Abstract
Tamoxifen (TAM) is a selective estrogen receptor modulator that is widely used in the prevention and treatment of estrogen receptor-positive (ER+) breast cancer. Its use has significantly contributed to a decline in breast cancer mortality, since breast cancer patients treated with TAM for 5 years exhibit a 30–50% reduction in both the rate of disease recurrence after 10 years of patient follow-up and occurrence of contralateral breast cancer. However, in patients treated with TAM there is substantial interindividual variability in the development of resistance to TAM therapy, and in the incidence of TAM-induced adverse events, including deep vein thrombosis, hot flashes, and the development of endometrial cancer. This article will focus on the UDP glucuronosyltransferases, a family of metabolizing enzymes that are responsible for the deactivation and clearance of TAM and TAM metabolites, and how interindividual differences in these enzymes may play a role in patient response to TAM.
Keywords: breast cancer, tamoxifen, UDP glucuronosyltransferase, glucuronidation, metabolism, pharmacogenetics
Introduction
Tamoxifen (TAM) (1-[4-(2-dimethylamino-ethoxy)phenyl]-1,2-diphenylbut-1(Z)-ene) is a nonsteroidal antiestrogen that has been commonly used for the treatment and prevention of estrogen-dependent breast cancer.1–4 First approved in 1977 by the FDA (U.S.) for the treatment of women with metastatic breast cancer, TAM is currently an established hormonal treatment for all stages of estrogen receptor (ER)-positive breast cancer. Adjuvant TAM treatment increases recurrence-free survival and overall survival in breast cancer patients with hormone receptor-positive tumors, irrespective of these patients’ nodal status, menopausal status, or age.3,4 TAM is also widely used as a chemopreventive agent in women at risk for developing breast cancer.1,2
In addition to its antiestrogenic properties, which have been related to menopause-like symptoms including hot flashes and vaginal bleeding,4 TAM also has seemingly tissue-dependent partial estrogen-agonistic effects that may be linked to reduced risk for is-chemic heart disease and osteoporosis,5,6 but may also increase the risk for endometrial cancer7,8 and venous thromboembolism.9 Although TAM is generally well tolerated, significant interindividual variability has been observed in the clinical efficacy as well as toxicity of TAM. For instance, about 30% of patients acquire TAM resistance and relapse.10 In addition, the relative risk of endometrial cancers in patients treated with TAM is estimated to be 2- to 3-fold that of controls, and the risk increases with both the duration and cumulative dose of TAM treatment.6,11–13
Tamoxifen Metabolism and Mechanism of Action
TAM acts by binding to the estrogen receptor, thereby competitively inhibiting the binding of estrogen in breast tissue.14 While TAM exists in both a trans and a cis configuration, the trans isomer of TAM is the pharmaceutically manufactured form of TAM used in the treatment and prevention of breast cancer.
TAM is metabolized via cytochrome P450 (CYP450)-mediated pathways into several metabolites after oral administration, including the metabolites N -desmethylTAM (DMT), 4-hydroxyTAM (4-OH-TAM), α-hydroxyTAM (α-OH-TAM), TAM-N-oxide, N, N -didesmethylTAM (DDMT), and 4-OH-N -desmethylTAM (endoxifen; see Fig. 1). CYP3A4 has been shown to be the main CYP450 enzyme involved in the metabolism of TAM to α-OH-TAM15,16 and to DMT,17,18 although CYPs 2D6, 1A1, 1A2, 1B1, 2C9, and 2C19 may also play a role in DMT formation.18,19 CYP2D6 appears to be the major CYP involved in the hydroxylation of trans-TAM to trans-4-OH-TAM17,19–24 and DMT to endoxifen.17 Endoxifen formation was observed to be highest in activity assays of recombinant CYP2D6 enzyme incubated with trans-4-OH-TAM, although CYP3A4 activity also correlates well with endoxifen formation in human liver microsomes (HLM17).
Figure 1.
Schematic of TAM metabolism.
DMT has been established as the primary metabolite of TAM as determined by in vitro assays performed with HLM17 and in vivo studies that have shown levels of steady-state plasma DMT to be greater than 70 times the levels of 4-OH-TAM in the serum25,26 and steady-state plasma levels of endoxifen are ~6-fold the levels observed for 4-OH-TAM in TAM-treated subjects.17,26,27 However, the major therapeutic contributors are hypothesized to be endoxifen and 4-OH-TAM based on evidence that indicates they exhibit up to 100-fold the levels of antiestrogenic activity as compared to TAM and other TAM metabolites,14,26,28–32 they exhibit the same relative levels of antiestrogenic activity,26,28 they inhibit expression of β-estradiol-induced ER-dependent target genes,28,32 they inhibit global estrogen-dependent gene expression31 and have high affinity for the ER (α and β) in RBA assays.28
Non-CYP450-mediated conjugation pathways also appear to be highly important in terms of TAM’s overall metabolism and activity profile. While the hydroxysteroid sulfotransferase, SULT1A2, is involved in sulfation of α-OH-TAM,33 this enzyme does not exhibit activity against either the trans or cis isomers of 4-OH-TAM.34,35 The phenol sulfotransferase, SULT1A1, appears to be the major sulfotransferase involved in the conjugation of both trans-and cis-4-OH-TAM in humans.35,36
Perhaps the most important route of elimination of TAM and its metabolites is via glucuronidation by the UDP-glucuronosyltransferases (UGTs). TAM is excreted predominantly through the bile, a process largely facilitated by TAM conjugation to glucuronic acid during the glucuronidation process,37 and TAM glucuronides have been identified in the urine and serum of TAM-treated patients.37–39 Most of the 4-OH-TAM and endoxifen found in the bile of TAM-treated patients was as a glucuronide conjugate37,38 and TAM, 4-OH-TAM, and endoxifen are glucuronidated with very high activity by HLM.17,40 Although TAM metabolites are often found in their unconjugated form in feces, this is likely due to β-glucuronidase-catalyzed removal of glucuronic acid within the microflora of the small intestine.37
Large interindividual variability in endoxifen plasma concentrations in women taking TAM have been observed and can be explained, in part, by CYP2D6 genotype.26,27 Recent evidence demonstrates that the CYP2D6*4 deletion allele has been associated with decreased time until breast cancer recurrence, relapse-free survival, disease-free survival, and overall survival in patients treated with TAM.41,42 In addition, variant alleles that result in low activity/expression in CYPs 2D6, 2B6, and 2C9 were correlated with levels of trans-4-OH-TAM formation in HLM from individual subjects.21 These data suggest that the levels of circulating active TAM metabolites may differ between individuals based upon metabolizing enzyme genotype.
The UDP-Glucuronosyltransferases
The UGTs are a superfamily of enzymes located primarily in the endoplasmic reticulum of cells that detoxify a diverse range of xenobiotics, as well as endogenous compounds, through their conjugation to glucuronic acid in a reaction with the hydrophilic co-substrate, UDPGA. The conjugated sugar alters the biological properties of the compound to enhance its excretion in the urine or bile and usually converts substrates into less pharmacologically active products.43–45 Based on differences in sequence homology, three main families of UGTs have been identified, each containing several UGT genes with high homology at their COOH− end.
The UGT family 1A gene complex has been localized to chromosome 2 in humans46 and consists of multiple isoform-specific exons that are expressed by alternate splicing of one unique exon (exon 1) to a domain consisting of four common exons (exons 2–5). This unique gene complex consists of at least 13 UGT family 1A-specific exons46,47 that are highly conserved between species, with the same gene structure observed in both humans46 and rodents.48 The UGT2A enzymes are found mainly in olfactory tissues, with UGT2A1 active against odorants, steroid hormones, and some drugs49; to date, the only substrates shown to be glucuronidated by UGT2A2 are β-estradiol and epiestradiol.50 There are at least six members of the UGT2B family with unique genes clustered on chromosome 4 in humans.51 Most of the UGT1A and 2B sub-families are expressed hepatically and extrahepatically, except for UGTs 1A7, 1A8, and 1A10, which are exclusively extrahepatic.52 Interestingly, a considerable number of prevalent, functional polymorphisms have been previously identified in several UGT genes, including 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A10, 2B4, 2B7, 2B15, and 2B1753–63 with several implicated as determinants of cancer risk or response to chemotherapy.
TAM Glucuronidation
Microsomes from HLM exhibit high glucuronidating activities toward TAM to form TAM-N +-glucuronide, and 4-OH-TAM to form N +- and of O-glucuronides of 4-OH-TAM.35,64,65 Both isomers of endoxifen are O-glucuronidated; however, unlike 4-OH-TAM, no N-glucuronidation of endoxifen isomers was detected in assays for either HLM or individually overexpressed UGTs,40 suggesting that the demethylation of the electrophilic amine on the 4-OH-TAM side chain to form endoxifen results in a lack of N-glucuronidation by UGTs.
One of the major UGTs involved in the glucuronidation of TAM and its metabolites is the hepatic enzyme, UGT1A435,64,65 which catalyzes the formation of a quaternary ammonium-linked glucuronide with TAM’s and 4-OH-TAM’s N, N-dimethylaminoalkyl side chain.64,65 This pattern of ammonium-linked glucuronidation is consistent with UGT1A4’s glucuronidation activity against primary, secondary, and tertiary amines present in a variety of carcinogenic compounds, androgens, progestins, and plant steroids.66–69 In addition to UGT1A4, UGTs 1A1, 1A3, 1A8, 1A9, 2B7, and 2B15 overexpressing baculosomes exhibited detectable activity against 4-OH-TAM.64 In a comprehensive characterization and kinetic analysis of the glucuronidating enzymes responsible for O-glucuronidation of TAM metabolites,40 UGTs 2B7 ≃ 1A8 > 1A10 exhibited the highest overall activity against trans-4-OH-TAM as determined by Vmax/KM, with the hepatic enzyme, UGT2B7, exhibiting the highest binding affinity and lowest KM (3.7 μM). UGTs 1A10 ≃ 1A8 > 2B7 exhibited the highest overall glucuronidating activities as determined by Vmax/KM for trans-endoxifen, with the extrahepatic enzyme UGT1A10 exhibiting the highest binding affinity and lowest KM (39.9 μM), but with UGT2B7 again demonstrating the highest activity of hepatic UGTs (Table 1). These data suggest that several UGTs, including UGTs 1A10, 2B7, and 1A8, could play an important role in the metabolism of 4-OH-TAM and endoxifen. The absence of activity that was observed for UGT1A4-overexpressing cell homogenates against endoxifen was consistent with the lack of N -glucuronidation observed for endoxifen and with UGT1A4’s primary enzymatic function to perform N -glucuronidation as observed for TAM and 4-OH-TAM.40,65
TABLE 1.
Kinetic Analyses of O-glucuronidation of trans-4-OH-TAM or trans-Endoxifen by UGTsa
|
trans-4-OH-TAM |
trans-Endoxifen |
|||||
|---|---|---|---|---|---|---|
| UGT | Vmax (pmol min−1 μg−1)b | KM (μM) | Vmax/KM (μl min−1 μg−1)b | Vmax (pmol min−1 μg−1)b | KM (μM) | Vmax/KM (μl min−1 μg−1)b |
| 1A1 | 3.4 ± 0.2 | 124 ± 16 | 0.028 ± 0.004 | 2.3 ± 0.3 | 333 ± 60 | 0.007 ± 0.0005 |
| 1A3 | 1.9 ± 0.3 | 94 ± 17.8 | 0.02 ± 0.001 | 2.9 ± 0.4 | 158 ± 29 | 0.02 ± 0.001 |
| 1A7 | 1.2 ± 0.2 | 166 ± 27 | 0.0074 ± 0.0002 | low activityc | ||
| 1A8 | 3.2 ± 0.2 | 23 ± 2.4 | 0.14 ± 0.02 | 11.6 ± 1.4 | 101 ± 13 | 0.12 ± 0.01 |
| 1A9 | 3.0 ± 0.1 | 319 ± 38 | 0.009 ± 0.001 | low activityc | ||
| 1A10 | 4.7 ± 0.3 | 96 ± 8.0 | 0.049 ± 0.006 | 5.7 ± 0.7 | 40 ± 3.0 | 0.14 ± 0.005 |
| 2B7 | 0.55 ± 0.18 | 3.7 ± 0.6 | 0.15 ± 0.03 | 3.0 ± 0.4 | 101 ± 17 | 0.03 ± 0.004 |
| 2B17 | 0.02 ± 0.001 | 41 ± 6 | 0.001 ± 0.0001 | no detectable activityd | ||
All data are the mean ± SD based on three independent experiments.
Data are expressed per μg UGT protein as determined by Western blot analysis.
Low activity describes the fact that although some glucuronidation activity was observed for a UGT enzyme against a particular TAM metabolite, the level of detection was below sensitivity for kinetic studies.
In addition to no detectable glucuronidation activity observed for homogenates of UGT2B15-overexpressing cells observed against trans-endoxifen, homogenates from cells overexpressing UGTs 1A6, 2B4, 2B10, 2B11, or 2B15 exhibited no detectable glucuronidating activity against trans-4-OH-TAM or trans-endoxifen.
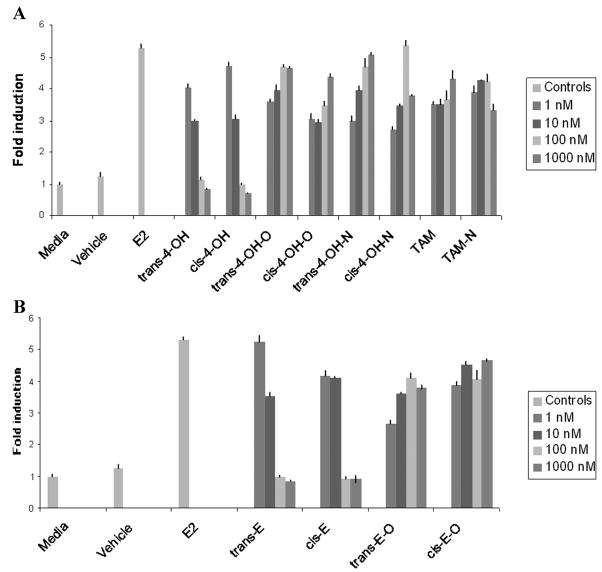
While high antiestrogenic activity has been reported for both 4-OH-TAM and endoxifen, studies examining the effects of glucuronide conjugation of these metabolites were only recently described.70 E2-mediated induction of the gene encoding the progesterone receptor (PGR) was determined in MCF-7 cells by real-time RT-PCR for individual TAM metabolites and isomers. While E2 (1 × 10−10M) induction of PGR mRNA was 6-fold after a 12-h incubation, unconjugated TAM metabolites (i.e., cis and trans isomers of 4-OH-TAM and endoxifen) inhibited this effect (Fig. 2). A similar dose-dependent inhibition of E2-induced PGR gene expression was found for both the trans and cis isomers of 4-OH-TAM and endoxifen, with maximal inhibition attained at 1 × 10−6M of TAM metabolite. In contrast, the glucuronide conjugates of all 4-OH-TAM and endoxifen isomers exhibited no effect on E2-mediated induction of PGR expression at all concentrations of TAM metabolite conjugates examined in this study. These data indicate that isomers of both 4-OH-TAM and endoxifen exhibit roughly equipotent antiestrogenic effects on E2-induced gene expression and that glucuronide conjugates of the same metabolites effectively negate this activity.
Figure 2.
Effect of TAM and TAM metabolites on PGR gene expression in MCF-7 cells. Cells were incubated with 1 × 10−10 M E2 for 12 h at 37°C in 5% CO2. Total RNA was extracted and PGR mRNA levels were determined by RT-PCR as described in the Methods section. PGR mRNA levels were expressed as the fold-induction of PGR mRNA levels observed for untreated cells (media). (A) PGR gene expression in E2-induced MCF-7 cells treated with TAM and 4-OH-TAM isomers or glucuronides; (B) PGR gene expression in E2-induced MCF-7 cells treated with endoxifen isomers or glucuronides. The E2 positive control, the media alone, and vehicle negative control are shown on both panels. E2, cells incubated with 1 × 10−10 M E2; vehicle, cells incubated with 0.1% DMSO; trans-4-OH, cells incubated with trans-4-OH- TAM; cis-4-OH, cells incubated with cis-4-OH-TAM, trans-4-OH-O, cells incubated with trans-TAM-4-O-glucuronide; cis-4-OH-O, cells incubated with cis-TAM-4-O-glucuronide; trans-4-OH-N, cells incubated with trans-4-OH-TAM-N+-glucuronide; cis-4-OH-N, cells incubated with cis-4-OH-TAM-N+-glucuronide; trans-E, cells incubated with trans-endoxifen; cis-E, cells incubated with cis-endoxifen; trans-E-O, cells incubated with trans-endoxifen-O-glucuronide; cis-E-O, cells incubated with cis-endoxifen-O-glucuronide; TAM, cells incubated with TAM; TAM-N, cells incubated with TAM-N-glucuronide. The figure legend describes the concentration of TAM or TAM metabolite used in this experiment. The mean ± standard error is shown for three experiments.
More recent studies have focused on ER-binding activities of TAM metabolites. Similar to that observed for PGR induction, the trans isomers of both 4-OH-TAM and endoxifen exhibit similar relative binding activities compared to E2, while their glucuronide counterparts exhibited 57–130-fold decreases in RBA as compared to their unconjugated counterparts (Fig. 3) (P. Lazarus, unpublished results). While a similar pattern was also observed for cis TAM metabolite isomers and their glucuronides, the RBA of these unconjugated cis isomers were 35–67-fold lower than their trans unconjugated counterparts. These data were similar to that which were observed previously for 4-OH-TAM and endoxifen,28 but was the first assessment of individual isomers and glucuronide conjugates. The trans-4-OH-TAM and trans-endoxifen isomers exhibited 30–37-fold higher RBA than trans-TAM in this study. Overall, these differences suggest that the trans isomers of 4-OH-TAM and endoxifen are the major active antiestrogenic metabolites of TAM, that glucuronides of TAM metabolites are relatively inactive, and that cis isomers may be inhibiting E2-induced activities by mechanisms other than competitive binding to the ER. This could have important implications in how metabolic pathways are targeted in terms of augmenting the therapeutic efficacy of TAM.
Figure 3.
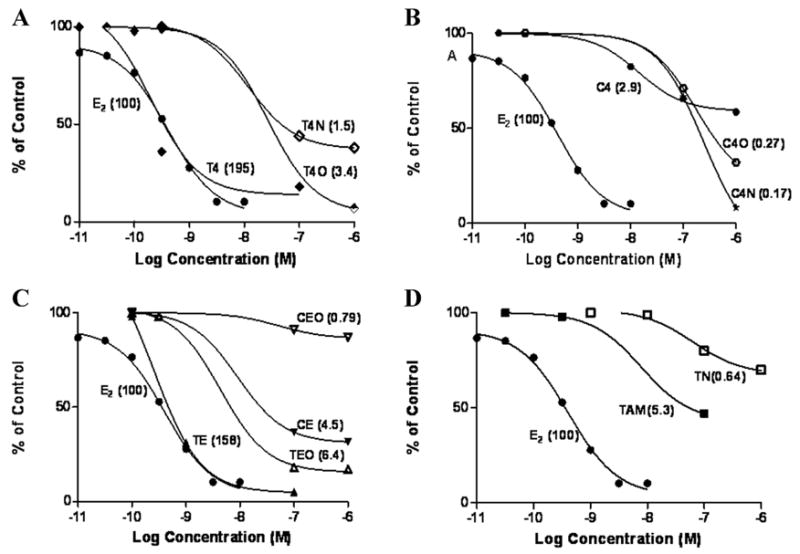
Competitive binding assay of TAM and metabolites of TAM. The cytosolic fraction of MCF-7 cells was incubated (500 μL total reaction) for 18 h at 4°C with the indicated concentration of competitor (10−9 to 10−6 M) or [3H]-labeled E2 (10−11 to 10−6 M). After incubation, 5% dextran-coated charcoal was added to adsorb unbound ligand for 15 min at 4°C, and radioactivity was measured in the supernatant. Data are expressed as the percentage of specific binding of [3H]-E2 for the ER when competitor was not present. Numbers in parentheses, RBA to ER for each test compound, with all data normalized to the RBA of E2 (set at 100). (A) T4, trans-4-OH-TAM; T4O, trans-TAM-4-O-glucuronide, T4N, trans-4-OH-TAM-N-glucuronide; (B) C4, cis-4-OH-TAM; C4O, cis-TAM-4-O-glucuronide; C4N, cis-4-OH-TAM-N-glucuronide; (C) TE, trans-endoxifen; TEO, trans-endoxifen-O- glucuronide; CE, cis-endoxifen; CEO, cis-endoxifen-O-glucuronide; (D) TN, TAM-N-glucuronide.
UGT Polymorphism Effects on TAM Glucuronidation Activities in UGT-Overexpressing Cell Lines
Known missense polymorphisms have been identified in the UGTs active against TAM metabolites, including nonsynonomous SNPs at codons 24 and 48 of the UGT1A4 gene;62 at codon 268 of the UGT2B7 gene,55 at codon 139 in the UGT1A10 gene that is present in African Americans,21 and at codon 173 and 277 of the UGT1A8 gene.72 To determine whether any of these SNPs result in differential activities against the trans isomers of 4-OH-TAM or endoxifen, in vitro kinetic analyses of HEK293 cells overexpressing the wild-type or variant isoforms of each of these three UGT enzymes was performed.65,72 The UGT1A8173Gly/277Cys variant exhibited no difference in overall glucuronidation activity (Vmax/KM) against trans-4-OH-TAM and exhibited a small (1.25-fold) but significant (P < 0.05) decrease in overall activity (manifested primarily by a higher KM) against trans-endoxifen as compared to wild-type UGT1A8173Ala/277Cys (Table 2). In contrast, the UGT1A8173Ala/277Tyr variant exhibited no detectable glucuronidation against the trans isomers of either 4-OH-TAM or endoxifen (Table 2).
TABLE 2.
Kinetic Analyses of O-glucuronidation of the trans Isomers of 4-OH-TAM and Endoxifen by UGT Variantsa
|
trans-4-OH-TAM |
trans-Endoxifen |
|||||
|---|---|---|---|---|---|---|
| UGT Variant | Vmax (pmol min−1 μg−1)b | KM (μM) | Vmax/KM (μl min−1 −g−1)b | Vmax (pmol min−1 μg−1)b | Km (μM) | Vmax/KM (μl min−1 μg−1)b |
| UGT2B7268His | 0.55 ± 0.18 | 3.7 ± 0.6 | 0.15 ± 0.03 | 3.0 ± 0.44 | 101 ± 17 | 0.03 ± 0.004 |
| UGT2B7268Tyr | 0.54 ± 0.09* | 8.7 ± 0.8** | 0.062 ± 0.01** | 0.55 ± 0.01** | 101 ± 15 | 0.006 ± 0.001** |
| UGT1A10139Glu | 4.7 ± 0.3 | 96 ± 8 | 0.05 ± 0.006 | 5.7 ± 0.7 | 40 ± 3 | 0.14 ± 0.005 |
| UGT1A10139Lys | 2.1 ± 0.2** | 52 ± 6** | 0.04 ± 0.006 | 1.9 ± 0.2** | 13 ± 2** | 0.14 ± 0.004 |
| UGT1A8173Ala/277Cys | 2.3 ± 0.1 | 23 ± 2 | 0.10 ± 0.02 | 5.4 ± 0.2 | 98 ± 9 | 0.06 ± 0.004 |
| UGT1A8173Gly/277Cys | 5.4 ± 0.2** | 43 ± 7** | 0.13 ± 0.03 | 5.9 ± 0.4 | 135 ± 26 | 0.04 ± 0.005* |
| UGT1A8173Ala/277Tyr | no detectable activity | no detectable activity | ||||
All data are the mean ±SD based on three independent experiments. Homogenates from cells overexpressing UGT1A8173Ala/277Tyr exhibited no detectable activity against trans-4-OH-TAM and trans-endoxifen.
Data are expressed per μg UGT protein as determined by Western blot analysis.
P = 0.05;
P < 0.01.
For UGT1A4, the codons 24 (Pro>Thr) and 48 (Leu>Val) SNPs were examined (Table 3). Kinetic analysis demonstrated that higher N -glucuronidation activities were observed for UGT1A424Pro/48Val-overexpressing microsomes as compared to cell microsomes from wild-type UGT1A424Pro/48Leu-overexpressing cells against TAM and trans- 4-OH-TAM, with a significantly (P ≤ 0.02) lower Km observed for trans-4-OH-TAM for the UGT1A424Pro/48Val variant. No significant effect on enzyme kinetics was observed for the UGT1A424Thr/48Leu variant against trans-4-OH-TAM or TAM. In addition, no difference in overall glucuronidation activity was observed for the UGT1A10139Lys variant versus wild-type UGT1A10 against the trans isomers of both 4-OH-TAM and endoxifen (Table 2).
TABLE 3.
Kinetic Analysis of UGT1A4-induced Glucuronidation of TAM, trans-4-OH-TAM and cis-4-OH-TAMa
| Substrate | UGT1A4 Variant | Vmax (pmol min−1 μg−1)b | Km (μM) | Vmax/Km (μl min−1 μg−1)b |
|---|---|---|---|---|
| trans-4-OH-TAM | UGT1A424Pro/48Leu | 62.4 ± 5.8 | 2.2 ± 0.4 | 29.3 ± 2.7 |
| UGT1A424Thr/48Leu | 54.9 ± 11.4 | 1.6 ± 0.1 | 33.2 ± 4.9 | |
| UGT1A424Pro/48Val | 49.3 ± 2.8 | 1.2 ± 0.1c | 40.8 ± 1.4d | |
| trans-TAM | UGT1A424Pro/48Leu | 68.0 ± 8.6 | 2.0 ± 0.51 | 35.2 ± 9.6 |
| UGT1A424Thr/48Leu | 62.1 ± 2.6 | 1.5 ± 0.20 | 41.0 ± 7.0 | |
| UGT1A424Pro/48Val | 52.1 ± 10.1 | 1.3 ± 0.10e | 40.1 ± 7.5 |
Data are expressed as mean ±SD for three independent experiments.
Data are expressed per μg UGT protein as determined by Western blot analysis. Values are significantly
P = 0.01,
P = 0.005 or
near-significantly (P = 0.053) different from that observed for microsomes from wild-type UGT1A424Pro/48Leu overexpressing cells.
For UGT2B7, kinetic analysis demonstrated that significantly higher glucuronidation activities were observed for the wild-type UGT2B7268His as compared to the UGT2B7268Tyr variant against the trans isomers of both 4-OH-TAM (P < 0.05) and endoxifen (P < 0.01; Table 2). This was manifested by a higher KM (2.4-fold) and a lower Vmax/KM (2.4-fold) for 4-OH-TAM, as well as a lower Vmax (5.5-fold) and lower Vmax/KM (5.0-fold) for endoxifen.
UGT Genotypes and TAM Glucuronidation Phenotype in HLM
To determine the rate of O- and N -glucuronidation of trans-4-OH-TAM and trans-endoxifen, glucuronidation assays were performed in a series of HLM and analyzed by ultra-pressure liquid chromatography (UPLC).72 The mean rate of formation of TAM-4-O-glucuronide, 4-OH-TAM-N +-glucuronide, and endoxifen-O-glucuronide in 111 HLM specimens was 141 ± 44.9, 175 ± 51.5, and 168 ± 65.5 pmol min−1 mg−1, respectively. 4.5-, 10-, and 17-fold ranges in glucuronide formation were observed for TAM-4-O-glucuronide, 4-OH-TAM-N +-glucuronide, and endoxifen-O-glucuronide, respectively. The range of the ratio of TAM-4-O-glucuronide:4-OH-TAM-N +-glucuronide in the HLM samples was 8.0-fold. These data suggest that significant differences in glucuronidation capacity exist among individual HLM. After stratifying by UGT2B7 codon 268 genotype, there was a near-significant (P = 0.059) 13% decrease in TAM-4-O-glucuronide formation in HLM with the UGT2B7 (His268Tyr) genotype and a significant (P < 0.001) 28% decrease in TAM-4-O-glucuronide formation in HLM with the UGT2B7 (Tyr268Tyr) genotype as compared to HLM with the UGT2B7 (His269His) genotype (Fig. 4A).72 A significant (P = 0.01) 17% decrease in TAM-4-O-glucuronide formation was observed in HLM with the UGT2B7 His268Tyr genotype versus HLM with the UGT2B7 (Tyr268Tyr) genotype. A significant trend of decreasing O-glucuronidation of trans-4-OH-TAM was observed in HLM with increasing numbers of the UGT2B7268Tyr allele (P < 0.001).
Figure 4.
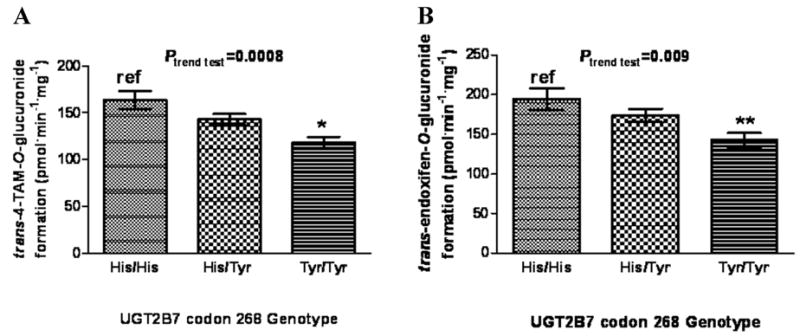
Analysis of glucuronidation activities against trans-4-OH-TAM and trans-endoxifen in HLM stratified by UGT2B7 genotypes. Glucuronidation assays were performed and 4-OH-TAM and endoxifen glucuronides separated by UPLC as described previously.72 (A) trans-4-OH-TAM and UGT2B7 codon 268 genotypes; (B) trans-endoxifen and UGT2B7 codon 268 genotypes. Comparative analysis was performed using the wild-type UGT2B7268His as the referent; *P < 0.001; **P < 0.002, and error bars represent standard error.
Similar to that observed for trans-4-OH-TAM, a significant (P = 0.002) 27% decrease in O-glucuronidation of trans-endoxifen was observed in HLM with the UGT2B7 (Tyr268Tyr) genotype as compared to HLM with the UGT2B7 (His268His) genotype (Fig. 4B). A significant trend of decreasing O-glucuronidation of trans-endoxifen was observed in HLM with increasing numbers of the UGT2B7268Tyr allele (P = 0.009). No N -glucuronidation of endoxifen was observed for any of the HLM specimens analyzed in these studies.
UGTs and TAM Pharmacogenetics
As discussed above, glucuronidation plays a major role in TAM metabolism, with specific UGT enzymes performing either N or O-glucuronidation of active TAM metabolites. UGT2B7 appears to be the most active hepatic UGT. Additionally, UGT2B7 expression has been detected in a variety of tissues including liver, the gastrointestinal tract, and breast47,52,73–76; therefore, variations in UGT2B7 function or expression could potentially significantly impact individual response to drugs or chemotherapeutic agents. The data presented here demonstrate that O-glucuronidation of both trans-4-OH-TAM and trans-endoxifen in HLM was significantly associated with UGT2B7 genotype, with lower activities correlated with increasing numbers of the UGT2B7268Tyr allele. These data were consistent with the observation that HEK293 cells that overexpressed the UGT2B7268Tyr variant exhibited lower activity in vitro against both TAM metabolites as compared to cells over-expressing wild-type UGT2B7268His. These results are also consistent with a functional role for this polymorphism against other substrates, including tobacco carcinogen metabolites like 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL).62
The extra-hepatic UGTs 1A10 and 1A8 exhibited the highest levels of activity in vitro against the trans isomers of 4-OH-TAM and endoxifen in previous studies.40 Of the SNPs examined in these genes, the UGT1A8277Tyr variant exhibited no detectable glucuronidating activity against both trans-4-OH-TAM and trans-endoxifen. This was consistent with previous data indicating that this variant exhibited dramatically reduced activity towards other substrates.71,77 While the prevalence of this polymorphism is low in the population (~2% in Caucasians),71 the observation that UGT1A8 is highly active against TAM metabolites and is well-expressed in the breast78,79 suggests that, like the UGT2B7 codon 268 polymorphism, the UGT1A8 codon 277 polymorphism potentially could be important in individual response to TAM.
Therefore, similar to what is described above for CYP2D6, functional SNPs in UGTs 2B7 and 1A8 potentially could affect overall patient response to TAM. Additional studies examining the effect of UGT1A8 and UGT2B7 genotypes on breast microsomal glucuronidation activity against TAM metabolites, plasma TAM metabolite levels in women taking TAM, and overall patient response to TAM are needed to further examine the role of UGT polymorphisms on the therapeutic efficacy of TAM.
Acknowledgments
These studies were supported by Public Health Service grant R01-DE13158 (Lazarus) from the National Institutes of Health, and a formula grant under the Pennsylvania Department of Health’s Health Research Formula Funding Program, State of PA, Act 2001–77 – part of the PA Tobacco Settlement Legislation (Lazarus).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Cuzick J, et al. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- 2.Fisher B, et al. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 3.Howell A, Howell SJ, Evans DG. New approaches to the endocrine prevention and treatment of breast cancer. Cancer Chemother Pharmacol. 2003;52(Suppl 1):S39–S44. doi: 10.1007/s00280-003-0645-5. [DOI] [PubMed] [Google Scholar]
- 4.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 5.McDonald CC, Stewart HJ. Fatal myocardial infarction in the Scottish adjuvant tamoxifen trial. The Scottish Breast Cancer Committee. BMJ. 1991;303:435–437. doi: 10.1136/bmj.303.6800.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutqvist LE, Mattsson A. Cardiac and thromboembolic morbidity among postmenopausal women with early-stage breast cancer in a randomized trial of adjuvant tamoxifen. The Stockholm Breast Cancer Study Group. J Natl Cancer Inst. 1993;85:1398–1406. doi: 10.1093/jnci/85.17.1398. [DOI] [PubMed] [Google Scholar]
- 7.Rutqvist LE, et al. Adjuvant tamoxifen therapy for early stage breast cancer and second primary malignancies. Stockholm Breast Cancer Study Group. J Natl Cancer Inst. 1995;87:645–651. doi: 10.1093/jnci/87.9.645. [DOI] [PubMed] [Google Scholar]
- 8.van Leeuwen FE, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–52. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- 9.Meier CR, Jick H. Tamoxifen and risk of idiopathic venous thromboembolism. Br J Clin Pharmacol. 1998;45:608–612. doi: 10.1046/j.1365-2125.1998.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: An overview of the randomised trials. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 11.Bergman L, et al. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Comprehensive Cancer Centres’ ALERT Group. Assessment of Liver and Endometrial Cancer Risk Following Tamoxifen. Lancet. 2000;356:881–887. doi: 10.1016/s0140-6736(00)02677-5. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein L, et al. Tamoxifen therapy for breast cancer and endometrial cancer risk. J Natl Cancer Inst. 1999;91:1654–1662. doi: 10.1093/jnci/91.19.1654. [DOI] [PubMed] [Google Scholar]
- 13.Fisher B, et al. Endometrial cancer in tamoxifen-treated breast cancer patients: Findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–537. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- 14.Furr BJ, Jordan VC. The pharmacology and clinical uses of tamoxifen. Pharmacol Ther. 1984;25:127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- 15.Boocock DJ, et al. Identification of human CYP forms involved in the activation of tamoxifen and irreversible binding to DNA. Carcinogen. 2002;23:1897–1901. doi: 10.1093/carcin/23.11.1897. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, et al. Alpha-hydroxylation of tamoxifen and toremifene by human and rat cytochrome P450 3A subfamily enzymes. Chem Res Toxicol. 2003;16:1138–1144. doi: 10.1021/tx0300131. [DOI] [PubMed] [Google Scholar]
- 17.Desta Z, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: Prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 18.Jacolot F, et al. Identification of the cytochrome P450 IIIA family as the enzymes involved in the N-demethylation of tamoxifen in human liver microsomes. Biochem Pharmacol. 1991;41:1911–1919. doi: 10.1016/0006-2952(91)90131-n. [DOI] [PubMed] [Google Scholar]
- 19.Crewe HK, et al. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: Formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:869–874. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Coller JK. Oxidative metabolism of tamoxifen to Z-4-hydroxy-tamoxifen by cytochrome P450 isoforms: An appraisal of in vitro studies. Clin Exp Pharmacol Physiol. 2003;30:845–848. doi: 10.1046/j.1440-1681.2003.03921.x. [DOI] [PubMed] [Google Scholar]
- 21.Coller JK, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54:157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crewe HK, et al. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol. 1997;53:171–178. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 23.Dehal SS, Kupfer D. CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 1997;57:3402–3406. [PubMed] [Google Scholar]
- 24.Hu Y, et al. CYP2D6-mediated catalysis of tamoxifen aromatic hydroxylation with an NIH shift: Similar hydroxylation mechanism in chicken, rat and human liver microsomes. Xenobiotica. 2003;33:141–151. doi: 10.1080/0049825021000042733. [DOI] [PubMed] [Google Scholar]
- 25.Lonning PE, et al. Clinical pharmacokinetics of endocrine agents used in advanced breast cancer. Clin Pharmacokinet. 1992;22:327–358. doi: 10.2165/00003088-199222050-00002. [DOI] [PubMed] [Google Scholar]
- 26.Stearns V, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 27.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 28.Johnson MD, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 29.Jordan VC, et al. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 30.Katzenellenbogen BS, et al. Bioactivities, estrogen receptor interactions, and plasminogen activator-inducing activities of tamoxifen and hydroxy-tamoxifen isomers in MCF-7 human breast cancer cells. Cancer Res. 1984;44:112–119. [PubMed] [Google Scholar]
- 31.Lim L, et al. Gene expression profiles of 4-hydroxy-N-desmethyltamoxifen (endoxifen) and 4-hydroxytamoxifen (4OHTAM) treated human breast cancer cells determined by cDNA microarray analysis. Clin Pharmacol Ther. 2004;75:49. [Google Scholar]
- 32.Lim YC, et al. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has antiestrogenic effects in breast cancer cells with potency similar to 4-hydroxytamoxifen. Cancer Chemother Pharmacol. 2005;55:471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 33.Apak TI, Duffel MW. Interactions of the stereoisomers of alpha-hydroxytamoxifen with human hydroxysteroid sulfotransferase SULT2A1 and rat hydroxysteroid sulfotransferase STa. Drug Metab Dispos. 2004;32:1501–1508. doi: 10.1124/dmd.104.000919. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, et al. 4-Hydroxytamoxifen sulfation metabolism. J Biochem Mol Toxicol. 2002;16:279–285. doi: 10.1002/jbt.10048. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama T, et al. Reverse geometrical selectivity in glucuronidation and sulfation of cis-and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol. 2002;63:1817–1830. doi: 10.1016/s0006-2952(02)00994-2. [DOI] [PubMed] [Google Scholar]
- 36.Falany CN. Enzymology of human cytosolic sulfotransferases. Faseb J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 37.Lien EA, et al. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- 38.Lien EA, et al. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48:2304–2308. [PubMed] [Google Scholar]
- 39.Poon GK, et al. Analysis of phase I and phase II metabolites of tamoxifen in breast cancer patients. Drug Metab Dispos. 1993;21:1119–1124. [PubMed] [Google Scholar]
- 40.Sun D, et al. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos. 2007;35:2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 41.Borges S, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Goetz MP, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 43.Bock KW. Roles of UDP-glucuronosyltransferases in chemical carcinogenesis. Crit Rev Biochem Mol Biol. 1991;26:129–150. doi: 10.3109/10409239109081125. [DOI] [PubMed] [Google Scholar]
- 44.Bock KW, et al. Functions and transcriptional regulation of PAH-inducible human UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31:411–422. doi: 10.1081/dmr-100101927. [DOI] [PubMed] [Google Scholar]
- 45.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 46.Owens IS, Ritter JK. Gene structure at the human UGT1 locus creates diversity in isozyme structure, substrate specificity, and regulation. Prog Nucleic Acid Res Mol Biol. 1995;51:305–338. doi: 10.1016/s0079-6603(08)60882-x. [DOI] [PubMed] [Google Scholar]
- 47.Strassburg CP, et al. Differential expression of the UGT1A locus in human liver, biliary, and gastric tissue: identification of UGT1A7 and UGT1A10 transcripts in extrahepatic tissue. Mol Pharmacol. 1997;52:212–220. doi: 10.1124/mol.52.2.212. [DOI] [PubMed] [Google Scholar]
- 48.Emi Y, Ikushiro S, Iyanagi T. Drug-responsive and tissue-specific alternative expression of multiple first exons in rat UDP-glucuronosyltransferase family 1 (UGT1) gene complex. J Biochem (Tokyo) 1995;117:392–399. doi: 10.1093/jb/117.2.392. [DOI] [PubMed] [Google Scholar]
- 49.Jedlitschky G, et al. Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J. 1999;340(Pt 3):837–843. [PMC free article] [PubMed] [Google Scholar]
- 50.Itaaho K, et al. The configuration of the 17-hydroxy group variably influences the glucuronidation of {beta}-estradiol and epiestradiol by human UDP-glucuronosyltransferases. Drug Metab Dispos. 2008;36:2307–2315. doi: 10.1124/dmd.108.022731. [DOI] [PubMed] [Google Scholar]
- 51.Monaghan G, et al. Localization of a bile acid UDP-glucuronosyltransferase gene (UGT2B) to chromosome 4 using the polymerase chain reaction. Genomics. 1992;13:908–909. doi: 10.1016/0888-7543(92)90188-x. [DOI] [PubMed] [Google Scholar]
- 52.Strassburg CP, et al. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. Biochem J. 1999;338(Pt 2):489–498. [PMC free article] [PubMed] [Google Scholar]
- 53.Burchell B, Hume R. Molecular genetic basis of Gilbert’s syndrome. J Gastroenterol Hepatol. 1999;14:960–966. doi: 10.1046/j.1440-1746.1999.01984.x. [DOI] [PubMed] [Google Scholar]
- 54.Ciotti M, et al. Genetic polymorphism in the human UGT1A6 (planar phenol) UDP-glucuronosyltransferase: pharmacological implications. Pharmacogenetics. 1997;7:485–495. doi: 10.1097/00008571-199712000-00007. [DOI] [PubMed] [Google Scholar]
- 55.Coffman BL, et al. The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y and UGT2B7H. Drug Metab Dispos. 1998;26:73–77. [PubMed] [Google Scholar]
- 56.Elahi A, et al. Detection of UGT1A10 polymorphisms and their association with orolaryngeal carcinoma risk. Cancer. 2003;98:872–880. doi: 10.1002/cncr.11587. [DOI] [PubMed] [Google Scholar]
- 57.Guillemette C, et al. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60:950–956. [PubMed] [Google Scholar]
- 58.Huang YH, et al. Identification and functional characterization of UDP-glucuronosyltransferases UGT1A8*1, UGT1A8*2 and UGT1A8*3. Pharmacogenetics. 2002;12:287–297. doi: 10.1097/00008571-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Iwai M, et al. Six novel UDP-glucuronosyltransferase (UGT1A3) polymorphisms with varying activity. J Hum Genet. 2004;49:123–128. doi: 10.1007/s10038-003-0119-y. [DOI] [PubMed] [Google Scholar]
- 60.Levesque E, et al. Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1997;7:317–325. doi: 10.1097/00008571-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 61.Levesque E, et al. Characterization and substrate specificity of UGT2B4 (E458): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1999;9:207–216. [PubMed] [Google Scholar]
- 62.Wiener D, et al. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–1196. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 63.Wilson W, 3rd, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Kaku T, et al. Quaternary ammonium-linked glucuronidation of tamoxifen by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol. 2004;67:2093–2102. doi: 10.1016/j.bcp.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Sun D, et al. Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res. 2006;8:R50. doi: 10.1186/bcr1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breyer-Pfaff U, et al. Comparative N-glucuronidation kinetics of ketotifen and amitriptyline by expressed human UDP-glucuronosyltransferases and liver microsomes. Drug Metab Dispos. 2000;28:869–872. [PubMed] [Google Scholar]
- 67.Green MD, Tephly TR. Glucuronidation of amines and hydroxylated xenobiotics and endobiotics catalyzed by expressed human UGT1.4 protein. Drug Metab Dispos. 1996;24:356–363. [PubMed] [Google Scholar]
- 68.Green MD, Tephly TR. Glucuronidation of amine substrates by purified and expressed UDP-glucuronosyltransferase proteins. Drug Metab Dispos. 1998;26:860–867. [PubMed] [Google Scholar]
- 69.Wiener D, et al. Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos. 2004;32:72–79. doi: 10.1124/dmd.32.1.72. [DOI] [PubMed] [Google Scholar]
- 70.Zheng Y, et al. Elimination of antiestrogenic effects of active tamoxifen metabolites by glucuronidation. Drug Metab Dispos. 2007;35:1942–1948. doi: 10.1124/dmd.107.016279. [DOI] [PubMed] [Google Scholar]
- 71.Huang YH, et al. Identification and funtional characterization of UDP-glucuronosyltransferases UGT1A8*1, UGT1A8*2 and UGT1A8*3. Pharmacognetics. 2002;12:287–297. doi: 10.1097/00008571-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 72.Blevins-Primeau, et al. Functional significance of polymorphic variants of UDP-glucuronosyltransferases (UGTs) active against tamoxifen metabolites. Cancer Res in press. [Google Scholar]
- 73.Nakamura A, et al. Expression of UGT1‘A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008;36:1461–1464. doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- 74.Ren Q, et al. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352–1360. [PubMed] [Google Scholar]
- 75.Turgeon D, et al. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- 76.Zheng Z, Fang JL, Lazarus P. Glucuronidation: An important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos. 2002;30:997–403. doi: 10.1124/dmd.30.4.397. [DOI] [PubMed] [Google Scholar]
- 77.Bernard O, et al. Influence of nonsynonymous polymorphisms of UGT1A8 and UGT2B7 metabolizing enzymes on the formation of phenolic and acyl glucuronides of mycophenolic acid. Drug Metab Dispos. 2006;34:1539–1545. doi: 10.1124/dmd.106.010553. [DOI] [PubMed] [Google Scholar]
- 78.Lehmann L, Wagner J. Gene expression of 17beta-estradiol-metabolizing isozymes: Comparison of normal human mammary gland to normal human liver and to cultured human breast adenocarcinoma cells. Adv Exp Med Biol. 2008;617:617–624. doi: 10.1007/978-0-387-69080-3_64. [DOI] [PubMed] [Google Scholar]
- 79.Thibaudeau J, et al. Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res. 2006;66:125–133. doi: 10.1158/0008-5472.CAN-05-2857. [DOI] [PubMed] [Google Scholar]