Abstract
The pacemaker is composed of specialized cardiomyocytes located within the sinoatrial node (SAN), and is responsible for originating and regulating the heart beat. Recent advances towards understanding the SAN development have been made on the genetic control and gene interaction within this structure. Here we report that the Shox2 homeodomain transcription factor is restrictedly expressed in the sinus venosus region including the SAN and the sinus valves during embryonic heart development. Shox2 null mutation results in embryonic lethality due to cardiovascular defects, including an abnormal low heart beat rate (bradycardia) and severely hypoplastic SAN and sinus valves attributed to a significantly decreased level of cell proliferation. Genetically, the lack of Tbx3 and Hcn4 expression, along with ectopic activation of Nppa, Cx40, and Nkx2-5 in the Shox2−/− SAN region, indicates a failure in SAN differentiation. Furthermore, Shox2 overexpression in Xenopus embryos results in extensive repression of Nkx2-5 in the developing heart, leading to a reduced cardiac field and aberrant heart formation. Reporter gene expression assays provide additional evidence for the repression of Nkx2-5 promoter activity by Shox2. Taken together our results demonstrate that Shox2 plays an essential role in the SAN and pacemaker development by controlling a genetic cascade through the repression of Nkx2-5.
Keywords: Shox2, Nkx2-5, Sinoatrial node, Pacemaker, Sinus valves, Heart development
Introduction
During embryonic heart development, the fused linear primitive heart tube contracts spontaneously, randomly and irregularly beginning at embryonic day 8.0 (E8.0). By E9.0 the heart beat becomes organized and regular with a caudal to cranial directionality (van Mierop, 1967; Nishii and Shibata, 2006). The regularity of the heart beat coincides with the “maturation” of the cardiac pacemaker located in the caudal region of the linear heart tube, the sinus venosus (van Mierop, 1967) and eventually becomes restricted to the right dorsal wall of the right atrium, structurally known as the sinoatrial node (SAN) (van Mierop, 1967; van Mierop and Gessner, 1970). Deficiencies in such pacemaking function lead to cardiac defects specially arrhythmias and in severe cases to cardiac arrest and sudden death (Durham and Worthley, 2002).
Genetically, the SAN is characterized by the expression or lack thereof of several molecular markers. The hyperpolarization-gated cyclic nucleotide cation-activated channel, Hcn4, a specific molecular marker for the pacemaker (Santoro and Tibbs, 1999) is expressed as early as E7.5 in the cardiac crescent, becoming restricted to the sinus horns of the sinus venosus and eventually to the SAN region by E12.5 (García-Frigola et al., 2003). Hcn4 mutant mice die at mid-gestation stage due to the lack of formation of a “mature” pacemaker, while its overexpression mimics pacemaker properties in cell cultures (Santoro and Tibbs, 1999; Santoro et al., 2000; Moosmang et al., 2001; Stieber et al., 2003; Liu et al., 2007). The T-box transcription factor Tbx3, has been reported to be expressed in the cardiac conduction system including the SAN (Hoogaars et al., 2004). Mutation of the Tbx3 gene causes limb and mammary deformities and embryonic lethality due to cardiovascular defects (Davenport et al., 2003), however, does not impair the early formation of the SAN in mice (Hoogaars et al., 2007). Moreover, Tbx3 was shown to repress the expression of Nppa (ANF) and Cx40 by directly binding to the promoter of each gene in cultured cells (Hoogaars et al., 2004). Indeed, the expression of Nppa and Cx40 is mutually exclusive to that of Tbx3 in the heart (van Kempen et al., 1996; Gaussin, 2004; Soufan et al., 2004; Stennard and Harvey, 2005). The early cardiac differentiation marker Nkx2-5 is expressed as early as the formation of the cardiac crescent (Komuro and Izumo, 1993; Moses et al., 2001). However, it is not expressed in the SAN or in the venous valves (Kasahara et al., 1998). Mice deficient in Nkx2-5 are embryonically lethal due to disruption in the differentiation of cardiac tissue (Lyons et al., 1995; Tanaka et al., 1999), while its overexpression induces nodal myocytes characteristics in rat myocardial cultures (Wang et al., 2007). Additionally, haploinsufficiency and point mutations in the human Nkx2-5 gene are related to arrhythmias and atrioventricular conduction deficiencies (Schott et al., 1998; Benson et al., 1999; Watanabe et al., 2002). Nkx2-5, Tbx3 and Hcn4 have been integrated in a genetic network involved in the development and patterning of the SAN (Hoogaars et al., 2007; Mommersteeg et al., 2007).
Shox2, formerly known as Og12 in mice (Rovescalli et al., 1996), Prx3 in rats (van Schaick et al., 1997) and SHOT in humans (Blaschke et al., 1998), is closely related to the short stature homeobox gene SHOX, both being present only in vertebrates; however, a SHOX ortholog is absent in the mouse. Mutations in either SHOX or Shox2 cause skeletal and other abnormalities, particularly in the long bones (Ellison et al., 1997; Rao et al., 2001; Belin et al., 1998; Shears et al., 1998; Zinn et al., 2002; Yu et al., 2005, 2007; Cobb et al., 2006; Gu et al., 2008). Shox2 has been shown to be expressed in the SAN; its null mutation causes embryonic lethality and ectopic expression of Nkx2-5 and Cx40 in the SAN region, suggesting a possible role in the differentiation of this structure (Blaschke et al., 2007). However the detailed relationship between Shox2 and the development of the SAN is still not complete and the Shox2 molecular niche in such a process is still not clear.
In this study we show that Shox2 expression is restricted to the sinus venosus region, and eventually to the SAN, including the pacemaker and sinus valves in the developing mouse heart. Shox2 null mutation leads to cardiac edema, vascular defects, hypoplasia of the SAN and sinus valves as well as severe bradycardia. Gene expression analyses of molecular markers reveal failure in the differentiation of pacemaker cells and absence of a defined SAN in the Shox2 mutant hearts. Shox2 overexpression represses the expression of Nkx2-5 in cell cultures, and in Xenopus embryos leads to cardiac defects similar to those seen in the Nkx2-5-deficient Xenopus, zebrafish and mouse models.
Materials and methods
Animals and embryos
Generation of Shox2 mutant mice has been described previously (Yu et al., 2005). Shox2 heterozygotes were maintained on both C57BL/6 and CD-1 genetic backgrounds. Histological analyses and comparisons were performed using wild type (WT), and Shox2−/− mice on both genetic backgrounds. Embryos were collected at different embryonic stages for examination of death rate. Embryonic age was determined by counting the appearance of vaginal plug as embryonic day 0.5 (E 0.5). Genotyping was performed by PCR using genomic DNA extracted from tail biopsies or from the amniotic membrane as described previously (Yu et al., 2005). Xenopus laevis eggs were obtained from hormone-stimulated females, fertilized in vitro, and cultured following a standard procedure (Sive et al., 2000). Embryos were reared in 0.1× Marc’s Modified Ringer’s solution (MMR) at 25 °C until desired stages. Embryo staging was determined according to Nieuwkoop and Faber (1967). Microinjection was performed in 3% Ficoll solution; mRNA encoding GR-Shox2 (50 pg) was injected into the marginal zone of two dorsal blastomeres at the 4-cell stage. At the early neurula stage, control and injected embryos were divided into two groups, with one group cultured in 0.2× MMR, while the other group cultured in 0.2× MMR containing 10 μM dexamethasone (Sigma) (Kolm and Sive, 1995). Embryos were harvested at the tadpole stage, processed and analyzed for gene expression by in situ hybridization.
Histology, in situ hybridization, detection of cell proliferation and apoptosis
Wild type and Shox2−/− embryos on C57BL/6 or CD-1 backgrounds were collected in DEPC-treated PBS, fixed in 4% paraformaldehyde (PFA) in DEPC-treated PBS overnight at 4 °C, dehydrated in a series of alcohol, cleared in xylene and embedded in paraffin wax. Embryos were sectioned at 7 μm–10 μm and subjected to either Hematoxilin/Eosin staining or in situ hybridization after deparaffination and rehydration as described previously (Alappat et al., 2005). Xenopus embryos were similarly treated; however, dehydration in methanol series was used for whole mount in situ hybridization. Samples for in situ hybridization were hybridized with antisense RNA probes for mouse Tbx3 (provided by Dr. Ken Muneoka, Tulane University), mouse Nkx2-5 and Nppa (provided by Dr. Eric Olson’s laboratory, Southwestern Medical Center, University of Texas), Xenopus Nkx2-5 and cTnI (provided by Dr. Frank Conlon’s laboratory, University of North Carolina-Chapel Hill). Other probes used were cloned by RT-PCR in our laboratory and sequence confirmed. Cell proliferation rate was detected by in vivo BrdU labeling using a BrdU Labeling and Staining Kit following the manufacturer’s instruction (Roche Applied Sciences, IN), and cell apoptosis was detected by using a Cell Death Detection Kit (Roche Applied Sciences, IN), as previously described (Yu et al., 2005; Alappat et al., 2005). To obtain consistent results, all the experiments, including in situ hybridization, BrdU labeling, and TUNEL assays were repeated at least three times. For BrdU labeling studies, BrdU-positive cells were counted and presented as percentage of total cells within the selected areas. Nine sections from three individual samples of either wild type or mutant were counted and applied to statistical calculation. The Student’s t-test was used to determine significant statistical difference.
Measurement of heart beat rate
For measurement of heart beat rate, embryonic hearts were isolated and cultured according to a standard method as previously described (Stieber et al., 2003). Briefly, embryos at E9.5 and E10.5 were dissected in pre-warmed ADS buffer containing glucose. Hearts were excised and cultured in DMEM containing 10% FBS and 1% antibiotics at 37 °C and 10% CO2 for 24 h. Heart beats were counted under inverted microscope for 10 s three times for each heart before the genotype was known, thus assuring a blind heart beat count. Heart beat counts were converted to heart beats per minute and statistical analysis was performed using the Student’s t-test for determination of statistical difference.
Plasmid constructs
The Shox2a coding sequence was amplified by PCR using Pfu high fidelity enzyme (Strategene) and inserted into the pcDNA3 vector (Invitrogen) containing a myc tag in order to create the myc-Shox2 construct. To make the dexamethasone inducible fusion protein version of Shox2 (GR-Shox2), the Shox2a coding sequence was cloned into the pCS2+_GR vector, which contains the glucocorticoid ligand-binding domain (amino acids 513–777). The 3.3-kb mouse Nkx2-5 upstream sequence driving the expression of the Luciferase gene (Nkx2-5-Luc) (Liberatore et al., 2002) was provided by Dr. Katherine Yutzey (Cincinnati Children’s Hospital Medical Center) and utilized in subsequent experiments. Site-directed mutagenesis was carried out on the Nkx2-5-luc construct by PCR using Pfu high fidelity enzyme following the manufacturer’s instructions (Stratagene) and the following mutagenesis primers (forward sequence shown): BS-I, 5′-GGGCTGATCGCTTTTCAATCAAGAAGAAGTTATTTACGCAGGATGCGC-3′, BS-II, 5′-GCAGCTTATCTTTCACTTCCTCATATATACTTTTCGCGGC-3′; BS-III, 5′-GACGTCTCCCCGGCGCATTCCTGACATTCCGGGTGATAGTTG-3′; BS-IV, 5′-CAATATAGCTCCCCCAATTCAACGGTCCTATTTCAGGCGTCAGC-3′. Truncated derivatives of the Nkx2-5-luc construct were obtained by digesting plasmid DNA with KpnI and BstEII (2.9-kb) or NruI (2.1-kb) restriction endonucleases, filled in by Klenow fragment and re-ligated using T4 ligase. The resulting constructs were verified by sequence analysis.
Cell cultures and reporter expression assays
Rat cardiomyocytes were isolated from neonatal Sprague–Dawley rat hearts using a cardiomyocyte isolation kit (Worthington Biochemical, NJ) following the manufacturer’s instructions. H9c2 embryonic rat-heart derived cell line was obtained from American Tissue Culture Collection (ATCC — Manassas, VA). Cell cultures were maintained in DMEM supplemented with 10%FBS, with or without antibiotics, at 37 °C and 5% CO2. H9c2 cell cultures were not allowed to reach confluency to avoid differentiation and myoblast formation. Transfections were performed in triplicate in each experiment using Fugene6 (Roche) or Lipofectamine (Invitrogen) reagents following the manufacturer’s instructions. Cell cultures were lysated 48 h post-transfection with Luciferase Reporter lysis buffer (Promega). CMV-LacZ reporter vector was used as an internal control and LacZ activity was used for normalizing transfection results. Experiments were repeated twice.
Results
Shox2 expression in the developing heart
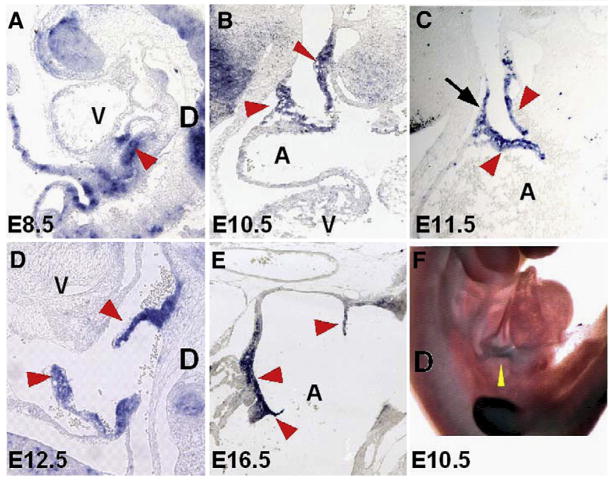
Shox2 expression has been reported to be restricted to the SAN, sinus valves and branches of the central conduction system in mouse and embryonic heart and (Blaschke et al., 1998, 2007; Semina et al., 1998; Clement-Jones et al., 2000). We analyzed a detailed Shox2 expression pattern in the early developing mouse heart by in situ hybridization. Shox2 expression was not detectable in the heart at E8.0 (data not shown), but was found in a restricted pattern at the junction formed by the common cardinal vein and the common atrial chamber, the sinus venosus, at E8.5 (Fig. 1A). Subsequently, cardiac heart looping and remodeling brings the sinus venosus to the right side of the heart and Shox2 expression extends to the sinus valves originating from the sinus node at E10.5 (Fig. 1B). At E11.5 the sinus valves develop to a definite structure, Shox2 expression becomes specifically restricted to the SAN and to the venous layers of the sinus valves (Fig. 1C). At E12.5 and later in development the expression pattern remains consistently in the SAN region and the sinus valves (Figs. 1D, E). Nucleotide sequence analysis suggests that the Shox2 gene generates two isoforms by alternative splicing, Shox2a and a shorter version Shox2b (Rovescalli et al., 1996; Blaschke et al., 1998). Both isoforms contain the sequence coding for the homeodomain, an SH3 binding domain, and P-loop binding domain. However, Shox2b lacks 363 nucleotides which correspond to the N-terminus unique to Shox2a. By using this sequence as a Shox2a specific probe, we were able to detect Shox2a expression in the sinus venosus region (Fig. 1F). It is noteworthy to mention that our analysis showed a very restricted Shox2 expression pattern, but could not confirm the presence of Shox2 in the primitive bundle branches of the cardiac conduction system, as reported by Blaschke et al. (2007).
Fig. 1.
Shox2 expression in the murine developing heart. (A) Shox2 expression is initially detected in the sinus venosus region (arrowhead) at E8.5 embryo. At E10.5 (B) and E11.5 (C), Shox2 expression gradually becomes restricted to the right side of the heart to the dorsal wall of the right atrium, the SAN region (arrow, C) and sinus valves (arrowheads, B, C). At E12.5 (D), restricted Shox2 expression in the SAN and sinus valves (arrowheads) is clearly observed. (E) Shox2 expression at E16.5 follows the same pattern as previous stages. (F) Whole mount in situ hybridization showing the expression of Shox2a isoform in the sinus venosus region (yellow arrowhead). A, atrium; V, ventricle; D, dorsal side.
Shox2-deficient mice die at mid-gestation and exhibit morphological abnormalities
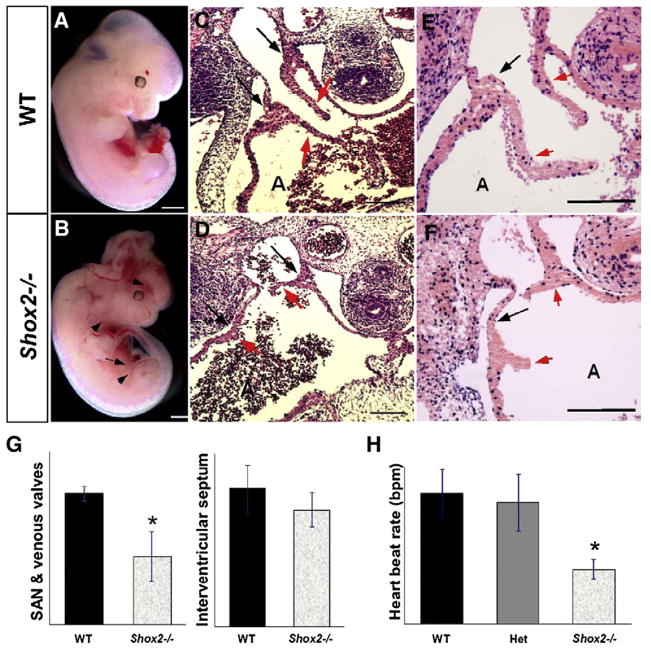
We have previously reported that on C57BL/6 background, 63% of Shox2 homozygous embryos died between E11.5 and E12.5, while few embryos survive up to E17.5 (Yu et al., 2005). Interestingly, we have found that on CD1 background, about 78% of the embryos die between E11.5 and E12.5 and no surviving embryos were found beyond E13.5. Embryonic mortality is attributable to cardiovascular defects evidenced by cardiac and vascular edema (Figs. 2A, B arrows). Histological analyses demonstrate that at E10.5 the Shox2−/− hearts from both genetic backgrounds show no obvious structural differences when compared to wild type embryos (WT). However, the mutant forming sinus valves show a slight difference (data not shown). At E11.5, the Shox2−/− hearts from both genetic backgrounds exhibit severe hypoplasia of the sinus valves, thinner atrial walls and enlargement of the right atrium in some specimens (Figs. 2C, D; and data not shown). In the most severe cases, the sinus valves were almost absent (Fig. 2D). In addition, the SAN region is also hypoplastic, as evidenced by its markedly reduced size (Figs. 2C, D, arrows). The cardiac defect observed in Shox2−/− mice on C57BL/6 background is relatively milder in comparison to the Shox2−/− hearts on CD-1 background (data not shown), which possibly accounts for the survival of some mutant embryos up to late gestation stages. For those Shox2−/− embryos on C57BL/6 background that survived to late developmental stages, severe cardiac abnormalities were observed, including defective myocardial wall and significantly enlarged right atrium (data not shown). For all subsequent experiments we chose to use Shox2−/− embryos on CD-1 background which exhibit more severe cardiac abnormalities.
Fig. 2.
Shox2 deficiency causes embryonic lethality and cardiac morphological and functional abnormalities. (A, B) Panoramic comparison between a wild type (A) and a Shox2−/− (B) embryos at E11.5. The mutant embryo exhibits cardiac edema (arrow) and vascular defects (arrowheads). Brain defects are observed in some specimens. (C, D) Histological comparison shows hypoplastic SAN and sinus valves in the Shox2−/− mutant heart (red arrowheads in panel D), as compared to the wild type controls (C), evidenced by the length of the sinus valves (red arrows in panels C and D) and the difference in size of the SAN region (black arrows in panel D). Additionally, a thinner atrial wall and an enlarged atrial chamber were observed in the Shox2−/− heart (D). (E, F) BrdU labeling shows a significantly decreased level of cell proliferation in the Shox2−/− SAN and sinus valves (F), as compared to the wild type (E). (G) Comparison of the percentage of BrdU-positive cells present in the SAN and sinus valves demonstrate a significant difference between wild type and Shox2−/− hearts (*: P<0.05); the interventricular septum region was chosen as a control region shows no significant difference. (H) Measurement of the contraction rate in isolated hearts at E10.5 after 24 h of culture shows a significantly slower beating rate in the Shox2−/− samples, as compared to the wild type (wt) and Shox2 heterozygote (het) samples (*: P<0.001). A, atrium; bpm=beats per minute. Scale bars represent 1 mm (A, B) or 50 μm (C–F).
Shox2−/− mice show reduced growth in the SAN region
Shox2 expression in the developing heart is restricted to the sinus venosus, SAN and sinus valves. Histological analyses demonstrate severe abnormalities in these structures of Shox2 mutants, consistent with Shox2 expression pattern. To identify the cellular defects that contribute to the formation of the hypoplastic SAN and sinus valves, we examined the levels of both cell proliferation and apoptosis in the heart of wild type and Shox2−/− embryos at E11.5 by BrdU labeling and TUNEL assays. As a control region, BrdU-positive cells were counted in the interventricular septum myocardium, which does not express Shox2 and does not present obvious structural differences when the control and Shox2−/− mutant hearts were compared. Although the level of apoptosis was not altered (data not shown), a dramatically reduced level of cell proliferation was observed in the mutant SAN and sinus valves, as compared to their wild type controls (Figs. 2E, F, arrows). Cell counting for BrdU-positive cells and total number of cells in the mentioned regions show that the percentage of proliferating cells is significantly different in the mutant hearts (9.9%±3.6%) when compared to the wild type controls (18.9%±1.0%). Similar analysis show no significant difference in the percentage of proliferating cells in the interventricular septum myocardium (wt=31.9%±5.5%; Shox2−/− =3.4%) (Fig. 2G).
Shox2−/− hearts present a slower heart beat rate
Morphological defects observed in the SAN region of the Shox2−/− hearts strongly suggest that the cardiac pacemaker is dysfunctional. We examined the heart beat rate, an index of the pacemaker function (Stieber et al., 2003) in isolated hearts individually cultured in vitro for 24 h. The heart beat rate of each sample was counted before the genotype was determined. Analysis on embryonic hearts of E9.5 embryos yielded no significant difference in heart beat rates observed in Shox2 mutants as compared to those of wild type and Shox2 heterozygotes (data not shown). However, analysis on embryonic hearts of E10.5 embryos revealed this defect in pacemaker function in the mutant. Although the cultured hearts contracted regularly, Shox2−/− hearts (n=8) were contracting slower than wild type (n=10) and heterozygote hearts (n=36) (Fig. 2H). The heart beat rate in the mutant (69.75±12.40 beats per minute—bpm) was less than 50% of that in the wild type (165±31.65 bpm) and heterozygote hearts (154±35.61 bpm), respectively. This cardiac abnormality observed in Shox2−/− hearts exhibit similar characteristics to human bradycardia cases (Watanabe et al., 2002; Milanesi et al., 2006) and to those found in the mouse Hcn4−/− model (Stieber et al., 2003).
Shox2 is a key regulator in pacemaker differentiation
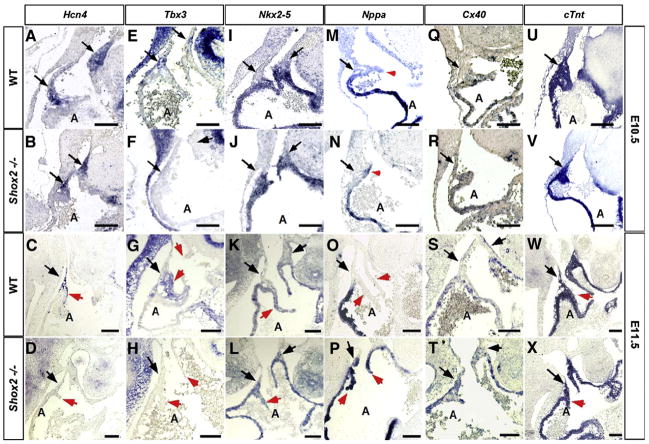
Genetically the SAN and pacemaker region in the developing embryo have been generally determined by a combination of the expression or lack thereof of several molecules to define its boundaries. Thus, to investigate the role of Shox2 in the development of the SAN, we examined the expression of several genes that have been used to identify the SAN or known to play a role in its formation and function (Soufan et al., 2004). These include Hcn4, a specific molecular marker for the pacemaker; Tbx3, a molecular marker for the cardiac conduction system including the SAN; the early cardiac differentiation marker Nkx2-5, expressed in the myocardium, but not in the SAN (Kasahara et al., 1998), Nppa (also known as atrial natriuretic factor—ANF), a chamber differentiation marker that is absent in the pacemaker; and Cx40 whose high expression represents areas of conductivity outside the SAN region. Our results reveal that expression of Hcn4 in the SAN, while remaining unchanged at E10.5 (Figs. 3A, B), was severely reduced at E11.5 in Shox2−/− hearts (Figs. 3C, D). Tbx3 was specifically down-regulated in the SAN region and the sinus valves in the Shox2−/− hearts (Figs. 3F, H), as compared to the wild type counterparts at both stages (Figs. 3E, G). Nkx2-5 was not expressed in the SAN of wild type embryos (Figs. 3I, K), but was ectopically expressed in the Shox2−/− SAN at E10.5 and E11.5 (Figs. 3J, L), in concordance to results obtained by Blaschke et al. (2007). Nppa and Cx40, the direct Tbx3 transcription targets, and whose expression patterns are mutually exclusive to that of Tbx3 (Rovescalli et al., 1996; van Kempen et al., 1996), were ectopically activated in the mutant SAN and sinus valves at low levels at E10.5 (Figs. 3N, R) and markedly expressed at E11.5 (Figs. 3P, T), as compared to the wild type controls (Figs. 3M, Q, O, S). In contrast, expression of the pan-marker for differentiated cardiomyocytes cardiac Troponin T (cTnt) (Wang et al., 1994; Jiao et al., 2003; Wang et al., 2001), was unaltered in the Shox2−/− hearts, as compared to the wild type samples (Figs. 3U–X). We also analyzed the expression of two other cardiac markers, MLC2a, a marker for differentiated atrial myocardium (Yutzey et al., 1994; Doevendans et al., 2000), and MLC2v, a marker for differentiated ventricular myocardium (O’Brien et al., 1993; Kubalak et al., 1994). These two genes showed no changes in expression in the mutants (data not shown). These observations demonstrate that cardiomyocyte differentiation is not affected in the absence of Shox2. The reduced expression of Hcn4 at E10.5 and the absence of Hcn4 expression at E11.5 in Shox2 mutants, along with the fact that Hcn4 is required for the regulation of a normal heart beat (Stieber et al., 2003) appear to account for the slow firing of the SAN which leads to an aberrant heart function, as evidenced by the bradycardia phenotype.
Fig. 3.
Altered gene expression in the Shox2−/− heart. Expression patterns of Hcn4 (A–D), Tbx3 (EH), Nkx2-5 (I–L), Nppa (M–P), Cx40 (Q–T) and cTnt (U–X) at E10.5 and at E11.5 in the wild type and Shox2−/− hearts are shown. Hcn4 is expressed in the dorsal right wall of the right atrium in the wild type (A, C) and is reduced or absent in the Shox2 mutant (B, D). Tbx3 is expressed in the dorsal right wall of the right atrium and in the sinus valves (E, G), however is absent in the Shox2 mutant (F, H) at both stages. Nppa, Cx40, and Nkx2-5 are ectopically expressed in the dorsal right wall of the Shox2−/− right atrium at both stages (J, N, R, L, P, T), as compared to the wild type controls (I, M, Q, K, O, S). cTnt shows comparable expression level in the Shox2−/− heart (V, X) and the wild type controls (U, W). In all panels, the arrows point to the SAN region, while the arrowheads point to the sinus valves. A, atrium. Scale bars represent 50 μm.
Histological, cellular and molecular results indicate failure in the differentiation process of pacemaker cells and lack of a defined SAN. Moreover, the ectopic Nkx2-5 expression and the down-regulated Tbx3 expression, along with the ectopic expression of Nppa and Cx40, suggest that the SAN in the mutant adopts an atrial working myocardial fate.
Shox2 acts upstream of Nkx2-5 to regulate SAN differentiation
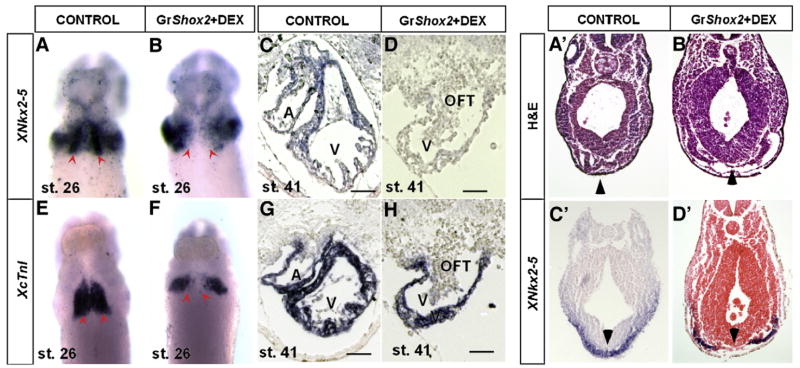
The early cardiac transcription factor Nkx2-5 is vital for the differentiation of primary heart field (PHF) cardiomyocytes (Komuro and Izumo, 1993; Tanaka et al., 1999; Moses et al., 2001) and the cardiac conduction system (Schott et al., 1998; Benson et al., 1999; Watanabe et al., 2002). Recent studies suggest that the absence of Nkx2-5 expression is necessary for the SAN formation (Mommersteeg et al., 2007; Blaschke et al., 2007). The fact that the lack of Shox2 causes an ectopic expression of Nkx2-5 in the SAN (Blaschke et al. 2007; and this study) prompted us to test if Shox2 functions to repress Nkx2-5. We took a gain-of-function approach by overexpressing Shox2 driven by the rat cTnT promoter in developing mouse embryos. This rat cTnT promoter has been shown to drive gene expression specifically in the developing heart as early as E7.5 (Wang et al., 1994, 2001; Jiao et al., 2003). Unexpectedly, of 65 total potential transient transgenic embryos that were collected at E8.5 and E9.5, only 6 were positive for the transgene but none of them exhibited expression of the transgene in the heart (data not shown), suggesting that the earlier transgenic expression of Shox2 might be lethal to the embryos. To overcome this obstacle, we carried out the over-expression experiment in Xenopus embryos using an inducible system. Frog embryos were collected and injected with mRNA synthesized from the GR-Shox2 construct. Nuclear translocation of the fusion protein was induced by addition of Dexamethasone (10 μM) into the embryo culture buffer. Induction was initiated at stage 14, and embryos were harvested at stage 27 (early cardiac formation) or stage 41 (late cardiac formation). At stage 27 injected but un-induced embryos resembled control ones, while injected induced embryos show morphological abnormalities including a reduced size of the head (data not shown). Whole mount in situ hybridization for XNkx2-5 in control embryos showed the laterally extended expression resembling wings connected at the ventral midline by two small masses which will form the heart tube (Fig. 4A). In contrast, XNkx2-5 expression in the injected induced embryos maintained the lateral extension; however, the expression at the midline was absent (Fig. 4B). Expression of cardiac troponin I (cTnI) in control embryos shows an overlapping expression pattern with that of XNkx2-5 at the midline indicating cardiac formation in this region (Fig. 4E). In the injected induced embryos, expression of cTnI at the midline was absent, but nevertheless it persisted in regions adjacent to the midline (Fig. 4F). Histological sections of Xenopus embryos clearly revealed that the loss of XNkx2-5 in the ventral midline section correlates to the loss of a population of cells which corresponds to mesodermal cardiac progenitors (Figs. 4A′–D′). This observation is consistent with previous results obtained with over-expression of XNkx2-5 dominant negative repressor derivatives in Xenopus embryos (Fu et al., 1998) and morpholino knockdown against nkx2.5 in zebrafish (Targoff et al., 2008). A summary of the number of normal and abnormal embryos collected at stage 26 after injection and in situ hybridization is shown in Table 1. Later in development, at stage 41, injected induced embryos exhibited a smaller tubular or slightly chambered heart as compared to control samples and very slow and irregular heart beat was observed at the moment of collection. XNkx2-5 expression was absent in these embryonic hearts, while cTnI expression was unaltered (Figs. 4C, D, G, H). These results resemble and are consistent with the phenotype of Nkx2-5 null mice that also present cardiac arrest after cardiac looping and poor cardiac development (Tanaka et al., 1999).
Fig. 4.
Shox2 overexpression downregulates Nkx2-5 expression. XNkx2-5 and XcTnI expression was analyzed by in situ hybridization in control embryos at stage 26 (A, E) and at stage 41 (C, G) and in GR-Shox2 mRNA injected induced embryos at same stages (B, F, D, H). XNkx2-5 expression is clearly reduced at the midline of injected induced embryos at stage 26 (B), as compared to control embryos (A). XcTnI expression is maintained in injected induced embryos laterally, but is absent at the midline at stage 26 (F), as compared to the controls (E). At stage 41, XNkx2-5 expression is absent in injected induced embryos (D) compared to control embryos (C). XcTnI expression in injected induced embryos (D) is comparable to that of control embryos (H). Altered gene expression is accompanied by the abnormal heart morphology when comparing control and injected induced embryos (C, D, G, H). Histological transverse sections of Xenopus embryos showing loss of mesodermal cardiac progenitor cells in the ventral midline in injected induced embryos (B′, D′, arrowheads), as evidenced by the loss of XNkx2-5 expression in the corresponding region when compared to control embryos (A′, C′, arrowheads). Scale bars represent 50 μm.
Table 1.
Summary of the number of normal and abnormal embryos collected at stage 26 after injection and in situ hybridization for XNkx2-5 and cTnI
| Normal expression | Abnormal expression | Total | % Normal | % Abnormal | |
|---|---|---|---|---|---|
| Control | 37 | 0 | 37 | 100 | 0 |
| Gr-Shox2 | 38 | 0 | 38 | 100 | 0 |
| Control DEX | 64 | 2 | 66 | 97 | 3 |
| Gr-Shox2 DEX | 36 | 45 | 81 | 44 | 56 |
Shox2 acts as a transcriptional repressor on the Nkx2-5 promoter
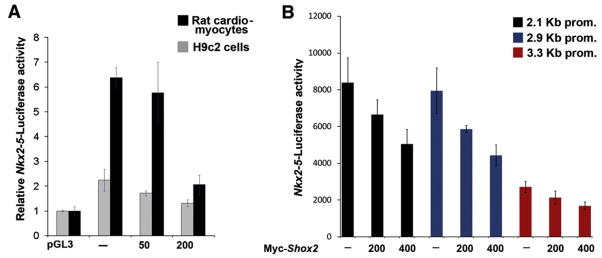
To determine if Shox2 has a direct or indirect effect on the expression of Nkx2-5, Luciferase reporter gene expression assays were carried out. A reporter construct harboring a 3.3-kb mouse Nkx2-5 upstream fragment linked to the luciferase reporter gene (Nkx2-5-Luc) was co-transfected with different concentrations of myc-Shox2 into neonatal rat cardiomyocytes or into H9c2 rat cardiogenic cells. H9c2 cells were induced to differentiate 24 h after transfection by exposure to DMEM culture medium supplemented with 1% horse serum. The induction of differentiation is necessary to induce the expression of the Nkx2-5 promoter (Lim et al., 2008). The reporter gene expression assays clearly demonstrated a concentration-dependent repression of the Nkx2-5 promoter activity by Shox2 in both types of cells (Fig. 5A). While the expression of reporter gene in H9c2 cells was markedly lower than that in rat cardiomyocytes, the results are consistent in both cell lines. Base on the criteria for homeobox genes binding sites, Shox2 might bind to a consensus “P” site, defined as a palindromic “ATTA(N)nTAAT” sequence in which “N” is any nucleotide and “n” is the number of nucleotides which separate the ATTA palindromic sequence (Rao et al., 2001), or to a consensus “TAATTA” site (Berger et al., 2008). Four putative Shox2 binding sites within the Nkx2-5 promoter were identified and labeled BS-I–IV. To determine if any of these sites mediates the repressive effect of Shox2 on the Nkx2-5 promoter, we eliminated these potential binding sites by site-directed mutagenesis, and by promoter deletion to engineer two shorter promoters, a 2.9 kb promoter which does not contain putative binding sites I–III and a 2.1 kb promoter which does not contain any putative binding site. Co-transfection of mutated or truncated promoters with different concentrations of Shox2 demonstrate that either mutant or truncated promoters responded in the same fashion as the 3.3-kb original promoter (Fig. 5B and data not shown). Promoter activities were down-regulated in a Shox2 concentration-dependent manner. Taken together, our results suggest that Shox2 indeed represses the Nkx2-5 promoter activity. However, Shox2 might not directly interact with the Nkx2-5 promoter.
Fig. 5.
Shox2 represses the activity of the Nkx2-5 promoter in cell cultures. (A) Neonatal rat cardiomyocytes and H9c2 cells were utilized. Co-transfection of with 100 ng of pGL3 basic vector as controls or Nkx2-5-Luc construct, with Myc-Shox2 expression vector at 50 ng, and 200 ng was performed. Reporter gene expression shows a progressive significant reduction in the activity of the Nkx2-5 promoter on both cell culture systems (* =P<0.001 at 200 ng). The Y axis represents activity of the Nkx2-5 promoter relative to the background after normalization. B) Reporter gene expression assay shows that the activity of the truncated Nkx2-5 promoters when co-transfected with myc-Shox2 (0, 200 and 400 ng/μl) follows a similar trend to the 3.3-kb promoter (*, significant difference, P<0.05).
Discussion
Histological analyses of the Shox2−/− heart revealed early cardiac structural malformation including hypoplastic SAN and sinus valves, defective atrial myocardial wall and at later developmental stages a significantly enlarged right atrium. These results are in concordance with results obtained by Blaschke et al. (2007). Since Shox2 expression in the heart is restricted to the sinus venosus, the SAN and the sinus valves, it indicates that the structural defects observed in other regions are most likely a secondary effect caused by the absence of a functional pacemaker and hypoplastic sinus valves. Malformation of the SAN and sinus valves is attributed to a significantly decreased level of cell proliferation, consistent with a role previously reported for Shox2 in the developing palate, limbs and the TMJ (Yu et al., 2005, 2007; Gu et al., 2008). The sinus valves are composed of two myocardial layers that have different physical characteristics (Gallego et al., 1997) with one being continuous with the atrial working myocardium and the other continuous with the venous dorsal wall of the atrium containing the pacemaker area. Reduced growth observed in both of the sinus valves layers and the SAN region of the Shox2−/− heart suggests that growth of the myocardial layer uses the venous layer as a template to elongate and “mature” before the recession and integration of the entire valve in the dorsal wall of the right atrium.
Reduction and possibly absence of a functional SAN leads to a reduced heart beat rate and embryonic lethality in the Shox2 mutant embryo. This phenotype resembles symptomatic sinus bradycardia related to sick sinus syndrome in humans which affects the capability of the heart to contract at a normal rate or below 55 beats per minute (Savalieva and Camm, 2008). Hcn1-4 molecules are responsible for the generation of the “funny” current which is characteristic of the pacemaker activity (Accili et al., 2002). Hcn4 expression in the SAN is the strongest and has been demonstrated to simulate pacemaker characteristics when overexpressed in cell cultures (Santoro and Tibbs, 1999; Santoro et al., 2000; Moosmang et al., 2001; Stieber et al., 2003; Liu et al., 2007). A mutation in the cAMP binding site of the human HCN4 gene was identified as the cause of sinus bradycardia and sick sinus syndrome in a familial case (Milanesi et al., 2006). Moreover, mice deficient in Hcn4 die between E9.5 and E11.5 presenting a similar electrophysiological phenotype with lower heart beat rate and slower pacemaker kinetics as compared to the wild type controls (Stieber et al., 2003). In this study we demonstrate that the expression of Hcn4 is down-regulated in the Shox2−/− heart beginning at E10.5. Taking this into consideration along with the fact that Hcn4 is required for the regulation of the normal intrinsic electric firing of the SAN, we conclude that the loss of Hcn4 is the cause for the slow firing of the SAN cells and the consequent slowing of the heart beat rate in Shox2−/−. This phenomenon is accompanied by the altered expression of several genes that have been used to demarcate the location of the SAN and pacemaker. Tbx3 is expressed in the cardiac conduction system including the SAN (Hoogaars et al., 2004). In contrast, the expression of Nkx2-5, Nppa and Cx40 is widely seen in the myocardium but not in the SAN region (Soufan et al., 2004; Liu et al., 2007). The absence of Hcn4 and Tbx3 expression and the ectopic expression of Nkx2-5, Nppa and Cx40 in the sinus node of the Shox2−/− heart indicate the lack of a definable SAN region. It has been reported that Tbx3 imposes a sinoatrial fate to myocardial cells (Hoogaars et al. 2007), however it was concluded that its function is to shield the SAN from becoming atrial working myocardium and preserve an embryonic phenotype. Additionally in Tbx3−/− hearts the formation of a primordial SAN is observed and expression of Hcn4 is maintained beyond the embryonic stage in which Shox2 mutation becomes lethal. These observations suggest that Shox2 might act earlier than Tbx3 in the formation of the SAN and the pacemaker. Alterations in gene expression pattern observed in the Shox2−/− heart are not likely attributed to a specific loss of potential SAN progenitor cells, since the mutant heart does not exhibit an increased level of apoptosis. However, it could be argued that a low level of cell proliferation might account for the lack of a definable SAN and the lack of expression of certain genes. Nevertheless, positive expression of Hcn4 in the Shox2−/− heart at E10.5 reveal that progenitor cells are indeed present and the genetic pattern of other genes is similar at E10.5 and at E11.5 when Hcn4 is absent. Our histological, functional and molecular results combined with observations detailed above indicate that cells within the SAN region of the Shox2−/− heart fail to differentiate into specialized pacemaker cardiomyocytes but rather adopt the fate of the atrial working myocardium which places Shox2 as a key and upstream intrinsic factor in a very specific and important niche in the molecular network that regulates the differentiation of SAN cells. It was proposed that Pitx2, which is expressed in the left side of the heart (Ryan et al., 1998; Yu et al., 2001), acts to determine the asymmetrical positioning of the sinus node to the right side, thereby preventing the left atrium to assume right side characteristics (Mommersteeg et al., 2007). Thus, expression of Pitx2 in the left side of the heart restricts Shox2 expression to the sinus node in the right atrium. This conclusion is supported by the observation that in Pitx2 mutants, an ectopic Shox2 expression domain is seen in the left atrium, mirror-imaging the original one in the sinus node in the right atrium and that Pitx2 expression in the Shox2−/− heart is unaltered (Espinoza-Lewis and Chen, unpublished observations). Together with previous studies that place Tbx3 and Hcn4 downstream of Nkx2-5 (Mommersteeg et al., 2007), we present a model integrating Pitx2, Shox2, Nkx2-5, Tbx3, and Hcn4 that operates to regulate the pacemaker development (Fig. 6). In this model, the expression of Pitx2 leads to the inhibition of the right side program in the left side, thus the absence of Pitx2 in the right side leads to the activation of Shox2 expression, which in turn acts to repress Nkx2-5. The repression of Nkx2-5 further allows for the activation of the differentiation program in the sinus node where Tbx3 further inhibits the expression of Nppa and Cx40.
Fig. 6.
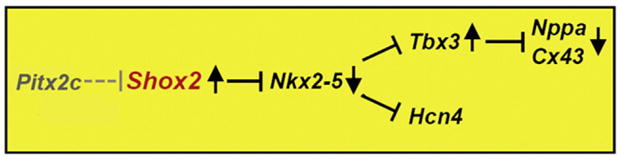
Shox2 regulates the SAN genetic program through repression of Nkx2-5. A genetic hierarchy model places Shox2 downstream of Pitx2 but upstream of Nkx2-5 in the regulation of SAN formation. Black arrows represent the up- or downregulation of expression depending on the expression or lack thereof of the upstream gene.
Early histological studies (van Mierop and Gessner, 1970; Challice and Virágh, 1973) and recent molecular analyses indicate that SAN cells are recruited into the venous pole of the heart, and that this population is Nkx2-5 negative and Tbx18 positive (Christoffels et al., 2006). Thus, the question pertaining to how these cells become Nkx2-5 negative arises. Here, gain-of-function studies in Xenopus embryos as well as in cell cultures reveal that Shox2 is able to repress the expression of Nkx2-5. In Xenopus embryos overexpression of Shox2 causes the down-regulation of XNkx2-5 in the ventral middle section, however the lateral expression remains. Later in development the resulting heart is small and tubular. These observations are consistent with several previous studies in which, 1) overexpression of dominant negative derivatives of XNkx2-5 results in the loss of mesodermal cardiac progenitor cells; 2) anti-nkx2.5 morpholino injection in zebrafish results in elongation abnormalities of the tubular heart, disorganized atrial cells and an enlarged ventricle; and 3) Nkx2-5 null mutation in mice leads to cardiac arrest after cardiac looping and the formation of an abnormal, small and tubular heart. Additionally, Luciferase reporter assays show that indeed Shox2 represses the activity of the Nkx2-5 promoter. Mutations at the putative Shox2 binding sites and truncations of the promoter to eliminate these binding sites have no effect in the repressive activity of Shox2 on the Nkx2-5 promoter. It is known that homeobox genes have transcriptional activity either by binding directly to the DNA or by binding to other proteins and acting as co-factors (del Bene and Wittbrodt, 2005). The Runx2 homeobox gene has been shown to regulate gene expression by direct binding to the Osteocalcin promoter (Ducy et al., 1997) or by acting as a co-factor to SRF (Serum Response Factor, a MADS box transcription factor) to prevent the Myocardin-induced myogenic genetic program and to promote the activation of the osteogenic genetic program (Tanaka et al., 2008). It is also likely that Shox2 interacts with other factors in order to repress the expression of Nkx2-5. This repressive activity of Shox2 on Nkx2-5 facilitates the expression of a genetic program to maintain the SAN fate and shield the SAN from becoming working atrial myocardium.
Our results demonstrate that Shox2 is essential for the proper formation and differentiation of the SAN, in addition to regulate a genetic network through the indirect repression of Nkx2-5 to maintain a SAN muscle fate. Additionally, this study provides one more insight for future genetic therapeutic approach against deficiencies in the SAN function in humans.
Acknowledgments
We thank Drs. Ken Muneoka, Frank Conlon, Eric Olson and Katherine Yutzey for providing constructs and probes for this study. We are also grateful to members in the Chen lab for their suggestions and discussion. This work was supported by an EIA grant from the AHA grant (0340166N) and NIH grant (R01 DE14044) to YPC, and an NIH grant (R01 DE 16329-01 to JFM).
References
- Accili EA, Proenza C, Baruscotti M, Di Francesco D. From funny current to HCN channels: 20 years of excitation. News Physiol Sci. 2002;17:32–37. doi: 10.1152/physiologyonline.2002.17.1.32. [DOI] [PubMed] [Google Scholar]
- Alappat S, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen YP. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Belin V, Cusin V, Viot G, Girlich D, Toutain A, Moncla A, Vekemans M, Le Merrer M, Munnich A, Cormier-Daire V. SHOX mutations in dyschondrosteosis (Leri–Weill syndrome) Nat Genet. 1998;19:67–69. doi: 10.1038/ng0198-67. [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Monaghan AP, Schiller S, Schechinger B, Rao E, Padilla-Nash H, Reid T, Rappold GA. SHOT, a Shox-related homeobox gene, is implicated in craniofacial, brain, heart, and limb development. Proc Natl Acad Sci USA. 1998;95:2406–2411. doi: 10.1073/pnas.95.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Schöler H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Challice CE, Virágh S. The architectural development of the early mammalian heart. Tissue Cell. 1973;6:447–462. doi: 10.1016/0040-8166(74)90037-8. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98:1555. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- Clement-Jones M, Schiller S, Rao E, Blaschke RJ, Zuniga A, Zeller R, Robson SC, Binder G, Glass I, Strachan T, Lindsay S, Rappold GA. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Gen. 2000;9:695–702. doi: 10.1093/hmg/9.5.695. [DOI] [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci USA. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Mjewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wittbrodt J. Cell cycle control by homeobox genes in development and disease. Sem Cell & Dev Biol. 2005;16:449–460. doi: 10.1016/j.semcdb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Doevendans PA, Bronsaer R, Lozano PR, Kubalak S, van Bilsen M. The murine atrial myosin light chain-2 gene: a member of an evolutionarily conserved family of contractile proteins. Cytogenet Cell Genet. 2000;90:248–252. doi: 10.1159/000056782. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Durham D, Worthley LI. Cardiac arrhythmias: diagnosis and management. The bradycardias Crit Care Resusc. 2002;4:54–60. [PubMed] [Google Scholar]
- Ellison JW, Wardak Z, Young MF, Gehron Robey P, Laig-Webster M, Chiong W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum Mol Genet. 1997;6:1341–1347. doi: 10.1093/hmg/6.8.1341. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- Gallego A, Durán AC, De Andrés AV, Navarro P, Muňoz-Chápuli R. Anatomy and development of the sinoatrial valves in the dogfish (Scyliorhinus canicula) Anat Rec. 1997;248:224–232. doi: 10.1002/(SICI)1097-0185(199706)248:2<224::AID-AR9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- García-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. GEP. 2003;3:777–783. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaussin V. Offbeat mice. Anat Rec. 2004;280A:1022–1026. doi: 10.1002/ar.a.20074. [DOI] [PubMed] [Google Scholar]
- Gu S, Wei N, Yu L, Fei J, Chen YP. Shox2-deficiency leads to dysplasia and ankylosis of the temporomandibular joint in mice. Mech Dev. 2008;125:729–742. doi: 10.1016/j.mod.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WMH, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiov Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Hoogaars WMH, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21 (9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BLM. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubalak SW, Miller-Hance WC, O’Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- Lim JY, Kim WH, Park SI. Induction of Id2 expression by cardiac transcription factors GATA4 and Nkx2.5. J Cell Biochem. 2008;103:182–194. doi: 10.1002/jcb.21396. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- Liu J, Dobrzynski H, Yanni J, Boyett MR, Lei M. Organization of the mouse sinoatrial node: structure and expression of HCN channels. Cardiov Res. 2007;73:729–738. doi: 10.1016/j.cardiores.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. New Eng J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- Mommersteeg MT, Hoogaars WMH, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur J Biochem. 2001;268:1646–1652. doi: 10.1046/j.1432-1327.2001.02036.x. [DOI] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis Daudin. North-Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- Nishii K, Shibata Y. Mode and determination of the initial contraction stage in the mouse embryo heart. Anat Embryol. 2006;21:95–100. doi: 10.1007/s00429-005-0065-x. [DOI] [PubMed] [Google Scholar]
- O’Brien TX, Lee KJ, Chien KR. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proc Natl Acad Sci USA. 1993;90:5157–5161. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao E, Blaschke RJ, Marchini A, Niesler B, Burnett M, Rappold GA. The Leri–Weill and Turner syndrome homeobox gene SHOX encodes a cell-type specific transcriptional activator. Hum Mol Gen. 2001;10:3083–3091. doi: 10.1093/hmg/10.26.3083. [DOI] [PubMed] [Google Scholar]
- Rovescalli AC, Asoh S, Nirenberg M. Cloning and characterization of four murine homeobox genes. Proc Natl Acad Sci USA. 1996;93:10691–10696. doi: 10.1073/pnas.93.20.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Peña J, Sabbagh W, Greenwald J, Choe S, Norris DP, Robertson EJ, Evans RM, Rosenfeld MG, Izpisúa-Belmonte JC. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Santoro B, Tibbs GR. The HCN gene family: molecular basis of the hyperpolarization-activated pacemaker channels. Annals NY Acad Sci. 1999;868:741–764. doi: 10.1111/j.1749-6632.1999.tb11353.x. [DOI] [PubMed] [Google Scholar]
- Santoro B, Chen S, Lüthi A, Pavlidis P, Shumyatsky GP, Tibbs GR, Siegelbaum SA. Molecular and functional heterogeneity of hyperpolarization-activated pacemaker channels in the mouse CNS. J Neurosci. 2000;20:5264–5275. doi: 10.1523/JNEUROSCI.20-14-05264.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savalieva I, Camm AJ. If inhibition with ivabradine: electrophysiological effects and safety. Drug Saf. 2008;31:95–107. doi: 10.2165/00002018-200831020-00001. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter RS, Murray J. A new human homeobox gene OG12X is a member of the most conserved homeobox gene family and is expressed during heart development in mouse. Hum Mol Gen. 1998;7:415–422. doi: 10.1093/hmg/7.3.415. [DOI] [PubMed] [Google Scholar]
- Shears DJ, Vassal HJ, Goodman FR, Palmer RW, Reardon W, Superti-Furga A, Scambler PJ, Winter RM. Mutation and deletion of the pseudoautosomal gene SHOX cause Leri–Weill dyschondrosteosis. Nat Genet. 1998;19:70–73. doi: 10.1038/ng0198-70. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. Early Development of Xenopus laevis. [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel Hcn4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci USA. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufan AT, van der Hoff MJB, Ruijter JM, de Boer PAJ, Hagoort J, Webb S, Anderson RH, Moormarn AFM. Reconstruction of the patterns of gene expression in the developing heart reveals an architectural arrangement that facilitates the understanding of atrial malformations and arrhythmias. Circ Res. 2004;95:1207–1215. doi: 10.1161/01.RES.0000150852.04747.e1. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Sato H, Doi H, Yoshida CA, Shimizu T, Matsui H, Yamazaki M, Akiyama H, Kawai-Kowase K, Iso T, Komori T, Arai M, Kurabayahi M. Runx2 represses myocardin-mediated differentiation and facilitates osteogenic conversion of vascular smooth muscle cells. Mol Cell Biol. 2008;28 (3):1147–1160. doi: 10.1128/MCB.01771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–321. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kempen MJA, Vermeulen JL, Moorman AF, Gros D, Paul DL, Lamers WH. Developmental changes of connexin40 and connexin43 mRNA distribution patterns in the rat heart. Cardiov Res. 1996;32:886–900. doi: 10.1016/0008-6363(96)00131-9. [DOI] [PubMed] [Google Scholar]
- van Mierop LH. Location of pacemaker in chick embryo heart at the time of initiation of heart beat. Am J Phisyol. 1967;212:407–415. doi: 10.1152/ajplegacy.1967.212.2.407. [DOI] [PubMed] [Google Scholar]
- van Mierop LHS, Gessner IH. The morphologic development of the sinoatrial node in the mouse. Am J Cardiol. 1970;25:204–226. doi: 10.1016/0002-9149(70)90580-1. [DOI] [PubMed] [Google Scholar]
- van Schaick HSA, Smidt MP, Rovescalli AC, Luijten M, van der Kleij AA, Asoh S, Kozak CA, Nirenberg M, Burbach JP. Homeobox gene Prx3 expression in rodent brain and extraneural tissues. Proc Natl Acad Sci USA. 1997;94:12993–12998. doi: 10.1073/pnas.94.24.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ye HI, Lin JJC. Characterization of cis-regulating elements and trans-activating factors of the rat cardiac troponin T gene. J Biol Chem. 1994;269:30595–30603. [PubMed] [Google Scholar]
- Wang Q, Reiter RS, Huang QQ, Jin JP, Lin JJ. Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat Rec. 2001;263:72–84. doi: 10.1002/ar.1078. [DOI] [PubMed] [Google Scholar]
- Wang Y, Morishima M, Zheng M, Uchino T, Mannen K, Takahashi A, Nakaya Y, Komuro I, Ono K. Transcription factors Csx/Nkx2.5 and GATA4 distinctly regulate expression of Ca2+ channels in neonatal rat heart. J Mol Cell Card. 2007;42:1045–1053. doi: 10.1016/j.yjmcc.2007.03.905. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Benson DW, Yano S, Akagi T, Yoshino M, Murray JC. Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet. 2002;39:807–811. doi: 10.1136/jmg.39.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, St Amand TR, Wang S, Li G, Zhang Y, Hu YP, Nguyen L, Qiu MS, Chen YP. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Development. 2001;128:1005–1013. doi: 10.1242/dev.128.6.1005. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen YP. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu H, Yan M, Yang J, Long F, Muneoka K, Chen YP. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev Biol. 2007;306 (2):549–559. doi: 10.1016/j.ydbio.2007.03.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Rhee JT, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Wei F, Zhang L, Elder FF, Scott CI, Jr, Marttila P, Ross JL. Complete SHOX deficiency causes Langer mesomelic dysplasia. Am J Med Genet. 2002;110:158–163. doi: 10.1002/ajmg.10422. [DOI] [PubMed] [Google Scholar]