Abstract
BACKGROUND: Ewing sarcoma is a paradigm of solid tumour -bearing chromosomal translocations resulting in fusion proteins that act as deregulated transcription factors. Ewing sarcoma translocations fuse the EWS gene with an ETS transcription factor, mainly FLI1. Most of the EWS–FLI1 target genes still remain unknown and many have been identified in heterologous model systems.
METHODS: We have developed a stable RNA interference model knocking down EWS–FLI1 in the Ewing sarcoma cell line TC71. Gene expression analyses were performed to study the effect of RNA interference on the genetic signature of EWS–FLI1 and to identify genes that could contribute to tumourigenesis.
RESULTS: EWS–FLI1 inhibition induced apoptosis, reduced cell migratory and tumourigenic capacities, and caused reduction in tumour growth. IGF-1 was downregulated and the IGF-1/IGF-1R signalling pathway was impaired. PBK/TOPK (T-LAK cell-originated protein kinase) expression was decreased because of EWS–FLI1 inhibition. We showed that TOPK is a new target gene of EWS–FLI1. TOPK inhibition prompted a decrease in the proliferation rate and a dramatic change in the cell's ability to grow in coalescence.
CONCLUSION: This is the first report of TOPK activity in Ewing sarcoma and suggests a significant role of this MAPKK-like protein kinase in the Ewing sarcoma biology.
Keywords: Ewing sarcoma, EWS–FLI1, stable shRNAi model, IGF-1/IGF-1R, TOPK
Ewing sarcoma is an aggressive neoplasm of the bone and soft tissues of children and young adults. Ewing sarcoma is characterised by the presence of chimeric proteins joining the NH2-terminal domain of the RNA-binding protein EWS with the DNA-binding portion of an ETS transcription factor, FLI1, in 90% of the cases (Delattre et al, 1992; Sorensen et al, 1994; May et al, 1997). There is strong evidence for the oncogenic potential of EWS–ETS fusions (May et al, 1997). EWS–ETS chimeric proteins act as deregulated transcription factors, both activating and repressing the expression of target proteins (Prieur et al, 2004). Several of these have already been identified, including transcription, apoptotic, or signal transduction factors (Dauphinot et al, 2001; Amsellem et al, 2005; Mendiola et al, 2006). The proliferation and survival of Ewing sarcoma is also determined by autocrine and paracrine activation of growth factor receptors and their ligands, such as IGF-1 (Scotlandi et al, 1998).
The cellular context contributes to the phenotype because the introduction of EWS–ETS fusions into different cellular models resulted in diverse outcomes ranging from the induction of cell-cycle arrest or apoptosis to dedifferentiation (Thompson et al, 1999; Deneen and Denny, 2001; Lessnick et al, 2002; Zwerner et al, 2003). The use of RNA interference in the Ewing sarcoma cell lines allows for a detailed molecular analysis of the targeted gene fusion and its downstream targets in the tumour context itself, precluding the use of heterologous model systems.
We have used a stable RNA interference system knocking down EWS–FLI1 in the Ewing sarcoma cell line, TC71, to establish a long-term model in which we can analyse in depth the molecular behaviour of the fusion protein. Our shRNAi model disclosed TOPK, a kinase involved in cell proliferation and motility, as a new target of EWS–FLI1. TOPK expression was downregulated because of fusion interference. EWS–FLI1 inhibition decreased tumourigenic potential, reduced the tumour volume in a xenograft model, impaired migration, and rendered cells more sensitive to both apoptosis and the action of inhibitors of the IGF-1/IGF-1R pathway, showing the interaction between these two key players in Ewing sarcoma tumourigenesis.
Materials and methods
Cell lines and drugs
The Ewing sarcoma cell line, TC71, was maintained as described earlier (Martins et al, 2006). NVP-AEW541 was obtained from Novartis Pharma AG (Basel, Switzerland). PD-98059 and LY-294002 were purchased from Stressgen (Victoria, BC, Canada), and G418 was acquired from Invitrogen (Carlsbad, CA, USA). Cell lines were tested for the absence of mycoplasma using the VenorGeM kit (Minerva Biolabs, Berlin, Germany).
shRNAi interference
An shRNAi approach was used based on the pSUPER-neo-GFP plasmid (Oligoengine, Seattle, WA, USA) (Brummelkamp et al, 2002). The shRNAi sequences were designed to target the fusion point between EWS and FLI1 following the manufacturer's instructions in order to avoid affecting the wild-type partners. The targeted sequences of EWS–FLI1 were 5′-CAGAGCAGCAGCTACGGGCAGCAGAACC-3′ for 7-6 shRNAi and 5′-GGGCAGCAGAACCCTTCTT-3′ for 7-6 M shRNAi.
The presence of the insert within the recombinant pSUPER vector was confirmed by sequencing (ABI Prism 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA).
TC71 cells were electroporated (125 V, 800 mA, 40–45 ms) using Gene Pulser II (Bio-Rad, Hercules, CA, USA), seeded, and allowed to recover for 24 h. G418 selection (500 μg ml−1) was carried out for 3 weeks, and individual selected clones were grown for 2 additional weeks and split into several plates to create a stock of the early stage. RNA interference was monitored by analysing EWS–FLI1 protein and mRNA levels using western blot and qRT–PCR, respectively. Up to 40 clones were analysed at both early and late stages (eight cellular passages later).
Validated TOPK siRNA oligos were purchased from Ambion (Austin, TX, USA). (sense sequence: 5′-GGUGAAAAGUCUCUAAAUGtt-3′; antisense sequence: 5′-CAUUUAGAGACUUUUCACCtc-3′.) TOPK siRNA oligos were transfected using jetPEI (Polyplus-transfection, Illkirch, France).
Western blot
Western blots were performed as described earlier (Martins et al, 2006). EWS–FLI1 expression was determined using the anti-FLI1 antibody (Santa Cruz BT, Santa Cruz, CA, USA). Other antibodies used were anti-phospho-p44/p42 MAPK (Thr202/Tyr204), anti-p44/42 MAPK, anti-phospho-AKT (Ser473), anti-AKT, anti-p57/Kip2, and anti-Caspase 9 (all from Cell Signaling, Danvers, MA, USA), anti-DAX1 (a gift from Dr Javier Alonso-IIB, Madrid, Spain), anti-PARP/Caspase 9 (Serotec Germany, Düsseldorf, Germany), anti-EWS, anti-IGF-1R and anti-NKX2.2 (Santa Cruz BT), anti-actin (Sigma, Saint Louis, MO, USA), anti-GAPDH (Stressgen), anti-IGF-1 (Abnova, Taipei, Taiwan), and anti-TOPK (BD, Franklin Lakes, NJ, USA). The cells were treated with the NVP-AEW541 for 15 min and with the inhibitors for 2 h, before IGF-1 stimulation (50 ng ml−1) for 15 min in serum-free conditions.
Quantitative real-time PCR
Quantitative real-time PCR was performed as described earlier (Martins et al, 2008). Primers for the selected genes were designed using primer3 software (MIT, Cambridge, MA, USA) (Supplementary Table 4).
Apoptosis and cell-cycle analysis
Assessment of the apoptotic index and cell-cycle analysis was made by flow cytometry using the Annexin V–Fitc kit (Immunostep, Salamanca, Spain), and IP staining (Sigma) as described (Martins et al, 2006).
Proliferation assay
To determine the rate of proliferation, we used the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method, as described earlier (Martins et al, 2006). Dose response under the influence of AEW571, LY294002, or PD98059 was analysed to determine the IC50 of proliferation. The cells were incubated with AEW541, LY294002, or PD98059 for 72 h on reaching 40% confluence.
Soft agar assay
Soft agar assay was performed as described earlier (Martins et al, 2006).
Immunohistochemistry
The immunohistochemical study was performed as described earlier (Mendiola et al, 2006), using a monoclonal anti-DAX1 antibody (clone 2F4; dilution 1 : 1000).
Motility assay
In all, 105 alive cells were counted using a Neubauer chamber (VWR, West Chester, PA, USA) and seeded per well with IMDM medium (Invitrogen) without serum in a transwell (BD) of 24-well plates (Corning, Acton, MA, USA). A volume of the medium (500 μl) was added to the bottom of the well and the plate was left overnight at 37°C and 5% CO2. On the next day, unmigrated cells in the upper part of the transwell were removed with a cotton swab and migrated cells in the transwells were fixed in a 4% paraformaldehyde–PBS solution for 15 min. The transwells were washed three times with PBS 1 × , dyed in blue violet solution (Sigma) for 30 min, and rinsed well with water and air-dried. Membranes of the transwell were cut and deposited individually in a well of 96-well plates with 300 μl of 10% glacial acetic acid (Merck, Darmstadt, Germany); subsequently, the absorbance was read at 570 nm (Tecan, Männedorf, Switzerland).
Gene expression assays and bioinformatic analysis
Total RNA was extracted, purified, and labelled according to Affymetrix (Santa Clara, CA, USA) instructions. RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). Labelled RNA was fragmented and hybridised to the HU-133A oligonucleotide arrays (Affymetrix). The arrays were scanned by using the Gene Array Scanner and analysed using the DNA-Chip Analyzer (Affymetrix). All experiments were performed in biological duplicates and for both, early (T0) and late passage (T8). The quality of cRNA was proved using the 3′/5′ ratios of housekeeping controls GAPDH and β-actin. Microarray data were collected and analysed using the bioinformatic software Genetrix (EpiCenter Software, Pasadena, CA, USA). Genetrix uses the Probe Profiler package and performs a GeneScreen analysis using Cox regression. GeneScreen outputs two values for each gene: the first one is the P-value, which measures the ‘significance of association’ (association with shRNAi or not); the second one is the t-statistic value, which measures the strength or ‘magnitude of association’. Lists of the top 300 genes most significantly associated with the shRNAi clone and the 300 genes that were not associated were obtained and combined into a single list of 600 probe sets used in a hierarchical clustering analysis and to clearly distinguish the shRNAi clone samples from the others. Genes with a P-value <0.05 were considered candidate genes.
Chromatin immunoprecipitation (ChIP)
The bioinformatics program, TFM-explorer (http://bioinfo.lifl.fr/TFME/), was used for searching EWS–FLI1 putative binding sites in the TOPK promoter and introns. ChIP was performed using the Magna ChIP G Chromatin Immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. The antibodies used for each sample were 1 μg of IgG-negative control, 1 μg anti-RNA polymerase II antibody-positive control (provided by the kit), 5.0 μg of EWS goat polyclonal IgG (Santa Cruz BT) – antibody of interest, no antibody–ChIP reaction containing pre-cleared chromatin, and DNA–protein complexes before immunoprecipitation, named ‘Input DNA’. As EWS contains a RNA-binding domain, the immunoprecipitated products corresponded only to the fusion protein (DNA-binding domain of FLI1) and not to wild-type FLI1. Cell extracts were sonicated four times with a 12 s constant burst (100 J energy, 20% amplitude) (SONICS, Newtown, CT, USA), following by 30–40 s rest on ice between each pulse. Purified DNA was subjected to PCR. The control human PCR primers used for ChIP were those described by Zupanska et al (2007). The primers used for probable TOPK-recognised sequences are detailed in Supplementary Table 5. The PCR program consisted of 38 cycles of amplification for 20 s at 94°C, 30 s at 58°C, and 30 s at 72°C. An earlier denaturing step of 3 min at 94°C and a final extension step of 2 min were added.
In vivo studies
Four to five-week old female NOD/SCID mice (Charles River, Barcelona, Spain) were used, following the Spanish and European Union guidelines (RD 1201/05 and 86/609/CEE, respectively). The study was approved earlier by the Bioethics Committee of our institution (CB-A4). Cell suspensions, containing 5 × 106 alive cells in 0.2 ml of 1 : 1 cellular medium (Matrigel Matrix (BD), were injected s.c. into the right flank of the mice. Cells were counted using a Neubauer chamber (VWR) and cellular viability was checked by trypan blue staining (Sigma). Mice were randomised into three controls (TC71wt, early mock, and late mock) and two treated groups (early and late shRNAi clone). Tumours were measured every 5 days with a caliper, and the diameters were recorded. Tumour volume was calculated as described earlier (Martins et al, 2008) by the formula a2b/6, in which a is the smallest diameter and b the biggest one. Mice were killed by anaesthesia overdosing 4 weeks after the cells injection, and tumours were collected for histopathology analysis.
Statistics
For in vivo studies, one-way ANOVA for independent samples was performed using the SPSS 15.0 software (SPSS Inc., Chicago, IL, USA) and mice with a tumour volume higher than 2.5 cm3 were excluded from the analyses. For comparisons between shRNAi and mock (early and late stages), we computed two-sided t-tests of independent samples for each condition.
Results
shRNAi permitted a sustained knockdown of EWS–FLI1
To analyse the effects of a stable suppression of EWS–FLI1 expression in its native context, we used a pSUPER vector that directs intracellular synthesis of siRNA-like transcripts.
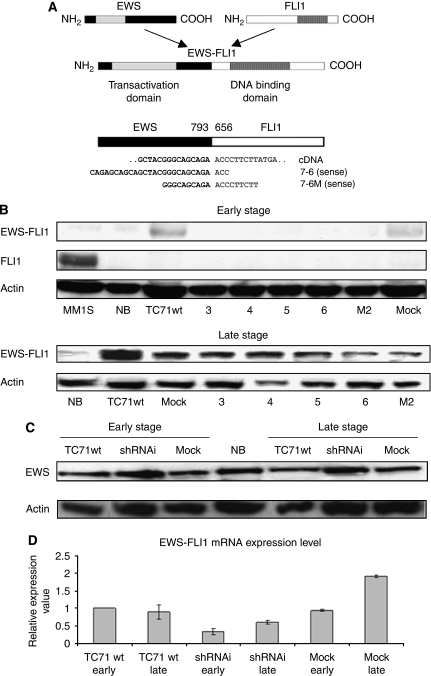
Two different and specific interfering shRNAi (named 7-6 and 7-6 M, respectively), both targeting the fusion point of type 1 EWS–FLI1 mRNA isoform (7-6), were tested in the Ewing sarcoma cell line, TC71 (Figure 1A). The loss-of-function phenotype was assessed in selected clones in early and late stages (eight cellular passages) to confirm the maintenance of the EWS–FLI1 ‘knockdown’, although a drop in the interference effect was detected (Figure 1B). Wild-type FLI1 was detected only in the multiple myeloma cell line MM1S, and its expression was not affected by the EWS–FLI1 shRNAi constructs. Expression of wild-type EWS protein was not changed by EWS–FLI1 RNAi, confirming the specificity of our shRNAi designs (Figure 1C). The effect of RNA interference was also measured at the mRNA level in the selected shRNAi clones resulting in 42–54% inhibition (P<0.01), and also at later passages (P<0.05) (Figure 1D).
Figure 1.
Targeting of EWS–FLI1 through shRNAi for stable protein and mRNA knockdown. (A) Diagram of wild-type EWS, wild-type FLI1, and EWS–FLI1 fusion type 1. The illustration of EWS–FLI1 cDNA and the location of the two shRNAi designs used, 7-6 and 7-6M, both against fusion break-point. (B) Analysis by western blotting showed that the EWS–FLI1 expression was reduced in several TC71 shRNAi clones. The EWS–FLI1 reduction is maintained partially in some shRNAi clones through the course of cellular passages (early and late stages). EWS–FLI1 68 kDa, FLI1 51 kDa, and actin 42 kDa. 3, 4, 5, 6: TC71 shRNAi clones transfected with shRNAi design type 7-6; M2: TC71 shRNAi clone transfected with shRNAi design type 7-6M. MM1S: multiple myeloma cell line. Positive control for FLI1. NB: neuroblastoma cell line SK-N-JD. Negative control for EWS–FLI1. Actin is shown as a loading control. (C) Analysis by western blotting showed that the EWS–FLI1 shRNAi designs are specific and did not alter the EWS expression in the early and late stages. The shRNAi clone corresponds to the TC71 shRNAi clone 6. Actin is shown as a loading control. (D) The shRNAi reduction in the EWS–FLI1 mRNA level assessed through qRT–PCR (SYBR green probes) in the early (T0) and late passage (T8). The shRNAi clone corresponds to the TC71 shRNAi clone 6. GAPDH was used as an internal control. Columns, mean of triplicates of three different replicates; bars, s.d.
These results confirmed the validity and the stable nature of the shRNAi model through the course of cellular passages.
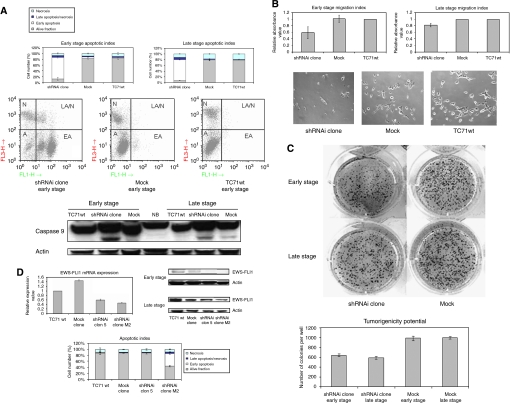
Stable reduction of EWS–FLI1 prompted an increase in the apoptotic fraction, and a reduction of the migratory capacity and tumourigenicity
Up to 40 different TC71 shRNAi clones were obtained. We selected the one, shRNAi clone 6, showing the most pronounced decrease in EWS–FLI1 expression, to further characterise and evaluate its molecular and cellular phenotype.
We observed a marked increase in the apoptotic index of up to 70% in both, early and late stages. Western blotting analysis confirmed the processing of caspase 9 in the interfered clone in both stages, although a slight activation could also be seen in the mock sample, which seemed insufficient for promoting apoptosis (Figure 2A). PARP was also processed in the shRNAi clone, confirming caspase cascade activation (data not shown).
Figure 2.
The long-term reduction in EWS–FLI1 provoked an increase in the apoptotic fraction, reduced migration capacity, and oncogenic transformation. There is a direct relationship between the extent of the interference of the fusion transcript and the degree of change observed. (A) Induction of apoptosis by reduction in the expression of EWS–FLI1 using specific shRNAi in the TC71 Ewing sarcoma cell line in an early and late stage. The shRNAi clone corresponds to the TC71 shRNAi clone 6. The cells were seeded on 24 well plates, 24 h after apoptotic index was determined by FACS using an annexin V detection kit assay. The means±standard deviations (error bars) of four independent experiments are shown. FACS Apoptotic Analysis Figures show the various cell populations. A, alive fraction; EA, early apoptosis; LA/N, late apoptosis/necrosis; N, necrosis. x asis: FL3-H Annexin V; y axis: FL1-H PI. Panel below shows the activation of Caspase 9 in the shRNAi clone in both stages by western Blot. (B) The shRNAi clone cells were less able to migrate through the matrix of Transwell in both stages, early and late. The shRNAi clone corresponds to the TC71 shRNAi clone 6. A total of 100 000 alive cells were counted using a Neubauer chamber and seeded over the transwell in 24 well-format plates. After 16 h, cells of the upper part of the transwell were scrapped, cells migrated to the bottom part of the transwell were fixed, stained with blue violet solution and discoloured with acetic acid. Absorbance was measured at 570 nm. Photographs of migrated cells taken with a Nikkon 10 × objective. Columns, mean of replicates of three different experiments; bars, s.d. (C) The diminished expression of EWS–FLI impairs the number and size of colonies in the shRNAi clone. The shRNAi clone corresponds to the TC71 shRNAi clone 6. Colonies were photographed after 2 weeks. Experiments were performed three times with several replicates. (D) The EWS–FLI1 shRNAi extension determined the degree of the changes that were not because of unspecific clonal ‘off-target’ effects. Analysis by western blotting showed that the EWS–FLI1 expression was reduced in the early stage in both shRNAi clones, 5 and M2, although only shRNAi clone M2 maintained partially EWS–FLI1 reduction in the late stage. Actin is shown as a loading control. The increase in the apoptotic index was directly correlated to the degree of EWS–FLI1 interference. The shRNAi clone number 5 showed no change in the apoptotic index compared with shRNAi clone M2 that had a more pronounced inhibition of EWS–FLI1 and showed a increase in its apoptotic fraction. The cells were seeded on 24 well plates, 24 h after apoptotic index was determined by FACS using an Annexin V detection kit assay. The means±standard deviations (error bars) of four independent experiments are shown. Columns, mean of triplicates of three separate replicates; bars, s.d.
To determine whether EWS–FLI1 knockdown affects the ability of TC71 cells to migrate, we performed a transwell experiment. The migratory capacity of shRNAi cells was found to be impaired, with around a 30% decrease in both stages (Figure 2B).
The tumourigenicity of the shRNAi clone was tested using a soft agar growth assay. The shRNAi cells were largely restricted in their tumourigenicity, giving rise to fewer and smaller colonies with a decrease of around 35–40% (P<0.0003) compared with the mock, which was maintained through the course of cellular passages (Figure 2C). Similar results were obtained with the shRNAi clone M2 (data not shown).
Analogous results regarding cell-cycle distribution and apoptotic index were obtained using other shRNAi clones in which interference of EWS–FLI1 expression was less dramatic. There was a direct relationship between the extent of interference of the fusion transcript and the degree of change observed, thus confirming that the effects appearing after EWS–FLI1 inhibition were due to its specific suppression of expression and not due to unspecific clonal ‘off-target’ effects (Figure 2D).
The growth rate of these interfered cells was reduced, as measured by MTT assay, to around 30% in the early passage (P<0.05). However, this inhibitory effect was lost in the late stage (Supplementary Figure 1A). Assessment of the cell-cycle pattern of the shRNAi clone showed no changes in cell-cycle distribution (Supplementary Figure 1B).
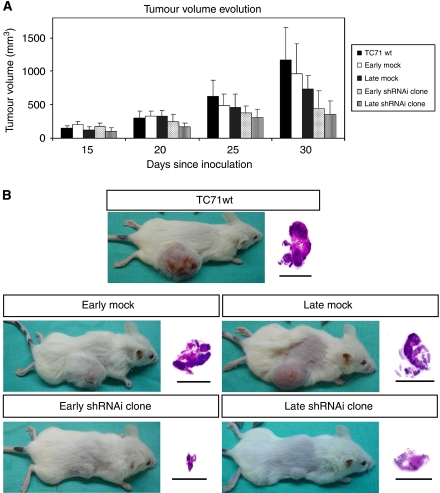
EWS–FLI1 interference reduced tumour growth
We injected in immunodeficient mice cells from the shRNAi clone, mock, and TC71wt, both in the early and late stages. Tumours appeared at the same time in all the animals, but mice harbouring cells from the shRNAi clone in both temporal stages developed tumours with a marked volume reduction (Figures 3A and B). Most of the animals injected with the early and late shRNAi clone cells developed tumours of <0.5 and 0.3 cm3, respectively, with no tumour reaching 1 cm3 in any case. However, tumours in control groups (injected with wild-type, early mock cells, and late mock cells) were markedly larger, most of them close to 1 cm3 or even larger (P<0.05). Moreover, five animals (two from the wild-type, two from the early mock, and one from the late mock) developed tumours between 2.5 and 4.9 cm3 and were not included in the statistical analyses.
Figure 3.
Tumour growth evolution. The EWS–FLI1 shRNAi reduced tumoural growth in vivo. (A) NOD-Scid mice were s.c. injected with 5 × 106 TC71 cells. The treated mice (early and late shRNAi clone) showed smaller tumours than the control groups (TC71wt, late and early mock) since the third week after cells injection. At the end of the study these differences were statistically significant (P<0.05). (B) Visual and histopathological evaluation of mice tumours. All of the tumours showed the same histopathological pattern: a large area of necrotic tissue, an area of proliferating cells, and a layer between them with cells in apoptosis.
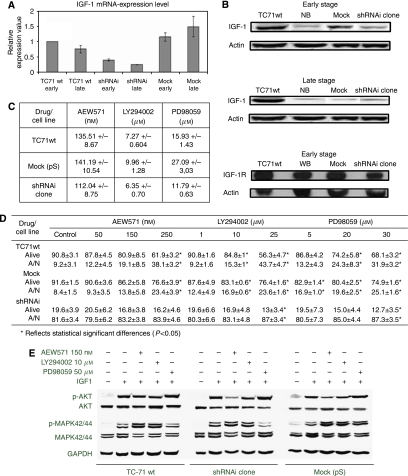
EWS–FLI1 interference downregulated IGF-1 and sensitised Ewing sarcoma cells to inhibitors of the IGF-1/IGF-1R signal transduction pathway
We next analysed the effects of EWS–FLI1 interference on the gene expression signature of the TC71 Ewing sarcoma cells. RNA from the shRNAi clone, mock, and TC71wt from the early and late stages was isolated and used to perform duplicate hybridisation experiments with the Affymetrix HG-U133A platform. EWS–FLI1 activated and repressed genes in similar numbers. Several of the genes identified were previously reported to be repressed (Supplementary Table 1) or induced (Supplementary Tables 2 and 3) by EWS–FLI1, including p57/Kip2, NXK2.2, and DAX1-NR0B1. Their expression was validated, confirming the original microarray data (Supplementary Figures 2A–C, respectively).
As IGF-1 was pointed as a relevant downregulated gene, we decided to focus on it. Western blot and qRT–PCR analyses confirmed IGF-1 downregulation (P<0.05) in the shRNAi clone during the course of the cellular passages compared with TC71wt and mock cells, although a decrease in IGF-1 expression could be detected in the late stage of the mock because of unknown reasons (Figures 4A and B). No change in IGF-1R expression was observed by WB (Figure 4B) or IHC (data not shown). We hypothesised that the shRNAi clone would be more sensitive to inhibitors of this signal transduction pathway. NVP-AEW541 is an inhibitor of the IGF-1 pathway (Garcia-Echeverria et al, 2004). PD98059 and LY294002 inhibit MEK and PI3-K, respectively, which are key genes in the IGF-1/IGF-1R pathway. We studied the influence of NVP-AEW541, PD98059, and LY294002 administration on AKT, p-AKT, MAPK42/44, and p-MAPK 42/44 levels by WB, and evaluated the induction of apoptosis and repression of proliferation. Knockdown of the EWS–FLI1 fusion protein rendered the shRNAi clone more sensitive to the action of NVP-AEW541, PD98059, and LY294002, resulting in a lower IC50 for all of them (Figure 4C). Although the shRNAi clone exhibited a very high apoptotic level before drug treatment, apoptosis could be further induced under all conditions by each of the inhibitors (Figure 4D). AKT and MAPK42/44 phosphorylation was diminished in the shRNAi clone under IGF-1 stimulation, whereas TC71wt and mock cells maintained the active phosphorylation status in both kinases (Figure 4E).
Figure 4.
EWS–FLI1 shRNAi impaired the IGF-1/IGF-1R survival pathway and its downstream targets. (A) Histogram representation of IGF-1 transcript level knockdown in the shRNAi clone as assessed by qRT–PCR normalised to GAPDH. The shRNAi clone corresponds to the TC71 shRNAi clone 6. SYBR probes were used. All experiments were performed in triplicate in the early and late stages. Columns, mean of triplicates of three different experiments; bars, s.d. (B) The IGF-1 protein level is reduced in the shRNAi clone as assessed through western blot whereas the IGF-1R protein level is not changed. The shRNAi clone corresponds to the TC71 shRNAi clone 6. Actin is shown as loading control. (C) IC50 of proliferation measured by MTT assay after the NVP-AEW541, LY294002, or PD98059 treatment (72 h of incubation). The shRNAi clone showed more sensitivity to the action of inhibitors of the IGF-1/IGF-1R pathway being statistically significant for AEW571 and PD98059 (P<0.05). The shRNAi clone corresponds to the TC71 shRNAi clone 6. (D) Apoptotic index after the NVP-AEW541, LY294002, or PD98059 treatment (72 h of incubation). Apoptosis could be further induced in the shRNAi clone under all conditions. The means±standard deviations (error bars) of four independent experiments are shown. The shRNAi clone corresponds to the TC71 shRNAi clone 6. P<0.05 values are considered as significant. (E) Effects of NVP-AEW541 combined with inhibitors LY294002 and PD98059 on the activation of the IGF-1/IGF-1R signalling pathway. The AKT and MAPK42/44 phosphorilation were diminished in the shRNAi clone under IGF-1 stimulation whereas TC71wt and mock cells maintained the active phosphorylation status in both kinases. All conditions were treated with AEW571 during 15 min and with the inhibitors for 2 h, before IGF-1 stimulation (50 ng ml−1) during 15 min (serum-free conditions). The shRNAi clone corresponds to the TC71 shRNAi clone 6.
TOPK is a new target of EWS–FLI1 involved in proliferation
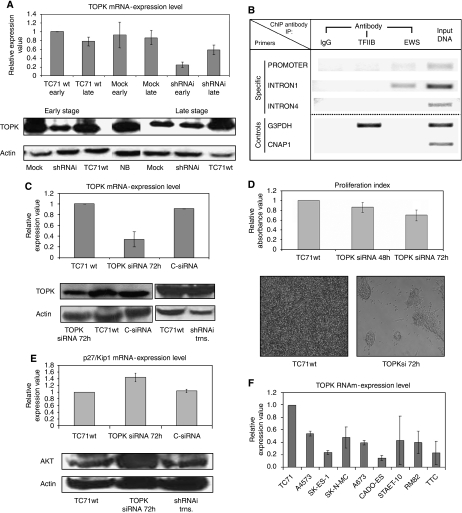
We focused our attention on a new potential target gene of EWS–FLI1, namely TOPK/PBK (T-lymphokine-activated killer cell-originated protein kinase). Our gene expression analyses indicated TOPK as a target gene downregulated as a consequence of EWS–FLI1 inhibition (score value −6.78 in Genetrix analysis). qRT–PCR and western blot analysis confirmed a decrease in TOPK at the mRNA (P<0.05) (Figure 5A) and protein levels (Figure 5A) which was more intense in the early stage. These results pointed out TOPK as a candidate EWS–FLI1 target gene.
Figure 5.
Identification and characterisation of TOPK as a new EWS–FLI1 target. (A) The histogram representation of TOPK mRNA level reduction in the shRNAi clone as assessed by qRT–PCR normalised to GAPDH. SYBR probes were used. All experiments were performed in triplicate in an early and late stage. Columns, mean of triplicates of three separate determinations; bars, s.d. The TOPK protein level is reduced in the shRNAi clone in the early and late stages as determined through western blot. Actin was used as a loading control. The shRNAi clone corresponds to the TC71 shRNAi clone 6. (B) EWS–ChIP. EWS–FLI1 binds TOPK intron 1. The figure shows amplicons resulting from immunoprecipitation with IgG (negative control), anti-RNA polymerase II antibody (positive control), anti EWS monoclonal antibody (specific), or input DNA (pre-cleared chromatin, no antibody) from mock clone. Primers for TOPK intron 4 were used as internal negative control. (C) The TOPK mRNA expression level decrease in TC71 TOPK siRNA cells (72 h after transfection) assessed by qRT–PCR normalised to GAPDH (TC71 C-siRNA as negative control). SYBR probes were used. Experiments were performed in triplicate. Columns, mean of triplicates of three separate determinations; bars, s.d. Protein level of TOPK determined by western blot. TOPK siRNA effectively diminished the TOPK protein levels with respect to the untransfected TC71wt cell line. Actin was used as a loading control. Western blot showing the TOPK protein levels restored after transfection of EWS–FLI1 type 1 fusion into the shRNAi clone. The shRNAi clone corresponds to the TC71 shRNAi clone 6. Actin was used as a loading control. (D) Proliferation index measured through MTT assay. The TOPK interference (48 and 72 h after transfection) caused a significant decrease in growth rate at 72 h compared with TC71wt and negative siRNA control (data not shown). Columns, mean of replicates of three different replicates; bars, s.d. The morphological changes of TC71 TOPKsi RNA cells respect the TC71wt (72 h after transfection). Cells were unable to reach confluence and formed clumps. (E) The p27/Kip1 mRNA expression level increases in TC71 TOPK siRNA cells assessed by qRT–PCR normalised to GAPDH (TC71 C-siRNA as negative control). SYBR probes were used. Experiments were performed in triplicate. Columns, mean of triplicates of three different experiments; bars, s.d. Western blot showing the AKT protein level increase in TOPKsi cells compared with TC71wt and shRNAi clone transfected with EWS–FLI1 type 1 fusion. Actin was used as a loading control. The shRNAi clone corresponds to the TC71 shRNAi clone 6. (F) Relative expression value of TOPK mRNA in nine different Ewing sarcoma cell lines determined by qRT–PCR normalised to GAPDH. SYBR probes were used. Experiments were performed in triplicate. Columns, mean of triplicates of three different experiments; bars, s.d.
To confirm this fact we looked for EWS–FLI1-binding sites in the TOPK proximal promoter and introns using the bioinformatics program TFM-explorer. Several consensus binding sites were found confirming TOPK as a putative interacting target of the fusion gene (TOPK proximal promoter location (−37:−248), P-value=2.20e−02; TOPK intron1 location (−2197:−2740), P-value=7.23e−03). Immunoprecipitation of chromatin with anti-EWS antibody from mock yielded amplification of the TOPK intron 1 selected region (Figure 5B). Immunoprecipitation with IgG or with anti-RNA polymerase II resulted in no amplification of the TOPK promoter proximal region, intron 1 and intron 4 (negative internal control). As a positive control, immunoprecipitation with anti-RNA polymerase II resulted in the amplification of the G3PDH promoter, without differences between the mock and the shRNAi clone. We can conclude that TOPK is a target of EWS–FLI1 because gene fusion binds TOPK intron 1.
We inhibited TOPK expression in TC71wt cells using siRNA oligos. Seventy-two hours after transfection a reduction in both TOPK RNA and protein levels was observed (Figure 5C). A noteworthy effect of TOPK inhibition was a mild decrease in proliferation rate as measured by MTT (Figure 5D) compared with TC71wt and negative siRNA control. A dramatic change in the growth pattern was detected after knocking down TOPK. TOPK siRNA cells grew in clumps and were not able to merge as compared with TC71wt cells (Figure 5D). The decrease in TOPK levels did not affect the cell-cycle pattern and did not cause an increase in apoptotic rate (data not shown). Transfection of EWS–FLI1 type 1 mRNA in the shRNAi clone restored EWS–FLI1 levels; TOPK protein expression was also significantly restored (Figure 5C). In addition, AKT levels also decreased, thus pointing to a possible connection between both protein kinases (Figure 5E). When the expression of TOPK was diminished an increase in the transcript level of p27/Kip1 was detected (Figure 5E). TOPK expression could be detected through qRT–PCR in nine Ewing sarcoma cell lines analysed regardless of the type of EWS–ETS fusion (Figure 5F).
Discussion
Ewing sarcoma fusion proteins act as aberrant transcription factors by interfering with different signalling pathways (Prieur et al, 2004). Identification of EWS–FLI1 targets represents a key aspect in the understanding of the molecular behaviour of Ewing sarcoma. RNA interference has been shown to be a powerful tool that allows targeting chimeric oncogenes and gaining a deeper knowledge of tumour biology in its own context (Sharp, 1999; Tijsterman et al, 2002).
Ectopic expression models for the study of gene fusions are disadvantageous as they do not account for the true cellular context. A wide variety of effects have been documented after EWS–FLI1 induction in heterologous models, ranging from dedifferentiation to growth arrest (Kovar, 2005).
shRNAi studies to suppress EWS–FLI1 in the Ewing sarcoma cells have been carried out using different systems (Siligan et al, 2005; Kinsey et al, 2006; Owen and Lessnick, 2006; Carrillo et al, 2007; Stegmaier et al, 2007). Our shRNAi approach, based on the pSUPER vector, targets at the junction point of EWS–FLI1 type 1 fusion transcripts in a reliable model, the Ewing sarcoma cell line, TC71. The effects due to EWS–FLI1 interference were observed in different clones and were related to the degree of fusion inhibition, indicating a direct and specific correlation. We reported a significant reduction in EWS–FLI1 expression, which was maintained through several cellular passages, confirming the validity of our approach, although a partial loss of RNA interference was detected through the course of cellular passages as also described by other group (Smith et al, 2006) in the Ewing sarcoma cell line A673.
The main consequence observed in our model because of the silencing of EWS–FLI1 was a strong induction of apoptosis, correlating with caspase 9 pathway activation and PARP processing. EWS–FLI1 interference has been documented to increase remarkably the number of apoptotic cells (Prieur et al, 2004). A decrease in the anchorage-independent capacity to grow has also been reported (Smith et al, 2006). The ability of cells to grow in semisolid media was also affected in our shRNAi model. Disruption of EWS–FLI1 provoked the appearance of fewer and smaller colonies mainly in the early stage. EWS–FLI1 reduction also impaired migration capacity. CCN3 (NOV) has been described as an important mediator of migration and invasion (Benini et al, 2005) and its expression was downregulated in our model; this could explain loss of function of interfered cells although it is not possible to rule out the contribution of early apoptosis to that observation.
NOD/SCID mice injected with shRNAi cells in both temporal stages developed tumours with a significant reduction in tumour volume with respect to mock and TC71wt. shRNAi cells had restricted their tumourigenicity and migratory capacity, which could explain the tumour growth reduction observed. Tumoural growth reduction in nude xenograft models has been described in several studies either using an shRNAi model (Smith et al, 2006) or through the inhibition of EWS–FLI1 targets such as IGF-1/IGF-1R (Scotlandi et al, 1998), VEGF (Guan et al, 2005), NR0B1/DAX1 (Kinsey et al, 2006), and cholecystokinin (Carrillo et al, 2007).
The strong decrease in IGF-1 gene expression observed after EWS–FLI1 silencing was analysed. The IGF-1/IGF-1R signal transduction pathway is over-expressed in many sarcomas (Sekyi-Otu et al, 1995) and has great relevance for the biology of Ewing sarcoma (Scotlandi et al, 1996). Blocking IGF-1/IGF-1R has shown its effectiveness in restricting tumoural growth (Martins et al, 2006). Our shRNAi clone showed an increased sensitivity to the action of IGF-1/IGF-1R pathway inhibitors NVP-AEW541, LY294002, and PD98059, and a reduction in the phosphorylation levels of AKT and MAPK 42/44. IGF-1R protein status is not affected, and therefore the effects observed were due to IGF-1 downregulation. This observation underlines the fact that IGF-1/IGF-1R signalling activation and EWS–FLI1 fusion are two key molecular features in Ewing sarcoma and suggests the need for blocking both of them in order to achieve therapeutical benefits.
Understanding the oncogenic properties of EWS–FLI1 requires identification of the downstream targets. The aim of our gene expression analysis was to identify potentially regulatory genes, whose transcript levels change in response to oncoprotein inhibition. Well-known EWS–FLI1 targets were among the genes identified, including NR0B1/DAX1, zyxin, p57/Kip2, and NKX2.2 (Dauphinot et al, 2001; Amsellem et al, 2005; Mendiola et al, 2006; Smith et al, 2006), confirming the validation of our stable shRNAi model.
We have mainly focused our attention on a new target gene of EWS–FLI1, TOPK/PBK. TOPK, T-lymphokine-activated killer cell-originated protein kinase, is a 36-kDa protein located on 8p21.2 belonging to the Ser/Thr protein kinase family. It was cloned as a new MAPKK-like protein kinase, highly conserved among vertebrates, specifically expressed in the testis, placenta, activated lymphoid cells, and neural lineages (Abe et al, 2000; Dougherty et al, 2005). TOPK has been involved in haematological malignancies (Simons-Evelyn et al, 2001; Nandi et al, 2004), in breast cancer (Park et al, 2006), in preventing apoptosis in a melanoma cell line (Zykova et al, 2006), as a proliferative agent related to DNA damage response through p38 MAPK, c-Jun-NH2-Kinase1, and H-Ras signalling (Ayllon and O'Connor, 2007; Oh et al, 2007), and as an enhancer of Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promoter of cytokinesis (Abe et al, 2007). Our gene expression analysis pointed out TOPK as a target gene downregulated because of EWS–FLI1 inhibition. TOPK reduction at protein and mRNA levels was confirmed through WB and qRT–PCR, respectively, and was maintained during cellular passages.
Using an in silico approach, we analysed the TOPK promoter and introns in order to find EWS–FLI1 binding sites. EWS–FLI1 and some ETS family members, such as wild-type FLI1, require a 9-bp consensus sequence harbouring a GGAA ‘core’. A 9-bp sequence, GAAGGAAGT, was found in the TOPK intron 1, which showed limited similarity to the high-affinity ETS-binding consensus (ACCGGAAGT) (Gangwal and Lessnick, 2008). It has been shown in promyelocytic leukaemia cells that the transcriptional control of TOPK promoter is mostly because of binding of transcription factors E2F and CREB/ATF to two distinct binding sites within it (Nandi and Rapoport, 2006). TOPK intron 1 showed an EWS–FLI1 binding site as validated by ChIP, probably corresponding to the above-mentioned ETS binding sequence. We also found E-box sequences that are high-affinity c-Myc-binding sites (CACATG at −574 and −3098), suggesting that c-Myc, an EWS–FLI1 target, could play a role in the transcriptional activation of the TOPK promoter.
Downregulation of TOPK activity was achieved in the Ewing sarcoma cell line, TC71, using RNAi oligos. TOPKsi cells showed a dramatic change in growth pattern caused presumably by TOPK inhibition; TOPK-interfered cells seemed to be disabled in their ability to reach confluence compared with TC71wt cells. The same observation was described earlier when TOPK was knocked down using siRNA oligos in the prostate carcinoma cell line, DU145 (Ayllon and O'Connor, 2007). The main consequence observed because of TOPK inhibition was a 35% reduction in the proliferation rate. The results were concordant with the significant suppression of cell growth caused by TOPK interference in human breast cancer and colorectal cell lines (Park et al, 2006; Zhu et al, 2007). Surprisingly, TOPK silencing raised AKT levels. When EWS–FLI1 expression was restored in the shRNAi clone, the TOPK level also recovered and AKT level decreased to the level observed in TC71wt, thus proving that AKT increase was due to the TOPK knockdown and not due to EWS–FLI1 interference. AKT is a relevant gene in the Ewing sarcoma biology that is involved in proliferation and survival, and the therapeutical benefit of its inhibition is clear (Martins et al, 2006); further research to clarify the relationship between TOPK and AKT needs to be carried out. Neither cell-cycle pattern nor apoptotic index was modified after TOPK silencing, as was the case in the MCF-10A breast epithelial cell line (Ayllon and O'Connor, 2007), although the levels of p27/Kip1, a gene described in Ewing sarcoma by its expression level inversely correlated with that of EWS–FLI1 (Matsunobu et al, 2006), were increased.
In this study, we have developed a stable RNAi model system. Our shRNAi model maintained the EWS–FLI1 knockdown through the course of the cellular passages, showing that it is a valid one for assessing the long-term effects of EWS–FLI1 interference. EWS–FLI1 interference caused a decrease in IGF-1 levels and rendered cells more sensitive to the action of inhibitors of the IGF-1/IGF-1R pathway, pointing to the importance of this survival pathway and the necessity to block it for therapeutic benefits. Our model could be a valuable tool for confirming targets already described and for discovering new ones such as TOPK, a kinase involved in proliferation and motility. This is the first time that TOPK expression is described in Ewing sarcoma, and the changes observed after TOPK interference were concurrent with those observed in other cellular models, confirming the relevant role of this gene in the Ewing sarcoma biology.
Acknowledgments
We thank Dr Iranzu González, Universidad de Navarra, Pamplona, Spain, for the TC71 cell line, Novartis Pharma AG, Basel, Switzerland, for providing us NVP-AEW541, Dr Miguel Alaminos, from the Department of Histology, School of Medicine-Universidad de Granada, Granada, Spain, for helpful comments regarding RNAi design, Dr Javier Alonso, from IIB, CSIC/UAM, Madrid, Spain, for giving us DAX1/NR0B1 antibody, Dr Juan Pedro Bolaños from INCYL, Universidad de Salamanca, Salamanca, Spain, for providing us the pSUPER plasmid, and the Diagnostic Molecular Pathology Laboratory of CIC for immunohistochemistry and tissue processing of the animal model. CIC-IBMCC and HHU belong to NoE Eurobonet, FP6-2004-Lifescihealth-5, proposal number 018814, European Commission. Work at CIC is also funded by Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation-FEDER (PI052524; RD06/0020/0059, CD6/00001). Herrero-Martín was a recipient of a pre-doctoral fellowship from the Departamento de Educación, Junta de Castilla y León, Spain. This work has been done within the Acción Transversal en Cáncer program and the cooperative agreement between ISCIII and FICUS.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
References
- Abe Y, Matsumoto S, Kito K, Ueda N (2000) Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J Biol Chem 275: 21525–21531 [DOI] [PubMed] [Google Scholar]
- Abe Y, Takeuchi T, Kagawa-Miki L, Ueda N, Shigemoto K, Yasukawa M, Kito K (2007) A mitotic kinase TOPK enhances Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promotes cytokinesis. J Mol Biol 370: 231–245 [DOI] [PubMed] [Google Scholar]
- Amsellem V, Kryszke MH, Hervy M, Subra F, Athman R, Leh H, Brachet-Ducos C, Auclair C (2005) The actin cytoskeleton-associated protein zyxin acts as a tumour suppressor in Ewing tumour cells. Exp Cell Res 304: 443–456 [DOI] [PubMed] [Google Scholar]
- Ayllon V, O'Connor R (2007) PBK/TOPK promotes tumour cell proliferation through p38 MAPK activity and regulation of the DNA damage response. Oncogene 26: 3451–3461 [DOI] [PubMed] [Google Scholar]
- Benini S, Perbal B, Zambelli D, Colombo MP, Manara MC, Serra M, Parenza M, Martinez V, Picci P, Scotlandi K (2005) In Ewing's sarcoma CCN3(NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene 24: 4349–4361 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I, Garcia-Miguel P, Pestana A, Gallego S, Segura D, Alonso J (2007) Cholecystokinin down-regulation by RNA interference impairs Ewing tumour growth. Clin Cancer Res 13: 2429–2440 [DOI] [PubMed] [Google Scholar]
- Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman BE, Delattre O (2001) Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene 20: 3258–3265 [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G (1992) Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359: 162–165 [DOI] [PubMed] [Google Scholar]
- Deneen B, Denny CT (2001) Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 20: 6731–6741 [DOI] [PubMed] [Google Scholar]
- Dougherty JD, Garcia AD, Nakano I, Livingstone M, Norris B, Polakiewicz R, Wexler EM, Sofroniew MV, Kornblum HI, Geschwind DH (2005) PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. J Neurosci 25: 10773–10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwal K, Lessnick SL (2008) Microsatellites are EWS/FLI response elements: genomic ‘junk’ is EWS/FLI's treasure. Cell Cycle 7: 3127–3132 [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao J, Brueggen J, Capraro HG, Cozens R, Evans DB, Fabbro D, Furet P, Porta DG, Liebetanz J, Martiny-Baron G, Ruetz S, Hofmann F (2004) In vivo antitumour activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell 5: 231–239 [DOI] [PubMed] [Google Scholar]
- Guan H, Zhou Z, Wang H, Jia SF, Liu W, Kleinerman ES (2005) A small interfering RNA targeting vascular endothelial growth factor inhibits Ewing's sarcoma growth in a xenograft mouse model. Clin Cancer Res 11: 2662–2669 [DOI] [PubMed] [Google Scholar]
- Kinsey M, Smith R, Lessnick SL (2006) NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res 4: 851–859 [DOI] [PubMed] [Google Scholar]
- Kovar H (2005) Context matters: the hen or egg problem in Ewing's sarcoma. Semin Cancer Biol 15: 189–196 [DOI] [PubMed] [Google Scholar]
- Lessnick SL, Dacwag CS, Golub TR (2002) The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 1: 393–401 [DOI] [PubMed] [Google Scholar]
- Martins AS, Mackintosh C, Martin DH, Campos M, Hernandez T, Ordonez JL, de Alava E (2006) Insulin-like growth factor I receptor pathway inhibition by ADW742, alone or in combination with imatinib, doxorubicin, or vincristine, is a novel therapeutic approach in Ewing tumour. Clin Cancer Res 12: 3532–3540 [DOI] [PubMed] [Google Scholar]
- Martins AS, Ordonez JL, Garcia-Sanchez A, Herrero D, Sevillano V, Osuna D, Mackintosh C, Caballero G, Otero AP, Poremba C, Madoz-Gurpide J, de Alava E (2008) A pivotal role for heat shock protein 90 in Ewing sarcoma resistance to anti-insulin-like growth factor 1 receptor treatment: in vitro and in vivo study. Cancer Res 68: 6260–6270 [DOI] [PubMed] [Google Scholar]
- Matsunobu T, Tanaka K, Nakamura T, Nakatani F, Sakimura R, Hanada M, Li X, Okada T, Oda Y, Tsuneyoshi M, Iwamoto Y (2006) The possible role of EWS-Fli1 in evasion of senescence in Ewing family tumours. Cancer Res 66: 803–811 [DOI] [PubMed] [Google Scholar]
- May WA, Arvand A, Thompson AD, Braun BS, Wright M, Denny CT (1997) EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumourigenic. Nat Genet 17: 495–497 [DOI] [PubMed] [Google Scholar]
- Mendiola M, Carrillo J, Garcia E, Lalli E, Hernandez T, de Alava E, Tirode F, Delattre O, Garcia-Miguel P, Lopez-Barea F, Pestana A, Alonso J (2006) The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumours. Int J Cancer 118: 1381–1389 [DOI] [PubMed] [Google Scholar]
- Nandi A, Tidwell M, Karp J, Rapoport AP (2004) Protein expression of PDZ-binding kinase is up-regulated in hematologic malignancies and strongly down-regulated during terminal differentiation of HL-60 leukemic cells. Blood Cells Mol Dis 32: 240–245 [DOI] [PubMed] [Google Scholar]
- Nandi AK, Rapoport AP (2006) Expression of PDZ-binding kinase (PBK) is regulated by cell cycle-specific transcription factors E2F and CREB/ATF. Leuk Res 30: 437–447 [DOI] [PubMed] [Google Scholar]
- Oh SM, Zhu F, Cho YY, Lee KW, Kang BS, Kim HG, Zykova T, Bode AM, Dong Z (2007) T-lymphokine-activated killer cell-originated protein kinase functions as a positive regulator of c-Jun-NH2-kinase 1 signaling and H-Ras-induced cell transformation. Cancer Res 67: 5186–5194 [DOI] [PubMed] [Google Scholar]
- Owen LA, Lessnick SL (2006) Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing's sarcoma. Cell Cycle 5: 2049–2053 [DOI] [PubMed] [Google Scholar]
- Park JH, Lin ML, Nishidate T, Nakamura Y, Katagiri T (2006) PDZ-binding kinase/T-LAK cell-originated protein kinase, a putative cancer/testis antigen with an oncogenic activity in breast cancer. Cancer Res 66: 9186–9195 [DOI] [PubMed] [Google Scholar]
- Prieur A, Tirode F, Cohen P, Delattre O (2004) EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 24: 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotlandi K, Benini S, Nanni P, Lollini PL, Nicoletti G, Landuzzi L, Serra M, Manara MC, Picci P, Baldini N (1998) Blockage of insulin-like growth factor-I receptor inhibits the growth of Ewing's sarcoma in athymic mice. Cancer Res 58: 4127–4131 [PubMed] [Google Scholar]
- Scotlandi K, Benini S, Sarti M, Serra M, Lollini PL, Maurici D, Picci P, Manara MC, Baldini N (1996) Insulin-like growth factor I receptor-mediated circuit in Ewing's sarcoma/peripheral neuroectodermal tumour: a possible therapeutic target. Cancer Res 56: 4570–4574 [PubMed] [Google Scholar]
- Sekyi-Otu A, Bell RS, Ohashi C, Pollak M, Andrulis IL (1995) Insulin-like growth factor 1 (IGF-1) receptors, IGF-1, and IGF-2 are expressed in primary human sarcomas. Cancer Res 55: 129–134 [PubMed] [Google Scholar]
- Sharp PA (1999) RNAi and double-strand RNA. Genes Dev 13: 139–141 [PubMed] [Google Scholar]
- Siligan C, Ban J, Bachmaier R, Spahn L, Kreppel M, Schaefer KL, Poremba C, Aryee DN, Kovar H (2005) EWS-FLI1 target genes recovered from Ewing's sarcoma chromatin. Oncogene 24: 2512–2524 [DOI] [PubMed] [Google Scholar]
- Simons-Evelyn M, Bailey-Dell K, Toretsky JA, Ross DD, Fenton R, Kalvakolanu D, Rapoport AP (2001) PBK/TOPK is a novel mitotic kinase which is upregulated in Burkitt's lymphoma and other highly proliferative malignant cells. Blood Cells Mol Dis 27: 825–829 [DOI] [PubMed] [Google Scholar]
- Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, Lessnick SL (2006) Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 9: 405–416 [DOI] [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT (1994) A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet 6: 146–151 [DOI] [PubMed] [Google Scholar]
- Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, Lessnick SL, Kung AL, Golub TR (2007) Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med 4: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AD, Teitell MA, Arvand A, Denny CT (1999) Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumourigenic phenotype on NIH3T3 cells. Oncogene 18: 5506–5513 [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Plasterk RH (2002) The genetics of RNA silencing. Annu Rev Genet 36: 489–519 [DOI] [PubMed] [Google Scholar]
- Zhu F, Zykova TA, Kang BS, Wang Z, Ebeling MC, Abe Y, Ma WY, Bode AM, Dong Z (2007) Bidirectional signals transduced by TOPK-ERK interaction increase tumourigenesis of HCT116 colorectal cancer cells. Gastroenterology 133: 219–231 [DOI] [PubMed] [Google Scholar]
- Zupanska A, Adach A, Dziembowska M, Kaminska B (2007) Alternative pathway of transcriptional induction of p21WAF1/Cip1 by cyclosporine A in p53-deficient human glioblastoma cells. Cell Signal 19: 1268–1278 [DOI] [PubMed] [Google Scholar]
- Zwerner JP, Guimbellot J, May WA (2003) EWS/FLI function varies in different cellular backgrounds. Exp Cell Res 290: 414–419 [DOI] [PubMed] [Google Scholar]
- Zykova TA, Zhu F, Lu C, Higgins L, Tatsumi Y, Abe Y, Bode AM, Dong Z (2006) Lymphokine-activated killer T-cell-originated protein kinase phosphorylation of histone H2AX prevents arsenite-induced apoptosis in RPMI7951 melanoma cells. Clin Cancer Res 12: 6884–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.