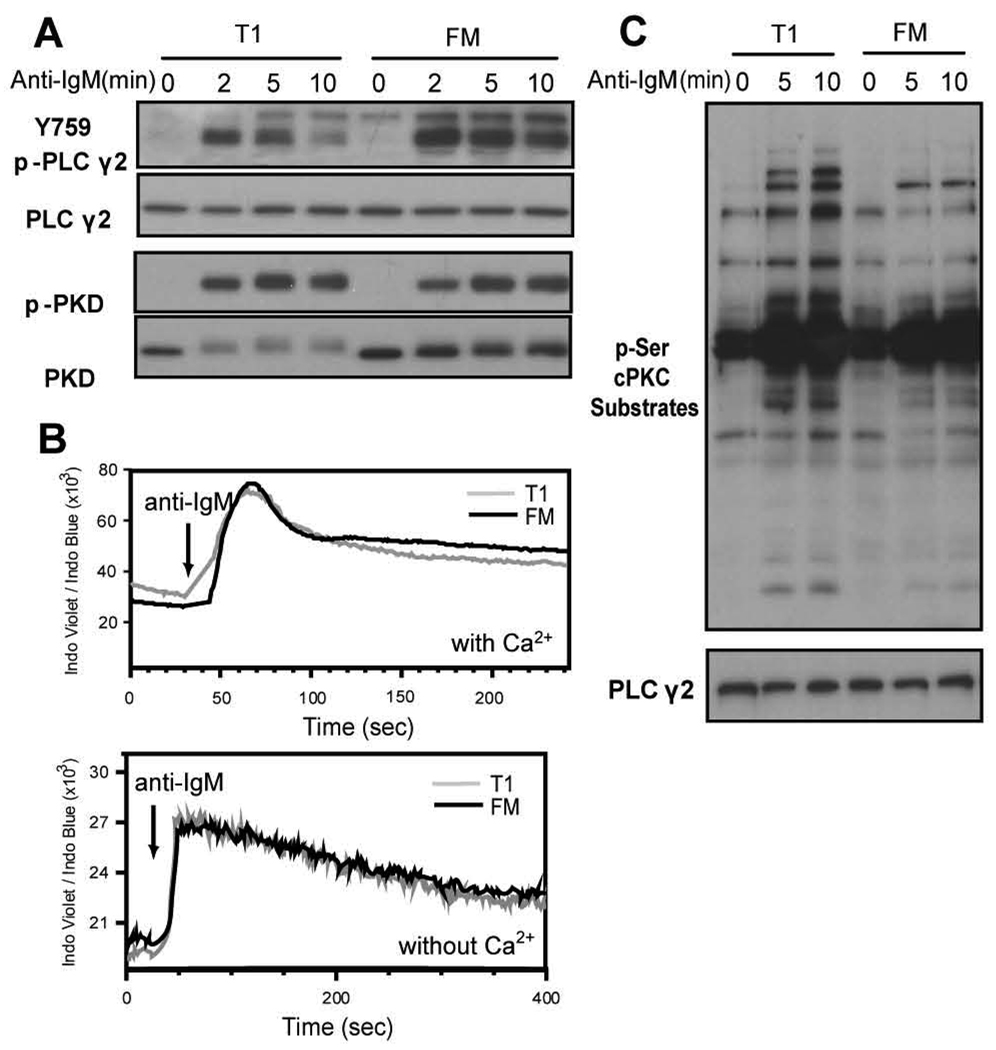
A1, c-Myc and Bcl-xL are each direct NF-κB target genes in BCR-triggered primary B cells (18–20, 28, 29). Therefore, to address whether BCR proximal events leading directly to PKCβ-mediated NF-κB activation are impaired in T1 B cells, we first assessed the relative activity of PLCγ2, which is responsible for the generation of DAG and InsP3, required for classical PKC (cPKC) activation. We used a phospho-specific antibody that recognizes pY759, a Btk-dependent phosphorylation site crucial for PLCγ2 activity (30). While phosphorylation at this site increased in response to BCR engagement in both T1 and FM B cells, the relative level of pY759 was consistently lower in T1 cells at all time points suggesting a partial deficit in PLCγ2 activation (Fig. 4A). In order to evaluate the initial release of Ca2+ from InsP3-receptor-gated ER stores, we measured the BCR Ca2+ response in the absence of extracellular Ca2+ and observed no appreciable differences (Fig. 4B, bottom panel). Ca2+ flux in the presence of extracellular Ca2+ was also similar between T1 and FM B cells (Fig. 4B, upper panel). Thus, BCR-induced InsP3 production in T1 cells triggers equivalent levels of ER Ca2+ release compared to FM B cells.
Figure 4.
PLCγ2 enzymatic activity is sufficient for downstream signaling events in T1 B cells. A, Activation of PLCγ2 and PKD. Sorted cells were stimulated with anti-IgM and whole cell lysates probed with antibodies recognizing phosphorylation of PLCγ2 at Y759 or PKD at the PKC trans-phosphorylation sites. Blots were stripped and reprobed with anti-PLCγ2 and anti-PKD to assess protein loading. B, Evaluation of calcium flux. Total splenic B cells were loaded with indo-1 and surface stained with CD21 and CD24 to identify T1 and FM B cell subsets. After acquiring a baseline for 30 seconds, anti-IgM was added as indicated. Ca2+ flux was measured in media containing 1.3mM Ca2+ with (top panel) or without the addition of excess levels of EGTA (bottom panel). C, Activation of cPKCs in T1 and FM B cells. Sorted cells were stimulated with anti-IgM and whole cell lysates probed with an antibody that recognizes phosphorylated serine residues preferentially phosphorylated by cPKCs. The blot was stripped and reprobed with anti-PLCγ2 to assess protein loading. All blots are representative of at least 3 independent experiments.
As a surrogate measure for proximal DAG dependent signaling, we evaluated activation of PKD. DAG directly binds to PKD promoting its membrane localization, and subsequent PKC-mediated PKD trans-phosphorylation (31). We observed equivalent levels of pPKD in T1 vs. FM B cells, indicating that initial, BCR-triggered, DAG-mediated signals are similar in T1 vs. FM B cells (Fig. 4A). Interestingly, while PKD levels were identical in resting cells, the protein level of the more slowly migrating pPKD species decreased in T1 B cells. While the significance of this latter finding remains unclear, these data strongly support the conclusion that initial, BCR-triggered, DAG-mediated signals are similar in T1 vs. FM B cells.
Because BCR-triggered Ca2+ flux and DAG signaling were intact and largely indistinguishable in T1 vs. FM B cells, we next sought to assess the relative activity of classical PKCs (cPKC) in these subsets. Based upon the limited cell numbers available in our experiments, we utilized an antibody that specifically recognizes consensus serine residues phosphorylated by cPKCs as a surrogate for cPKC enzymatic activity. Activated T1 cells consistently exhibited a greater number of phosphorylated species, as well as higher relative signal intensity, compared with FM B cells (Fig. 4C). These findings indicate that activation of cPKCs is intact in T1 B cells. Further, because cPKC expression levels are identical in these subsets (data not shown), and Ca2+ flux and DAG signaling are also similar, the increase in cPKC substrate phosphorylation in T1 B cells may reflect differences in relative phosphatase activity. A similar mechanism might also explain the increase in JNK activity in this subset.
In summary, these data indicate that while PLCγ2 site-specific phosphorylation is modestly reduced in T1 cells, PLCγ2 activity is sufficient to promote subsequent downstream signals including activation of cPKCs.
Activation of the NF-κB signaling cascade is intact in T1 B cells
As cPKC activation was not impaired in T1 B cells, we next directly evaluated the status of protein modifications required for canonical NF-κB signaling. Interestingly, phosphorylation of both IKKα/β and IκBα was higher and more accelerated in T1 B cells compared to FM B cells (Fig. 5A). Consistent with this, IκBα degradation proceeded more rapidly in T1 compared to FM B cells (Fig. 5A), though the basal level of total IκBα was consistently lower in T1 B cells. Despite this increase in IKKα/β and IκBα activity, trans-phosphorylation of p65 at S536, an IKK-mediated modification that enhances p65 transcriptional activity (32, 33), was similar in T1 and FM B cells (Fig. 5A). These data indicate that the composite signals required to release NF-κB subunits from sequestration within the cytosol are comparable, or somewhat increased, in T1 vs. FM B cells.
Figure 5.
Activation of the NF-κB signaling cascade in T1 and FM B cells. A, Sorted cells were stimulated with anti-IgM for the indicated times and whole cell lysates were probed with antibodies recognizing phosphorylation of IKKα/β, IκBα and p65. Blots were then stripped and reprobed with anti-IKKα/β, anti-IκBα and anti-p65 to assess protein loading. B, Nuclear import of NF-κB subunits. Cytoplasmic and nuclear extracts were isolated at the times indicated from sorted subsets following anti-IgM stimulation and probed with antibodies specific for c-Rel, p65 and IκBα. PLCγ2 and HDAC1 protein levels are shown as loading controls for the cytoplasmic and nuclear fractions, respectively. C, DNA-binding of NF-IκB subunits. An EMSA was performed using a radiolabeled probe containing a κB binding site and nuclear extracts from sorted WT cells incubated with or without anti-IgM for 3 h. D, Specific p65 or cRel binding was measured by supershift assay. Blots and EMSA are representative of at least 3 independent experiments.
We next evaluated whether any differences existed with regard to nuclear translocation of p65 or cRel in these developmental subsets. As shown in Fig. 5B, both the kinetics of nuclear import and the relative levels of c-Rel and p65 were indistinguishable in T1 and FM B cells using cells from Hel-Ig Tg (Fig. 5B) or WT mice (data not shown). We then measured the DNA-binding ability of nuclear p65 and cRel in anti-IgM stimulated T1 and FM B cells from WT mice. An electrophoretic mobility shift assay (EMSA) showed equivalent protein binding to a κB binding site upon anti-IgM stimulation in T1 vs. FM B cells (Fig. 5C). Supershift studies also demonstrated equal binding of both cRel and p65 (Fig. 5D). Thus, surprisingly, despite the lack of transcription of NF-κB target genes in T1 B cells, the BCR-driven signaling events leading to NF-κB subunit nuclear import and DNA binding are equivalently activated in T1 and FM B cells.
T1 B cells have lower BCR-induced NFAT activity
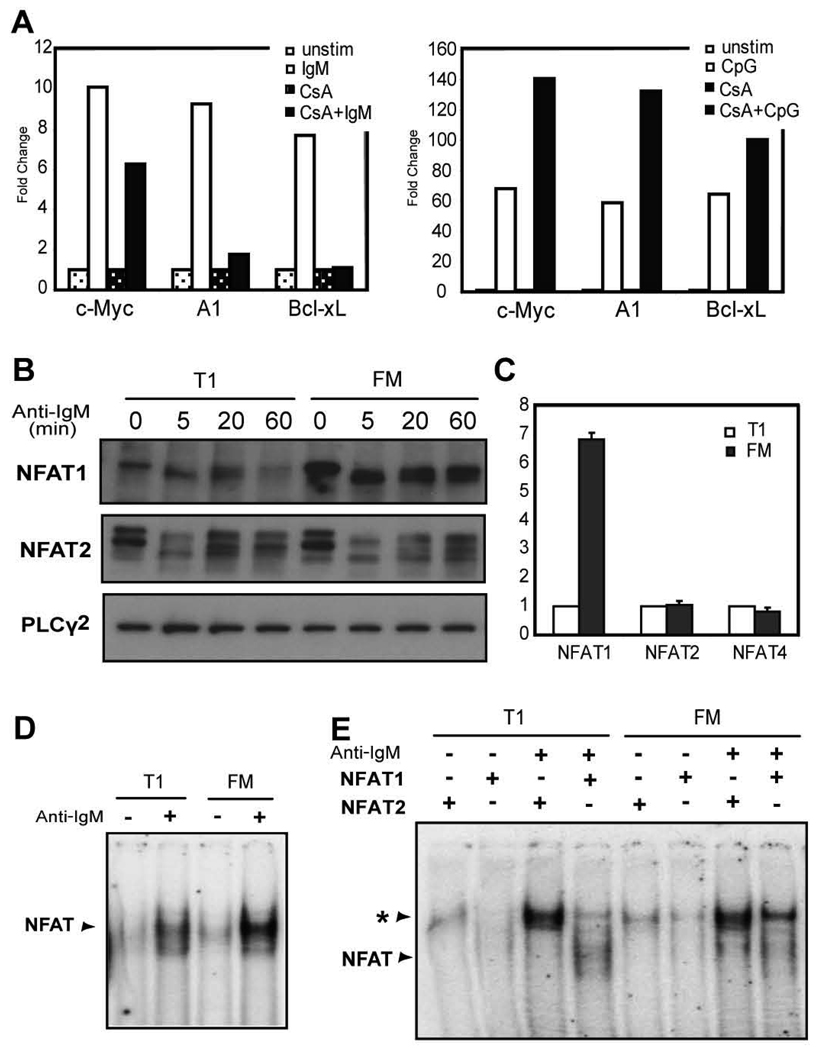
Although the expression of A1, Bcl-xL and c-Myc each requires canonical NF-κB signaling, the lack of transcription in the face of an apparently intact NF-κB signaling cascade, suggested that additional transcription factors might impact these events. In silico analysis of the promoter region of each of these genes identified conserved binding sites for the transcription factor NFAT (data not shown) and NFAT activity has recently been shown to be critical for A1 transcription in specific cell lineages (34). Indeed, treatment of FM B cells with the calcineurin inhibitor, cyclosporin A (CsA), led to reduced BCR-induced transcription of each of these genes; with nearly complete inhibition of both A1 and Bcl-xL compared to untreated cells (Fig. 6A, left panel). In contrast, CsA treatment did not limit CpG-induced gene expression (Fig. 6A, right panel).
Figure 6.
Expression and activation of NFAT family members in T1 and FM B cells. A, Sorted FM B cells were stimulated with anti-IgM (left panel) or CpG (right panel) with or without 2µM CsA for 3 h. Gene transcript levels were determined and shown as relative fold increase with anti-IgM stimulation relative to unstimulated cells; or cells treated with CsA alone. Representative of 4 independent experiments. B, NFAT1 and NFAT2 activation and protein levels. Whole cell lysates from unstimulated, or anti-IgM stimulated, sorted B cell subsets were probed with an antibody recognizing NFAT1 or NFAT2. PLCγ2 protein levels are shown as a loading control. C, NFAT family member transcript levels. mRNA levels of NFAT family members were determined by real-time PCR from sorted, unstimulated B cell subsets. Shown is the fold change between subsets where transcript levels in T1 B cells were set as 1. Mean with SD from 3 experiments. D, DNA binding of NFAT proteins. Sorted WT cells were incubated with or without anti-IgM for 3 h and an EMSA was performed with nuclear extracts using a radiolabeled probe containing the A1 promoter NFAT binding site. E, Specific binding of NFAT1 or NFAT2 was determined by supershift assay. The asterisk indicates supershifted complex. Representative of 3 independent experiments.
Given the apparent BCR-specific role for Ca2+ signaling in A1, Bcl-xL and to a lesser extent in c-Myc transcription, we hypothesized that NFAT activation might be impaired in T1 B cells. Consistent with the nearly identical calcium signals, T1 and FM B cells exhibited similar rates of BCR-triggered NFAT1 and 2 dephosphorylation (Fig. 6B); while NFAT activation was not observed upon CpG stimulation (data not shown). However, while both T1 and FM B cells expressed similar NFAT2 levels, NFAT1 expression was significantly lower in T1 B cells. This correlated with a ~6–8 fold reduction in NFAT1 transcript levels, consistent with stage-specific transcriptional regulation rather than altered processing or protein stability (Fig. 6C). NFAT4 transcript levels were equivalent in both subsets (Fig. 6C).
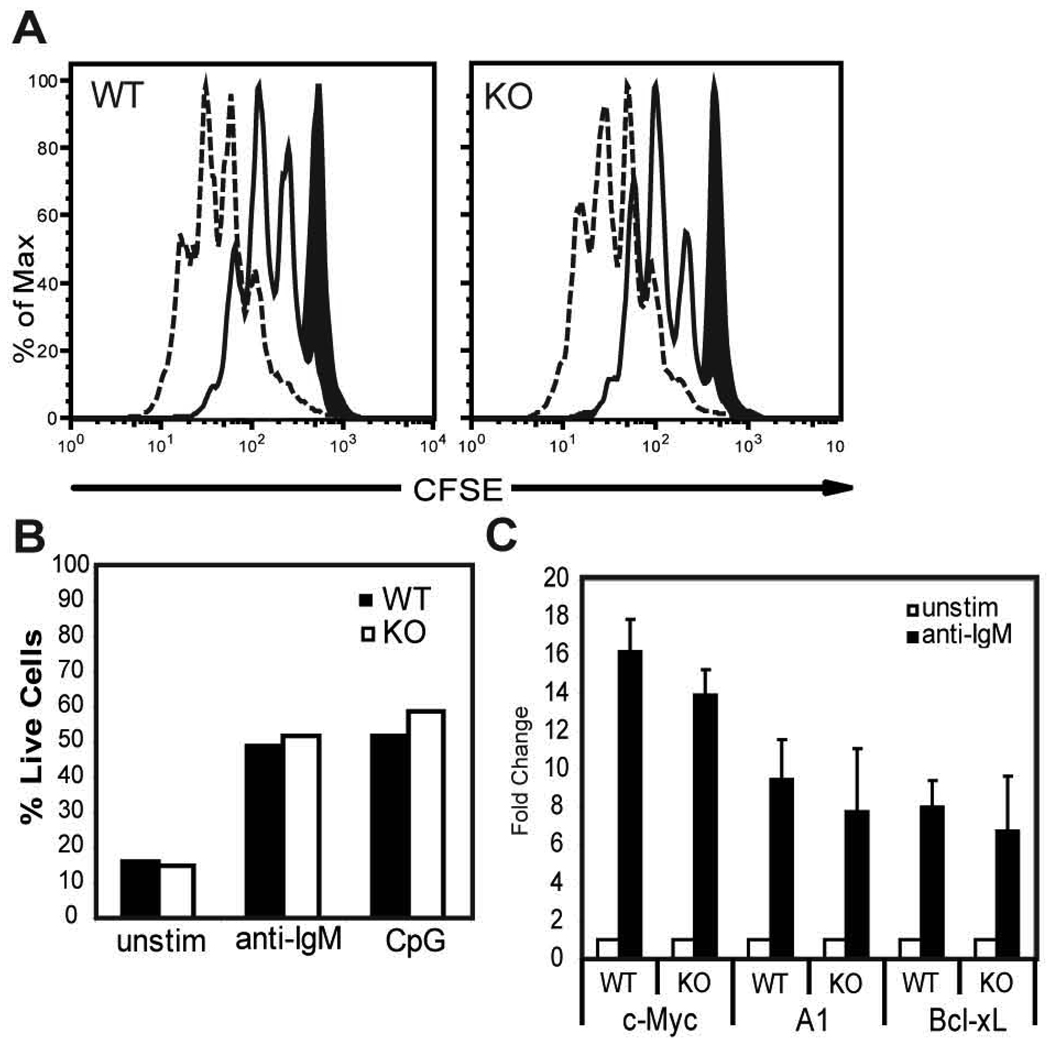
To evaluate the potential functional significance of reduced NFAT1 in T1 B cells, we first measured relative NFAT binding via EMSA. Using a probe containing the NFAT binding site within the A1 promoter, we observed significantly decreased NFAT binding in WT T1 B cells (Fig. 6D). Supershift studies demonstrated equivalent levels of NFAT2 containing DNA binding complexes (Fig. 6D). In contrast, NFAT1 complexes were almost undetectable in T1 B cells. Next, to determine whether reduced NFAT1 activity might account for the transcriptional defect in T1 B cells, we analyzed the function and transcriptional profile of NFAT1 KO B cells. Sorted WT and KO FM B cells exhibited similar proliferation and survival responses upon anti-IgM or CpG stimulation (Fig. 7A–B); and similar levels of A1, Bcl-xL and c-Myc transcription upon BCR engagement (Fig. 7C). Together, these observations indicate that despite altered NFAT1 expression and DNA binding, this difference alone is unlikely to limit BCR-dependent transcription of A1, Bcl-xL and c-Myc in T1 B cells.
Figure 7.
NFAT1 ko B cells exhibit intact transcriptional and functional responses to BCR engagement. A, FM B cells were FACS-sorted from WT or NFAT1 KO mice, labeled with CFSE and incubated for 72 h with no stimulus (black filled in), anti-IgM (solid line), or CpG (dashed line). B, Graph representing percent live cells under each condition based on incorporation of 7AAD. Representative of two independent experiments. C, Fold increase in transcript levels at 3 h following activation with anti-IgM compared to unstimulated cells from purified WT or KO FM B cells. Mean data with SD from 3 experiments.
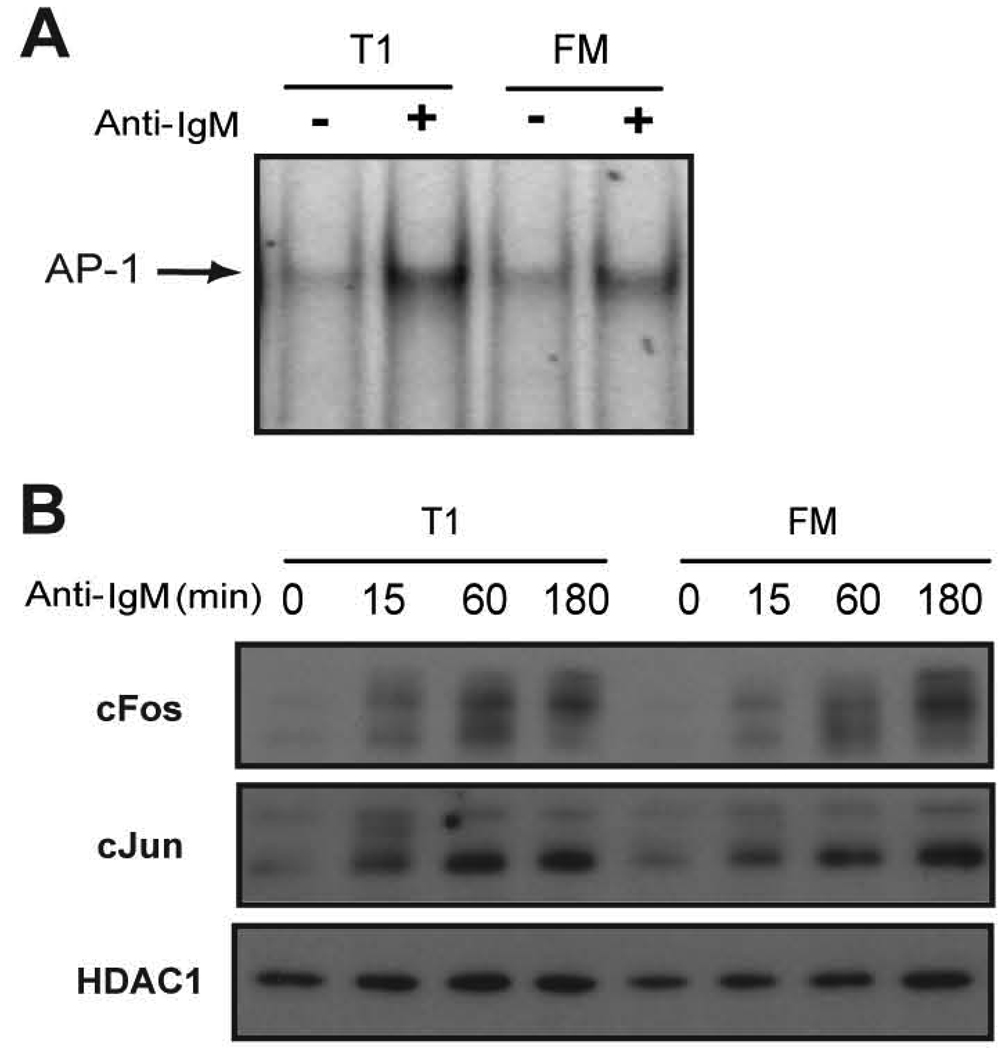
AP-1 has also been strongly implicated in mediating transcription of A1 (35). We therefore measured AP-1 activation via EMSA as well. WT T1 and FM B cells exhibited equivalent BCR-dependent AP-1 binding activity (Fig. 8A). In addition, BCR stimulation led to similar nuclear levels of both cJun and cFos (Fig. 8B). Thus, BCR-triggered AP-1 activity is equivalent in these developmental subsets.
Figure 8.
Activation of AP-1 in T1 and FM B cells. A, DNA binding of AP-1. Sorted WT cells were incubated with or without anti-IgM for 2 h and an EMSA was performed with nuclear extracts using a radiolabeled probe containing an AP-1 binding site. B, Nuclear expression of cJun and cFos. Sorted T1 and FM B cells were stimulated with anti-IgM for the indicated times and nuclear extracts were probed for antibodies specific to cJun and cFos. HDAC1 levels are also shown as a loading control.
Deficient RNA polymerase II (Pol II) recruitment at the A1 promoter in T1 B cells
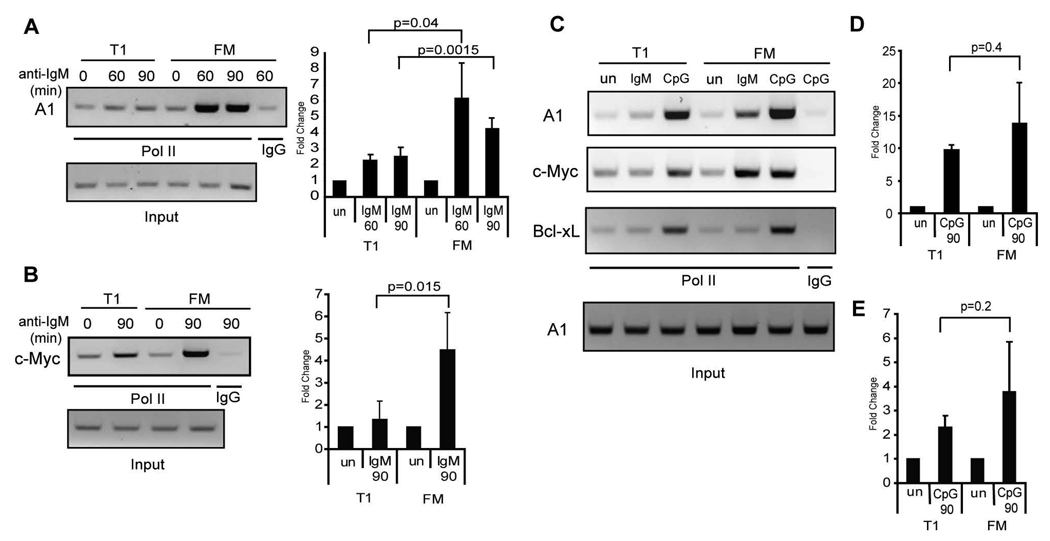
Transcriptional activation via NF-κB requires its association with histone acetyltransferases including, most notably, p300/CBP (36, 37). This greatly facilitates recruitment of the Pol II transcription complex and subsequent target gene transcription. This process has been extensively characterized for mitogen-induced transcriptional activation at the A1 promoter (35). Given the fact that lower NF-κB, NFAT or AP-1 activity cannot explain the transcriptional defect observed in T1 B cells, we wished to directly test whether the deficit in BCR-driven transcription in T1 cells correlated with alterations in Pol II recruitment. To do so, we used a chromatin immuno-precipitation (ChIP)-based assay to evaluate protein recruitment to the A1, c-Myc, and Bcl-xL gene promoter in WT T1 vs. FM B cells. BCR engagement lead to a significant (5–8 fold) increase in Pol II binding at both the A1 and c-Myc promoters in FM B cells upon BCR engagement (Fig. 9A–B). Strikingly, significantly less recruitment was observed in T1 B cells (~2 fold). We were unable to detect any recruitment of Pol II to the Bcl-xL promoter in T1 or FM B cells with anti-IgM stimulation (Fig. 9C). In contrast, CpG stimulation led to a strong accumulation of Pol II at the A1, c-Myc and Bcl-xL promoters in both T1 and FM B cells (Fig. 9C–E). Thus, despite equivalent activation of NF-κB, NFAT and AP-1 in BCR-stimulated T1 B cells, these cells exhibit a marked, signal-specific deficit in the capacity to assemble transcriptional complexes essential for A1, c-Myc and Bcl-xL expression.
Figure 9.
Deficient transcriptional activation at the A1 and c-Myc promoter in T1 B cells upon BCR crosslinking. A–B, Chromatin from WT T1 and FM B cells stimulated 60 or 90 min with anti-IgM was immunoprecipitated with antibodies that recognize Pol II or polyclonal rabbit IgG as a control. PCR was performed on DNA isolated from immunoprecipitates or 5% of input DNA (loading control) to amplify a DNA region within the A, A1 promoter, or B, c-Myc promoter. Shown is a representative figure of 3 independent experiments (left panel) and mean with SD of the fold increase in PCR product over unstimulated cells (right panel). C–E, Chromatin from WT T1 and FM B cells stimulated 90 min with anti-IgM or CpG was immunoprecipitated as above and PCR was performed as indicated. C, Representative figure of 4 independent experiments. D–E, Mean with SD of the fold increase in PCR product over unstimulated cells obtained using primers recognizing the D, A1 promoter, or E, c-Myc promoter.
DISCUSSION
Immature B cells are developmentally programmed to undergo deletion in response to strong receptor cross-linking via antigen, a response essential for the maintenance of B cell tolerance. Despite efforts by many investigators, the mechanisms that orchestrate the differential responsiveness to BCR engagement in early transitional vs. mature B cells remain incompletely defined. In the current study, we provide data that challenge existing models and suggest a new view of these events.
Our data clearly demonstrate that T1 B cells manifest a BCR-specific defect in transcriptional activation. T1 B cells essentially fail to activate proliferative and anti-apoptotic genes in response to BCR engagement; and this deficit proceeds independently of the apoptotic program initiated in response to BCR engagement. In contrast, efficient transcription of the identical gene products is triggered in response to the TLR ligand CpG. This observation prompted us to attempt to identify a signal-specific control mechanism that might be operative specifically within T1 B cells. Strikingly, our findings demonstrate that T1 B cells manifest a BCR-specific deficit in the capacity to assemble active transcriptional machinery at the BCR target genes A1 and c-Myc. T1 B cells exhibit this deficit despite normal proximal BCR signal transduction, as assessed via multiple biochemical parameters; and, much to our surprise, despite intact, inducible activation of key transcription factors including NF-κB, NFAT, and AP-1. Thus, our combined findings imply that T1 B cells are specifically programmed for nuclear non-responsiveness in the face of a strong antigen cross-linking signal.
Previous studies have suggested that altered activation of AKT, MAPK and/or PLCγ2 signals might account for the differential responsiveness of transitional B cell following anti-IgM stimulation (9, 10, 12–14, 22–24). In addition, both InsP3 (12, 24) and DAG levels (13) have been shown to be significantly lower in immature B cells following BCR stimulation. These various data, in association with indirect findings based upon pharmacological manipulation of PKC, have led to a long-standing hypothesis that immature B cells fail to activate the DAG dependent, proximal PKCβ/NF-κB signaling cascade, thereby explaining their reduced survival and aborted cell cycle entry (13, 15).
We directly tested this hypothesis using highly purified T1 vs. FM B cells that were identically matched for relative sIgM expression levels. In contrast to previous reports, we identified only minor differences in proximal BCR signaling in T1 vs. FM B cells. In our system, AKT and MAPK signaling was intact in T1 B cells; and despite a modest reduction in Btk-dependent PLCγ2 phosphorylation, subsequent PLCγ2-driven signals were nearly identical in T1 and FM B cells. Our findings are also supported by data derived from previously published work. As shown herein and in other studies (13, 14), immature B cells exhibit no defect in the release of InsP3-receptor regulated intracellular Ca2+ stores in response to BCR triggering. In addition, the PLCγ2 substrate, PIP2, is depleted at similar rates in activated T1 vs. FM B cells (13). Finally, work by Benschop et al suggests that the reported deficits in InsP3 and DAG in activated immature B cells likely reflect enhanced metabolism, rather than deficient production of these key second messengers (12). Importantly, and in agreement with our data suggesting intact cPKC signaling, T1 B cells demonstrated no defects in downstream IKK or IκBα phosphorylation, IκBα degradation, nuclear translocation of NF-κB subunits, or nuclear NF-κB binding activity. This latter finding directly contradicts an earlier report showing reduced nuclear NF-κB activity in post-irradiation immature B cells (22); and likely reflects use of more heterogenous populations. Together, our findings essentially rule out defective PKCβ activation as a cause for the altered transcriptional responses in T1 B cells.
We demonstrate that T1 B cells manifest a BCR-specific defect in transcriptional activation. While T1 B cell failed to activate proliferative and anti-apoptotic genes in response to BCR engagement, efficient transcription of these same genes was triggered in response to CpG. While c-Myc, A1 and Bcl-xL each comprise well-characterized NF-κB target genes (18–20, 28, 29), additional transcription factors are likely to coordinately regulate their expression in T1 B cells. Consistent with this idea, BCR-, but not TLR-, triggered Bcl-xL, c-Myc and A1 transcription were each inhibited by CsA. Notably, calcineurin b1 deficient B cells fail to proliferate in response to anti-IgM stimulation yet retain proliferative responses to LPS, consistent with a signal-specific requirement for Ca2+ signaling in BCR-driven proliferation (38). While both calcium flux and dephosphorylation of NFAT1 and NFAT2 were intact in T1 B cells, NFAT1 binding to an exogenous A1 promoter was markedly reduced in T1 cells and correlated with a specific deficit in NFAT1 transcript and protein levels in this developmental subset. To our knowledge, this is the first report of differential NFAT family member expression within B2 B cell subsets.
A1 gene transcription is preferentially regulated by NFAT1 in mast cells (34), suggesting that reduced NFAT1 levels might also account for deficient A1 transcription in T1 B cells. Contrary to this idea, NFAT1 deficient FM B cells efficiently transcribed A1, Bcl-xL and c-Myc; and exhibited no appreciable defects in BCR-induced survival or proliferation. Altered NFAT2 or 4 expression was also unlikely to compensate for NFAT1 deficiency in these cells as we detected no increase in transcript or protein levels in purified B cells (data not shown). While it remains formally possible that NFAT1 KO B cells manifest other compensatory events (or that T1 and FM B cells have differential requirement for NFAT1), our combined data suggest that the T1 B cell transcriptional deficit is largely independent of altered NFAT1 activity and that NFAT1 is dispensible for BCR-dependent A1, c-Myc and Bcl-xL transcription. AP-1 has also been shown to bind to the A1 promoter following PMA/Ionomycin treatement in Jurkat T cells (35). However, DNA binding of this transcription factor was similar between T1 and FM B cells upon BCR stimulation. Thus, differential activation or expression of AP-1 components also fails to explain the transcriptional defects seen in T1 B cells.
Despite considerable efforts, it proved technically impossible to measure BCR-dependent NF-κB or NFAT binding at endogenous promoter sites by ChIP in primary B cells. Therefore, we focused our efforts on assessment of Pol II recruitment to the promoter. Pol II recruitment was markedly reduced in T1 B cells following anti-IgM stimulation, yet the levels of Pol II recruitment in response to CpG in this subset was similar to that observed for FM B cells. Thus, despite similar ‘proximal’ and more ‘distal’ BCR-dependent signaling events in T1 and FM B cells, transitional B cells exhibit a marked defect in assembly of an active transcriptional complex at the promoter site of a key survival gene; and, based upon our transcriptional profiling are anticipated to manifest similar defects at additional BCR-dependent promoter elements.
Several factors, not addressed herein, might account for the signal-specific nuclear transcriptional defect identified in this study. First, phosphorylation and acetylation of NF-κB subunits, p65 in particular, are necessary for proper association with co-activators CBP/p300 and subsequent transactivation (37, 39). Importantly, mutations of several key phosphorylation/acetylation sites can alter p65 transactivation potential without reducing its nuclear translocation or DNA binding capacity. Second, developmentally controlled expression of another nuclear transcriptional regulator might impact these events. In particular, the calcineurin-regulated transcription factor, Mef2c, is expressed at lower levels in transitional B cells and is essential for BCR-induced proliferation and survival (40). These data, in concert with the blockade in BCR-dependent A1 and Bcl-xL transcription mediated by CsA, suggest that reduced Mef2c levels might significantly limit gene expression in T1 B cells. Assessment of the effect of enforced Mef2c expression within T1 B cells on BCR transcriptional activity will be required to address this idea. Third, BCR engagement could be activating transcriptional repressors in T1 B cells as opposed to a failure in activating transcription factors. However, this seems unlikely as the addition of anti-IgM to CpG-stimulated T1 B cells did not diminish transcript levels compared to cells stimulated with CpG alone (data not shown). Fourth, epigenetic differences in T1 vs. FM B cells might prevent gene transcription. This explanation seems unlikely based upon the rapid (3hr) transcriptional activation of these genes observed in T1 B cells with the TLR ligand CpG, but still remains a possibility. Finally, it has been proposed that initial activation of NF-κB signaling in T1 B cells might be enhanced relative to that present in FM B cells. This might lead to higher initial levels of IκBα transcription, and consequently, more rapid IκBα-mediated removal of NF-κB subunits thereby limiting NF-κB signaling (41). However, while initial IKK and IκBα phosphorylation was increased in T1 B cells, we observed no increase in IκBα transcription in T1 B cells even at very early time points post BCR engagement nor did we see differences in p65 nuclear/cytoplasmic shuttling up to 3 h post-stimulation. Thus, our data argue strongly for a defect in initiation, rather than premature termination, of transcription.
In summary, our findings provide important new insight into what controls the differential response to BCR engagement observed during peripheral B cell development. In contrast to T1 B cells, late transitional B cell exhibit increased survival and proliferative responses to BCR ligation with and without costimulatory signals (8, 42) and may serve as a target for antigen-driven positive selection (8). These observations, combined with the current study, suggest that transitional cells may gain BCR-transcriptional competence during this developmental window. Identification of the micro-environmental signals that mediate this response change, and/or those capable of bypassing it, will lead to a better understanding of the events that shape the mature B cell repertoire.
ACKNOWLEDGEMENTS
We thank Socheath Khim and Noel Blake for technical assistance with animal studies, and flow cytometry, respectively. We also would like to thank Troy Torgerson, Chris Wilson, Andrew Scharenberg and Edward Clark for critical reading of this work and members of the Rawlings lab for technical assistance and thoughtful discussions.
Footnotes
Support for this work has included funds from: Cancer Research Institute Immunology training grant (S.F.A); and NIH grants HD37091 and CA81140.
REFERENCES
- 1.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 2.Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Hippen KL, Schram BR, Tze LE, Pape KA, Jenkins MK, Behrens TW. In vivo assessment of the relative contributions of deletion, anergy, and editing to B cell self-tolerance. J Immunol. 2005;175:909–916. doi: 10.4049/jimmunol.175.2.909. [DOI] [PubMed] [Google Scholar]
- 4.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 5.Russell DM, Dembic Z, Morahan G, Miller JF, Burki K, Nemazee D. Peripheral deletion of self-reactive B cells. Nature. 1991;354:308–311. doi: 10.1038/354308a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Feng J, Qi CF, Li Z, Morse HC, 3rd, Clarke SH. Transitional B cells lose their ability to receptor edit but retain their potential for positive and negative selection. J Immunol. 2007;179:7544–7552. doi: 10.4049/jimmunol.179.11.7544. [DOI] [PubMed] [Google Scholar]
- 7.Carman JA, Wechsler-Reya RJ, Monroe JG. Immature stage B cells enter but do not progress beyond the early G1 phase of the cell cycle in response to antigen receptor signaling. J Immunol. 1996;156:4562–4569. [PubMed] [Google Scholar]
- 8.Meyer-Bahlburg A, Andrews SF, Yu KO, Porcelli SA, Rawlings DJ. Characterization of a late transitional B cell population highly sensitive to BAFF-mediated homeostatic proliferation. J Exp Med. 2008;205:155–168. doi: 10.1084/jem.20071088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petro JB, Gerstein RM, Lowe J, Carter RS, Shinners N, Khan WN. Transitional type 1 and 2 B lymphocyte subsets are differentially responsive to antigen receptor signaling. J Biol Chem. 2002;277:48009–48019. doi: 10.1074/jbc.M200305200. [DOI] [PubMed] [Google Scholar]
- 10.Su TT, Rawlings DJ. Transitional B lymphocyte subsets operate as distinct checkpoints in murine splenic B cell development. J Immunol. 2002;168:2101–2110. doi: 10.4049/jimmunol.168.5.2101. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes-Panana EM, Bannish G, van der Voort D, King LB, Monroe JG. Ig alpha/Ig beta complexes generate signals for B cell development independent of selective plasma membrane compartmentalization. J Immunol. 2005;174:1245–1252. doi: 10.4049/jimmunol.174.3.1245. [DOI] [PubMed] [Google Scholar]
- 12.Benschop RJ, Brandl E, Chan AC, Cambier JC. Unique signaling properties of B cell antigen receptor in mature and immature B cells: implications for tolerance and activation. J Immunol. 2001;167:4172–4179. doi: 10.4049/jimmunol.167.8.4172. [DOI] [PubMed] [Google Scholar]
- 13.Hoek KL, Antony P, Lowe J, Shinners N, Sarmah B, Wente SR, Wang D, Gerstein RM, Khan WN. Transitional B cell fate is associated with developmental stage-specific regulation of diacylglycerol and calcium signaling upon B cell receptor engagement. J Immunol. 2006;177:5405–5413. doi: 10.4049/jimmunol.177.8.5405. [DOI] [PubMed] [Google Scholar]
- 14.Koncz G, Bodor C, Kovesdi D, Gati R, Sarmay G. BCR mediated signal transduction in immature and mature B cells. Immunol Lett. 2002;82:41–49. doi: 10.1016/s0165-2478(02)00017-2. [DOI] [PubMed] [Google Scholar]
- 15.King LB, Monroe JG. Immunobiology of the immature B cell: plasticity in the B-cell antigen receptor-induced response fine tunes negative selection. Immunol Rev. 2000;176:86–104. doi: 10.1034/j.1600-065x.2000.00609.x. [DOI] [PubMed] [Google Scholar]
- 16.King LB, Norvell A, Monroe JG. Antigen receptor-induced signal transduction imbalances associated with the negative selection of immature B cells. J Immunol. 1999;162:2655–2662. [PubMed] [Google Scholar]
- 17.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 18.Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol Cell. 2002;10:1283–1294. doi: 10.1016/s1097-2765(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Arsura M, Wu M, Duyao M, Buckler AJ, Sonenshein GE. Role of Rel-related factors in control of c-myc gene transcription in receptor-mediated apoptosis of the murine B cell WEHI 231 line. J Exp Med. 1995;181:1169–1177. doi: 10.1084/jem.181.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brezski RJ, Monroe JG. B cell antigen receptor-induced Rac1 activation and Rac1-dependent spreading are impaired in transitional immature B cells due to levels of membrane cholesterol. J Immunol. 2007;179:4464–4472. doi: 10.4049/jimmunol.179.7.4464. [DOI] [PubMed] [Google Scholar]
- 22.Feng B, Cheng S, Hsia CY, King LB, Monroe JG, Liou HC. NF-kappaB inducible genes BCL-X and cyclin E promote immature B-cell proliferation and survival. Cell Immunol. 2004;232:9–20. doi: 10.1016/j.cellimm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Kovesdi D, Paszty K, Enyedi A, Kiss E, Matko J, Ludanyi K, Rajnavolgyi E, Sarmay G. Antigen receptor-mediated signaling pathways in transitional immature B cells. Cell Signal. 2004;16:881–889. doi: 10.1016/j.cellsig.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Yellen AJ, Glenn W, Sukhatme VP, Cao XM, Monroe JG. Signaling through surface IgM in tolerance-susceptible immature murine B lymphocytes. Developmentally regulated differences in transmembrane signaling in splenic B cells from adult and neonatal mice. J Immunol. 1991;146:1446–1454. [PubMed] [Google Scholar]
- 25.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 26.Gray A, Van Der Kaay J, Downes CP. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem J. 1999;344(Pt 3):929–936. [PMC free article] [PubMed] [Google Scholar]
- 27.Astoul E, Watton S, Cantrell D. The dynamics of protein kinase B regulation during B cell antigen receptor engagement. J Cell Biol. 1999;145:1511–1520. doi: 10.1083/jcb.145.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HH, Dempsey PW, Parks TP, Zhu X, Baltimore D, Cheng G. Specificities of CD40 signaling: involvement of TRAF2 in CD40-induced NF-kappaB activation and intercellular adhesion molecule-1 up-regulation. Proc Natl Acad Sci U S A. 1999;96:1421–1426. doi: 10.1073/pnas.96.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphries LA, Dangelmaier C, Sommer K, Kipp K, Kato RM, Griffith N, Bakman I, Turk CW, Daniel JL, Rawlings DJ. Tec kinases mediate sustained calcium influx via site-specific tyrosine phosphorylation of the phospholipase Cgamma Src homology 2-Src homology 3 linker. J Biol Chem. 2004;279:37651–37661. doi: 10.1074/jbc.M311985200. [DOI] [PubMed] [Google Scholar]
- 31.Wang QJ. PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol Sci. 2006;27:317–323. doi: 10.1016/j.tips.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Mattioli I, Sebald A, Bucher C, Charles RP, Nakano H, Doi T, Kracht M, Schmitz ML. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J Immunol. 2004;172:6336–6344. doi: 10.4049/jimmunol.172.10.6336. [DOI] [PubMed] [Google Scholar]
- 33.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 34.Ulleras E, Karlberg M, Moller Westerberg C, Alfredsson J, Gerondakis S, Strasser A, Nilsson G. NFAT but not NF-{kappa}B is critical for transcriptional induction of the prosurvival gene A1 after IgE receptor activation in mast cells. Blood. 2008;111:3081–3089. doi: 10.1182/blood-2006-10-053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edelstein LC, Lagos L, Simmons M, Tirumalai H, Gelinas C. NF-kappa B-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1/A1. Mol Cell Biol. 2003;23:2749–2761. doi: 10.1128/MCB.23.8.2749-2761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 37.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winslow MM, Gallo EM, Neilson JR, Crabtree GR. The calcineurin phosphatase complex modulates immunogenic B cell responses. Immunity. 2006;24:141–152. doi: 10.1016/j.immuni.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 40.Wilker PR, Kohyama M, Sandau MM, Albring JC, Nakagawa O, Schwarz JJ, Murphy KM. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol. 2008;9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen R. Control of B lymphocyte apoptosis by the transcription factor NF-kappaB. Immunity. 2006;25:871–883. doi: 10.1016/j.immuni.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Chung JB, Sater RA, Fields ML, Erikson J, Monroe JG. CD23 defines two distinct subsets of immature B cells which differ in their responses to T cell help signals. Int Immunol. 2002;14:157–166. doi: 10.1093/intimm/14.2.157. [DOI] [PubMed] [Google Scholar]
- 43.Shinners NP, Carlesso G, Castro I, Hoek KL, Corn RA, Woodland RT, Scott ML, Wang D, Khan WN. Bruton's tyrosine kinase mediates NF-kappa B activation and B cell survival by B cell-activating factor receptor of the TNF-R family. J Immunol. 2007;179:3872–3880. doi: 10.4049/jimmunol.179.6.3872. [DOI] [PubMed] [Google Scholar]
- 44.Timmerman LA, Healy JI, Ho SN, Chen L, Goodnow CC, Crabtree GR. Redundant expression but selective utilization of nuclear factor of activated T cells family members. J Immunol. 1997;159:2735–2740. [PubMed] [Google Scholar]
- 45.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]