Abstract
The p53 tumor suppressor gene can inhibit proliferation transiently, induce permanent cell-cycle arrest/senescence, or cause apoptosis depending on the cellular context. The mitogen-activated protein kinase (MAPK) cascade is known to play a crucial role in cell proliferation and differentiation. Moreover, the duration and intensity of MAPK activation can profoundly influence the biological response observed. We demonstrated that a sustained activation of MAPK cascade could be induced by wild-type p53 expression but not by p21Waf1/Cip1. Furthermore, exposure of normal cells to DNA-damaging agents induced MAPK activation in a p53-dependent manner. Tumor-derived p53 mutants defective in DNA binding failed to activate MAPK, implying that p53 transcriptional activity is essential for this function. Finally, activation of MAPK by p53 was inhibited by expression of dominant-negative Ras (N17Ras) and Raf1 mutants, indicating that MAPK activation by p53 is mediated at a level upstream of Ras. All of these findings establish a biochemical link between p53 signaling and the Ras/Raf/MAPK cascade.
Mitogen-actived protein kinases (MAPK; extracellular signal-regulated kinase or ERK) are involved in proliferative signaling triggered by tyrosine kinases, cytokines, and serpentine receptors, which generally are coupled through Ras activation to this pathway (1–4). MAPKs function in many cell types, are regulated by a diverse group of extracellular stimuli, and mediate a variety of cellular responses (5–7). There is accumulating evidence suggesting that the duration and intensity of MAPK activation can profoundly influence the biological response observed (1–6). For example, PC12 cells undergo proliferation after transient MAPK activation in response to epidermal growth factor (EGF), but these cells undergo growth arrest after sustained MAPK activation in response to nerve growth factor treatment (3, 7, 8). Whereas low levels of activated Raf, a downstream effector of Ras, cause cell-cycle progression, high levels cause cell-cycle arrest and p21Waf1/Cip1 induction in a p53-independent manner (9–12). In addition, recent findings show that activated Ras or sustained activation of MAPK accelerates the onset of senescence in some cells (13) and induces growth arrest in others (11, 12, 14).
There is evidence for cross talk between the proliferation/differentiation pathways activated by Ras/Raf/MAPK and growth arrest functions of tumor suppressor genes including p53, p16, and Rb (15, 16). Thus, sustained Ras or Raf signaling has been reported to activate p53 and/or p21 as well as p16 expression leading to growth arrest (17, 18). Given the similarities in the cellular response to MAPK activation and p53 induction, we sought evidence for activation of the MAPK cascade in response to p53 expression. We show that tumor suppressor p53 induction activates the MAPK cascade, that this function depends on p53 transcriptional activity, and that MAPK activation by p53 is a consequence of Ras and Raf activation.
Materials and Methods
Cell Culture.
EJ-p53, EJ-CAT, PC3-p53, PC3-p21, and PC3-Luciferase cells were cultured in the presence or absence of tetracycline (tet; 1 μg/ml) in DMEM containing 10% (vol/vol) FBS as described (19). MEK inhibitor (PD98059, New England Biolabs) was dissolved in DMSO, and cells were treated in 20 μM of the concentration as indicated. A rat embryo fibroblast (REF) line containing temperature-sensitive p53 mutant (p53Val135) was cultured as described (20); 293T cells were maintained in DMEM plus 10% (vol/vol) FBS. P53+/+ and p53−/− mouse embryo cells were maintained as described (21).
Plasmids and Cell Transfections.
Four p53 mutants (V143A, R175H, R248H, and P281G) affecting transcriptional activation of p53 as well as wild-type (wt) p53 and p21 were used for transfection studies (16, 22). Dominant-negative (DN) mutants of Ras (H-RasN17) (23) and Raf (K375 M) (24) were subcloned into pcDNA3 and transfected into 293T cells. tet-regulatable p53 was made, transfected, and selected in EJ or PC3 human tumor cells as described (19). The tet-regulatable chloramphenicol acetyltransferase (CAT) gene for EJ cells and the luciferase gene for PC3 were also generated as negative control cell lines, and 293T cells (≈50% confluent) were transfected by the calcium phosphate method as described (25).
Immunoblot Analysis.
Cells were lysed in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). After protein concentrations were determined, 100 μg of proteins were resolved on SDS/PAGE gels and transferred to nitrocellulose membranes. Membranes were probed with the appropriate antibodies, and immunoreactive protein complexes were detected by enhanced chemiluminescence (Amersham Pharmacia). Phospho-specific or regular antibodies against ERK, MEK, and p38 for Western blot analysis were purchased from New England Biolabs or Promega. Anti-ERK1/2 antibodies for immunoprecipitation experiments were from Santa Cruz Biotechnology or Transduction Laboratories (Lexington, KY). P21 (AB1, AB5) and p53 (1801) antibodies were obtained from Oncogene Science.
Treatment with DNA-Damaging Agents.
Cells were grown to 50% confluence before the exposure to DNA damage. Low-passage human normal diploid fibroblast (NDF; 501T) cells were irradiated with UV light (30 J/m2) or mitomycin C (MMC, 10 μg/ml), and cell lysates were prepared at the indicated times and immunoblotted with p53, phosphorylated ERK (pp-ERK), and total ERK antibodies. Primary mouse embryonic fibroblasts (MEFs) derived from wt or p53-null mice embryos were exposed to DNA-damaging agents (10 μg/ml MMC and 5 ng/ml actinomycin D) for the indicated times.
MAPK Activity.
Whole-cell lysates were prepared in RIPA buffer. Whole-cell lysates (100 μg) were incubated with anti-ERK overnight at 4°C. The immune complexes were immunoprecipitated with protein A Sepharose beads for 1 h, and the immune complexes bound to the beads were washed with the same lysis buffer. Kinase assays were performed with myelin basic protein (Sigma). The immunoprecipitates and substrates were incubated in a volume of 40 μl containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 0.125 μCi/μl [γ-32P]ATP at 30°C for 20 min. The reaction was stopped by the addition of 40 μl of 2× Laemmli buffer, and the mixture was boiled for 3 min and resolved by SDS/12.5% PAGE. Phosphorylated proteins were visualized by autoradiograph.
Results
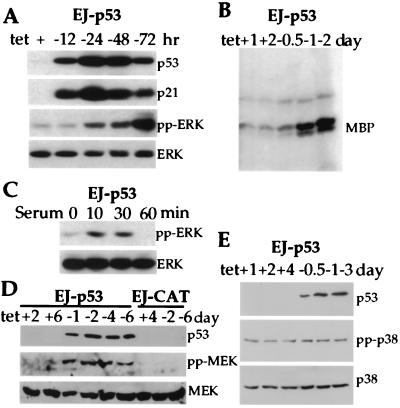
In an effort to investigate signaling pathways activated by p53, we used a system for tet-regulatable expression of wt p53 in the human bladder carcinoma cell line EJ, containing p53 that was nonfunctional because of a mutation in exon 5 (19). As shown in Fig. 1A, as early as 12 h after tet removal from the culture medium, p53 was readily detectable, and p53 expression achieved peak levels at 24 h. A strong induction of the p21 protein in response to p53 was observed as well (Fig. 1A). By using antibodies that specifically recognize the phosphorylated, active forms of MAPKs (ERK1 and ERK2), MEK/MAPK kinase, and p38/HOG (3, 4), we were able to monitor the status of these kinases after p53 induction. Of note, pp-ERK, which was barely detectable in EJ-p53 cells without p53 expression (with tet), increased markedly in response to p53 induction (without tet). Levels of pp-ERK continued to increase over the entire 72-h time course of the experiment (Fig. 1A). That pp-ERK was enzymatically activated was confirmed by its ability to phosphorylate the substrate myelin basic protein (Fig. 1B). These findings were even more remarkable when compared with a more modest and transient increase in pp-ERK after serum starvation. Under this condition, EJ-p53 cells were starved in serum-free medium (DMEM + 0.1% BSA) for 24 h and stimulated with serum for the indicated time periods in the presence of tet (no p53). As shown in Fig. 1C, pp-ERK peaked at 10 min and disappeared after 60 min of serum stimulation in the absence of p53 induction, whereas p53-induced ERK activation was higher and was sustained over the 72-h time course of the experiments.
Figure 1.
Activation of MAPK/MEK pathway by inducible expression of p53. (A) Increased phosphorylation of ERK (pp-ERK) in response to wt p53 induction. EJ-p53 cell lysates were analyzed by immunoblot analysis for the induction of p53 protein with (+) or without (−) tet. The expression of p21, pp-ERK, and total ERK proteins was also monitored. (B) Enzymatic activity of ERK in EJ-p53 cells with (+) or without (−) tet. Whole-cell lysates were immunoprecipitated with anti-ERK antibody, and kinase activity was measured with myelin basic protein (MBP) as substrate. (C) Phosphorylation of ERK in response to serum stimulation. Starved EJ-p53 cells were stimulated with serum in the presence of tet. (D) Activation of MEK1 after p53 induction. Lysates from EJ-p53 cells grown with (+) or without (−) tet for different time periods were immunoblotted with anti-p53, anti-phospho-specific MEK1, and anti-MEK1 antibodies. (E) No change in active p38 and total p38 MAPK after p53 induction. Lysates from EJ-p53 cells grown with (+) tet or without (−) tet for different time periods were immunoblotted with anti-p53, anti-phospho-specific p38, and anti-p38 antibodies.
We next investigated whether the immediate upstream regulator of MAPK/ERK, MEK/MAPK kinase, was also activated in response to p53 induction by using a phospho-specific MEK antibody. Fig. 1D shows that the phosphorylated form of MEK was induced in a sustained manner over the entire 6-day experiment. Total MEK protein expression remained constant. We also analyzed the response of p38 MAPK to p53 induction in EJ-p53 cells. The p38 MAPK pathway as well as the MEK and ERK pathways are MAPK signaling cascades, but they involve different effectors (1–3). There was no change in phosphorylated p38 in p53-induced or -uninduced cells (Fig. 1E). These results imply that p53-induced activation of ERK is mediated by MEK and that activation of both ERK and MEK is sustained. Moreover, the results indicate that there is specificity in this response to p53 induction, because not all of the MAPK cascades are affected.
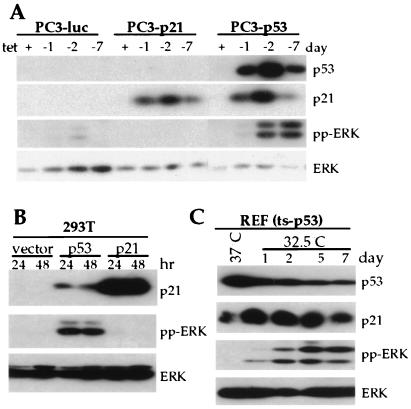
To investigate whether p53-mediated MAPK activation was a general phenomenon, another tet-regulated p53-inducible system in PC3 cells, a p53-null human prostate cancer cell line, was generated. wt p53 expression by removal of tet resulted in an increase of pp-ERK in PC3-p53 cells (Fig. 2A). Of note, p21 overexpression with tet-regulatable p21 in the same cells had no effect on the level of activated ERK activation (Fig. 2A). In addition, human 293T cells (26) were transfected with wt p53 (pCMVBam-p53), p21 (pCEP-p21), or vector alone (pCMVBam or pCEP). Transfection of p53 into 293T cells led to a dramatic increase in p53 above the endogenous levels (data not shown). As shown in Fig. 2B, ectopic p53 function was confirmed by the induction of p21. Immunoblots of the same cell lysates probed with antibody against pp-ERK showed a sustained induction of the activated forms of ERK, which was not observed with ectopic overexpression of p21 (Fig. 2B). The p53-induced response of ERK was corroborated further with a REF line (REF temperature-sensitive p53) containing a temperature-sensitive p53 mutant (p53Val135). The p53Val135 behaves as authentic wt p53 at 32.5°C and as mutant at 37°C. As shown in Fig. 2C, p21 induction occurred at 32.5°C along with ERK activation, but little p21 and no pp-ERK were observed at 37°C. Altogether, these results demonstrate that p53-mediated ERK activation is a generalized phenomenon and that expression of p21 alone was not responsible for ERK activation.
Figure 2.
Activation of ERK by p53 in various cell types. (A) Activation of ERK by inducible expression of p53 but not by p21 in PC3 cells. Western blots were performed with the lysates from PC3 cells with the tet-regulated expression of p53, p21, or luciferase. (B) Activation of ERK by p53 but not by p21 in 293T cells. Cells were transiently transfected with pcDNA3, p53, or p21. Lysates were made from cells after 24 and 48 h of transfection and were separated by SDS/PAGE. Immunoblotting was carried out with antibodies against p53, p21, pp-ERK, and total ERK. (C) Activation of ERK by temperature-sensitive p53 mutants (REF ts-p53). REFs were grown at 37°C and shifted to 32.5°C for the indicated time periods (1–7 days). Western blots were performed with p53, p21, pp-ERK, and total ERK antibodies.
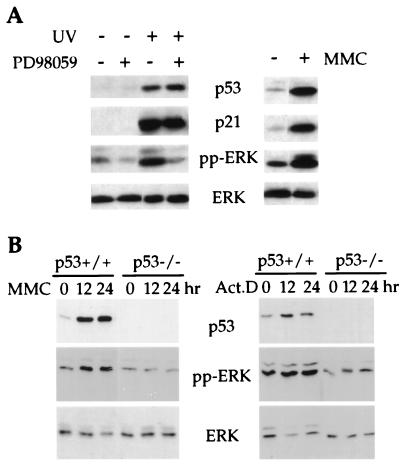
It is now well established that p53 functions to integrate cellular responses to stress such as DNA damage (27, 28). Therefore, we analyzed whether ERK was activated in human NDFs in response to DNA damage. As shown in Fig. 3A, the activated form of ERK was increased as was p53 in 501T NDFs exposed to UV irradiation or MMC. The p53 dependence of this response was established by using wt and p53-null MEFs. As shown in Fig. 3B, MMC induced a marked increase in pp-ERK in wt MEFs but not in p53−/− MEFs. pp-ERK was also increased in a p53-dependent manner after actinomycin D treatment when normalized to total ERK levels (Fig. 3B). These results demonstrate that functional p53 is required for ERK activation in response to cellular stress induced by DNA-damaging agents.
Figure 3.
p53-dependent ERK activation in response to DNA damage. (A Left) Activation of ERK and inhibition by MEK-specific inhibitor PD98059 in 501T cells in response to UV irradiation. Human NDFs (501T) were treated as described in Materials and Methods in the presence or absence PD98059 (20 μM) for 24 h. As a control, cells were treated with the same amounts of DMSO, the solvent for the inhibitor. Cells were harvested for analysis of protein expression, and Western blots were carried out with antibodies against p53, p21, pp-ERK, or total ERK. (A Right) Activation of ERK in 501T cells in response to MMC treatment. 501T cells were treated with 10 μg/ml MMC for 36 h. Lysates were made, and Western blots were performed as described above. (B) Activation of ERK in primary MEFs derived from wt and p53-null mice embryos. Cell lysates were prepared after treatment with DNA-damaging agents [10 μg/ml MMC or 5 ng/ml actinomycin D (Act. D)] for the indicated time periods. Western blots were performed with antibodies against p53, pp-ERK, and total ERK, respectively.
To assess MEK involvement in this response, we tested the effect of a specific MEK inhibitor, PD98059 (15), on UV-induced ERK activation in 501T NDFs. As shown in Fig. 3A, treatment of 501T cells with PD98059 after UV exposure had no effect on the characteristic increase in p53 and the associated induction of p21. However, treatment with this MEK inhibitor resulted in a marked reduction in the magnitude of ERK activation as assessed by the phospho-specific ERK antibody (Fig. 3A). PD98059 did not alleviate the growth arrest phenotype detectably (data not shown). This result may reflect the fact that the growth arrest phenotype can be induced independently by p21 (29–31).
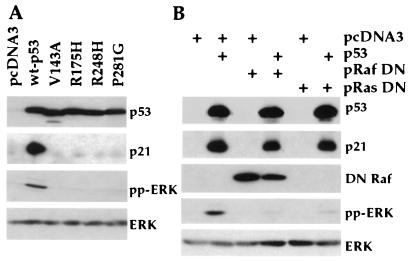
In an effort to determine whether p53-mediated MAPK signaling requires transcriptionally active p53 as assessed by p21 induction, tumor-derived p53 mutants affecting p53 transcriptional activity and containing hot spot mutations in the DNA binding domain (16, 22) were transfected into 293T cells and compared with wt p53 and vector alone (pcDNA3). All four mutants (V143A, R175H, R248H, and P281G) (22, 27) exhibited loss of both p21 induction and MAPK activation functions (Fig. 4A), implying that p53 transcriptional activity is required for MAPK activation.
Figure 4.
(A) Activation of ERK requires transcriptionally active p53. Four p53 mutants (V143A, R175H, R248H, and P281G) affecting transcriptional activation of p53 as well as wt p53 and control vector were individually transfected into 293T cells. At 48 h after transfection, cells were lysed and subjected to Western blot analysis with anti-p53, anti-p21, anti-pp-ERK, or anti-ERK antibodies. (B) Inhibition of p53-induced ERK activation by DN Raf and DN Ras. wt p53 was transfected into 293T cells with vector, DN Raf, or DN Ras as shown on top of each lane. Western blot analysis was performed with antibodies against p53, p21, flag tag (DN Raf), pp-ERK, or total ERK.
Finally, we sought to identify upstream effectors involved in activation of ERK signaling cascade by p53 through the use of DN mutants of Ras and Raf, whose products block the functions of the MAPKs within the cascade (23, 24). By transient cotransfection of 293T cells with p53 and either a DN mutant form of Ras (N17Ras) or Raf1 (DN Raf Flag), a direct downstream effector of Ras, activation of ERK by p53 was abolished (Fig. 4B). These results indicate that p53-mediated ERK activation requires functional Ras and Raf.
Discussion
Our present studies demonstrate that p53 induces sustained MAPK (ERK) activation, establishing a p53 signaling response. We observed this response initially by means of a tet-regulatable p53 system in EJ bladder carcinoma cells, which contain a ras oncogene and lack functional p53. However, this response was also observed in other continuous cell lines (PC3 and 293T cells) that lack ras oncogenes. We demonstrated further that MAPK activation depended on p53 transcriptional function, because tumor-derived, transcriptionally inactivated p53 mutants lacked this activity. In human NDFs, MAPK activation occurred as a physiologic response to DNA-damaging agents, and we showed its p53 dependence by using p53−/− MEFs. Thus, our evidence implies that this p53 signaling response must have a physiological role.
Activation of the MAPK signaling cascade by mitogenic stimulation is well characterized (1–4). However, recent studies have shown that overexpression of Ras or Raf, which causes activation of MAPK, can induce p53 and/or p21-mediated growth arrest (5, 6, 9, 10, 32, 33) as well as acceleration of permanent growth arrest/senescence (17, 18). Other reports have indicated that induction of p21 by Ras may be regulated through Rho signaling (34). Sustained activation of MAPK also causes arrest of nuclei in G1/S or mitosis in Xenopus oocyte extracts in the absence of active cdc2–cyclin B (35). Thus, if MAPK induces p53 (17, 18), our present studies demonstrating the reciprocal interconnection of these signaling pathways may imply a positive feedback loop in which permanent growth arrest could be augmented by sufficient up-regulation of either p53 or MAPK pathways.
Our efforts to identify the mechanisms responsible for MAPK activation by p53 led to evidence through use of DN Ras and Raf mutants that the signal initiates upstream of Ras in the MAPK cascade. Ras is known to be activated by a variety of signals, including growth factors acting through receptor tyrosine kinases (5–7). Differential screening of gene expression changes induced by p53 in tet-regulatable EJ-p53 cells has led to our detection of increased expression of certain growth factors (unpublished results). One such p53-inducible growth factor, heparin-binding EGF-like growth factor (HB-EGF), is induced more than 7-fold in response to p53. MB-EGF is known to trigger the EGF receptor, which acts through Ras to activate MAPK (36–38). Thus, it is possible that p53-induced growth factors such as MB-EGF may be involved in a cellular compensatory mechanism to alleviate adverse effects of DNA-damaging agents and other cellular stresses. For example, in the context of the whole organism, p53 induction of autocrine- or paracrine-acting growth factors might play a role in the renewal of damaged tissues.
In summary, our findings establish that the p53 tumor suppressor gene activates MAPK signaling through transcriptional activation of target genes and that the MAPK cascade has the potential to integrate the actions of diverse signals to dictate p53-induced cellular responses. A key to further understanding this p53 response may involve further characterization of those factors upstream of Ras involved in activation of the MAPK signaling cascade and assessing whether such factors contribute to p53-mediated tumor suppression or cell survival.
Acknowledgments
We thank B. Vogelstein for p53 and p21 cDNA, A. Levine for p53 mutants, A. Bradley for p53+/+ and p53−/− cells, M. Oren for REF temperature-sensitive p53 cells, J. Leung and L. Miller for technical support, and P. Arizti and C. Reimer for critical comments. K.P.L. is a Leukemia Society of America Scholar. L.F. is a fellow of the Forchheimer Foundation. This work was supported by grants from the National Institutes of Health–National Cancer Institute (to S.W.L., T.O., and S.A.A), a grant from the National Institutes of Health–General Medical Sciences (to K.P.L.), a research enhancement award from Mount Sinai School of Medicine (to T.O.), and a grant from T. J. Martell Foundation for Leukemia, Cancer, and AIDS Research (to S.A.A.).
Abbreviations
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
- EGF
epidermal growth factor
- wt
wild type
- MMC
mitomycin C
- MEF
mouse embryonic fibroblast
- REF
rat embryo fibroblast
- pp-ERK
phosphorylated ERK
- tet
tetracycline
- NDF
normal diploid fibroblast
- DN
dominant-negative
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150024397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150024397
References
- 1.Davis R J. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 2.Cobb M H, Goldsmith E J. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 3.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 4.Robinson M J, Cobb M H. Curr Opin Cell Biol. 1997;2:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 5.Marshall C J, Nigg E A. Curr Opin Genet Dev. 1998;8:11–13. doi: 10.1016/s0959-437x(98)80055-5. [DOI] [PubMed] [Google Scholar]
- 6.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 7.Cowley S, Paterson H, Kemp P, Marshall C J. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 8.Pang L, Sawada T, Decker S J, Saltiel A R. J Biol Chem. 1995;270:13585–13588. doi: 10.1074/jbc.270.23.13585. [DOI] [PubMed] [Google Scholar]
- 9.Kyriakis J M, App H, Zhang X F, Banerjee P, Brautigan D L, Rapp U R, Avruch J. Nature (London) 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 10.Marshall C J. Nature (London) 1996;383:127–128. doi: 10.1038/383127a0. [DOI] [PubMed] [Google Scholar]
- 11.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sewing A, Wiseman B, Lloyd A C, Land H. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd A C, Obermuller F, Staddon S, Barth C F, McMahon M, Land H. Genes Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 15.Aless D R, Cruenda A, Cohen P, Dudley D, Safiel A. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 16.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 17.Lin A W, Barradas M, Stone J C, van Aelst L, Serrano M, Lowe S. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Woods D, McMahon M, Bishop J M. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugrue M M, Shin D Y, Lee S W, Aaronson S A. Proc Natl Acad Sci USA. 1997;94:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michalovitz D, Halevy O, Oren M. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 21.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 22.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 23.Lange-Carter C A, Johnson G L. Science. 1994;265:1458–1461. doi: 10.1126/science.8073291. [DOI] [PubMed] [Google Scholar]
- 24.Dent P, Reardon D B, Morrison D K, Sturgill T W. Mol Cell Biol. 1995;15:4125–4135. doi: 10.1128/mcb.15.8.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang L, Lee S W, Aaronson S A. J Cell Biol. 1999;147:823–830. doi: 10.1083/jcb.147.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altun-Gultekin Z F, Chandriani S, Bougeret C, Ishizaki T, Narumiya S, de Graaf P, Van Bergen en Henegouwen P, Hanafusa H, Wagner J A, Birge R B. Mol Cell Biol. 1998;18:3044–3058. doi: 10.1128/mcb.18.5.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 28.Ko L J, Prives C. Gene Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 29.Fang L, Igarashi M, Leung J, Sugrue M M, Lee S W, Aaronson S A. Oncogene. 1999;18:2789–2797. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- 30.Sekiguchi T, Hunter T. Oncogene. 1997;16:369–379. doi: 10.1038/sj.onc.1201539. [DOI] [PubMed] [Google Scholar]
- 31.Erhardt J A, Pittman R N. Oncogene. 1998;16:443–451. doi: 10.1038/sj.onc.1201577. [DOI] [PubMed] [Google Scholar]
- 32.Zou X, Rudchenko S, Wong K, Calame K. Genes Dev. 1997;11:654–662. doi: 10.1101/gad.11.5.654. [DOI] [PubMed] [Google Scholar]
- 33.Pumiglia K M, Decker S J. Proc Natl Acad Sci USA. 1997;94:448–452. doi: 10.1073/pnas.94.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson M F, Paterson H F, Marshall C J. Nature (London) 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 35.Guadagno T M, Ferrell J E. Science. 1998;282:1312–1315. doi: 10.1126/science.282.5392.1312. [DOI] [PubMed] [Google Scholar]
- 36.Higashiyama S, Abraham J A, Miller J, Fiddes J C, Klagsbrun M. Science. 1991;251:936–939. doi: 10.1126/science.1840698. [DOI] [PubMed] [Google Scholar]
- 37.McCarty S A, Samuels M L, Pritchard C A, Abraham J A, McMahon M. Genes Dev. 1995;9:1953–1964. doi: 10.1101/gad.9.16.1953. [DOI] [PubMed] [Google Scholar]
- 38.Raab G, Klagsbrun M. Biochim Biophys Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]