Abstract
Oxysterol binding protein-related protein 2 (ORP2) is a member of the oxysterol binding protein family, previously shown to bind 25-hydroxycholesterol and implicated in cellular cholesterol metabolism. We show here that ORP2 also binds 22(R)-hydroxycholesterol [22(R)OHC], 7-ketocholesterol, and cholesterol, with 22(R)OHC being the highest affinity ligand of ORP2 (Kd 1.4 × 10−8 M). We report the localization of ORP2 on cytoplasmic lipid droplets (LDs) and its function in neutral lipid metabolism using the human A431 cell line as a model. The ORP2 LD association depends on sterol binding: Treatment with 5 μM 22(R)OHC inhibits the LD association, while a mutant defective in sterol binding is constitutively LD bound. Silencing of ORP2 using RNA interference slows down cellular triglyceride hydrolysis. Furthermore, ORP2 silencing increases the amount of [14C]cholesteryl esters but only under conditions in which lipogenesis and LD formation are enhanced by treatment with oleic acid. The results identify ORP2 as a sterol receptor present on LD and provide evidence for its role in the regulation of neutral lipid metabolism, possibly as a factor that integrates the cellular metabolism of triglycerides with that of cholesterol.
Keywords: cholesteryl ester, oxysterol, oxysterol binding protein, triglyceride
Oxysterols are oxygenated derivatives of cholesterol formed either enzymatically, mainly by the action of cytochrome P450 sterol hydroxylases or through nonenzymatic cholesterol autoxidation [reviewed in (1–4)]. Oxysterols are present in cells at very low amounts compared with cholesterol, but they are suggested to execute important signaling functions. Certain oxysterols activate liver X receptors, key transcriptional regulators of cholesterol homeostasis (5), and inhibit the processing of sterol regulatory element binding proteins (SREBPs) via binding to the Insig proteins, which retain SREBP/SCAP complexes in the endoplasmic reticulum (ER) (6). The cytosolic oxysterol receptor, oxysterol binding protein (OSBP), was identified in the 1980s (7). Families of OSBP-related proteins (ORPs) have recently been identified in practically all eukaryotic organisms studied (8). Most of the information on the ORP proteins has been obtained using yeast (Saccharomyces cerevisiae) or mammalian cells. The yeast ORPs (Osh proteins) are suggested to play major roles in the intracellular transport of sterols (9), in vesicle budding from the Golgi apparatus (10, 11), and in the establishment of cell polarity (12, 13), while mammalian ORPs have been suggested to participate in the regulation of lipid metabolism, vesicle transport, and cellular signaling (8).
All ORPs contain in their C-terminal part a structure designated OSBP-related domain (ORD), which is homologous to the oxysterol binding domain of OSBP (8). In addition to the ORD, most ORPs contain an N-terminal region involved in their subcellular targeting. The N-terminal extensions containing a pleckstrin homology domain target ORP1L to late endosomes (14), ORP3, -6, and -7 to the plasma membrane (15), and OSBP (16, 17) and ORP9 (18) to the Golgi complex. OSBP is targeted to Golgi complex in response to treatment of cells with its high-affinity ligand 25-hydroxycholesterol (25OHC). In several cases, the ORP pleckstrin homology domains are known to mediate the binding of phosphoinositides, but also the ORDs of ORP1, -2, -9, and -10 seem to be capable of binding these negatively charged phospholipids (19–21). A motif called FFAT (two phenylalanines in an acidic track), which mediates the binding to the ER resident protein VAP-A (for vesicle-associated membrane protein-associated protein), specifies association of several ORPs with the ER membranes (22).
Lipid droplets (LDs) are intracellular organelles composed of a neutral lipid core surrounded by a monolayer of phospholipids and cholesterol (23–25). In adipocytes, their main function is to store energy in the form of triglycerides (TGs), while in steroidogenic cells, LDs store cholesteryl esters (CEs) to be used as precursors for steroid hormone biosynthesis. In other cell types, LDs serve as storage organelles for fatty acids and cholesterol needed for cell membrane renewal and signaling purposes.
Whereas the role of ORPs in cholesterol metabolism has been addressed in a number of studies, their possible roles in TG metabolism have gained less interest. In the study of Yan and Olkkonen (8), overexpression of OSBP in mouse liver resulted in an increased amount of TG in liver tissue as well as enhanced secretion of TG into the circulation, apparently due to elevated activity of the lipogenic transcription factor SREBP-1c. On the other hand, overexpression of ORP2, a ubiquitously expressed ORP family member (26), was shown to decrease the amount of both TG and CE in Chinese hamster ovary (CHO) cells (27). In this study, we examine further the role of ORP2 in neutral lipid metabolism in the human A431 cell system. We show that ORP2 binds several oxysterols and localizes on the surface of intracellular LDs. Silencing of ORP2 affects the rate of TG hydrolysis and the degree of cholesterol esterification, thus suggesting that ORP2 acts as a sterol receptor involved in the integrated control of cholesterol and TG metabolism.
EXPERIMENTAL PROCEDURES
cDNA constructs, site-directed mutagenesis, short interfering RNAs, and antibodies
cDNAs for transient transfections were subcloned into the pcDNA4HisMaxC expression vector (Invitrogen, Carlsbad, CA), which encodes hexahistidine and Xpress™ epitope tags fused at the N terminus of the insert proteins. ORP2 cDNA engineered by PCR to remove the stop codon was also inserted in the BamHI site of pHcRed-N1 (BD Biosciences Clontech, San Jose, CA). For protein production in Escherichia coli, cDNAs were subcloned in the BamHI site of pGEX-1λT (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The full-length ORP open reading frames used in this study were ORP1L (AF323726), ORP2 (BC000296), and ORP3 (NM_015550). The glutathione S-transferase (GST)-MLN64 START domain construct was a kind gift from Prof. Elina Ikonen (Institute of Biomedicine, University of Helsinki, Finland).
The sequences of ORP2 short interfering RNA (siRNA) duplexes (Sigma Genosys, St. Louis, MO) were as follows: ORP2 siRNA 1, GGGAAGAUUUAGGAUUCAGtt (sense) and CUGAAUCCUAAAUCUUCCCtg (antisense); ORP2 siRNA 2, CGUAUGAAUUAAUCAGGGAtt (sense) and UCCCUGAUUAAUUCAUACGtt (antisense); ORP2 siRNA 3, GGACACAUUCAAGACAAAAtt (sense) and UUUUGUCUUGAAUGUGUCCtt (antisense); and ORP2 siRNA 4, GGACCGGCAAACCAUUUAAtt (sense) and UUAAAUGGUUUGCCGGUCCtc (antisense). The sequences of control siRNA duplexes were UAGCGACUAAACACAUCAATT (sense) and UUGAUGUGUUUAGUCGCUATT (antisense). The rabbit ORP2 antibody was described in (26). A monoclonal mouse antibody (mab) against the Xpress™ epitope tag was purchased from Invitrogen, the mab against GM-130 from BD Biosciences-Transduction Laboratories (San Jose, CA), the mab against transferrin receptor from Zymed (San Francisco, CA), the mab (clone H4A3) against Lamp1 from Developmental Studies Hybridoma Bank (Iowa City, IA), and the mab against GAPDH from Novus Biologicals (Littleton, CO). Alexa-labeled fluorescent goat secondary antibody conjugates were from Invitrogen.
Cell culture
The human epithelial carcinoma cell line A431 was grown in a 5% CO2 atmosphere in DMEM with 4.5 g/l glucose and sodium pyruvate (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (Invitrogen-Gibco BRL, Carlsbad, CA), 100 IU/ml penicillin,and 100 μg/ml streptomycin at 37°C for a maximum of 30 passages. The cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNA-mediated gene silencing was carried out with HiPerFect™ (Qiagen) according to the manufacturer's instructions using the fast-forward transfection protocol. LDs were induced in A431 cells by overnight treatment with 350 μM oleic acid-BSA complexes prepared according to (28). Stably transfected ORP2 HeLa TRex cells were cultured and induced with doxycyclin as previously described (21).
Fluorescence microscopy
A431 cells were fixed with 3% (w/v) paraformaldehyde in PBS for 30 min. Permeabilization of the cells and the blocking of nonspecific binding of antibodies were done simultaneously in PBS containing 0.5% BSA and 0.1% saponin (blocking buffer) for 30 min. Primary antibodies and secondary antibody conjugates were diluted in blocking buffer and incubated for 1 h at room temperature. When LDs were stained, 50 μM Bodipy 493/503 (Invitrogen-Molecular Probes, Carlsbad, CA) was added in the secondary antibody dilution. Washes after the antibody incubations were done three times with blocking buffer. Finally, the cover slips were washed three times with PBS and once with water, after which they were mounted in Mowiol 4-88 (Sigma-Aldrich) containing 2.5% 1,4-diazabicyclo[2.2.2]octane (Sigma-Aldrich). When antibody incubations were not required, cells were stained with 50 μM Bodipy 493/503 and with 4′,6-diamidino-2-phenylindole for 30 min before fixation. Images were acquired with Zeiss Meta or Leica TCS SP1 confocal microscopes.
Immunoelectron microscopy
For immunoelectron microscopy, A431 cells were transfected with ORP2-pcDNA4HisMaxC and loaded with 350 μM oleic acid-BSA complexes overnight. The cells were then fixed with 4% paraformaldehyde, scraped, and pelleted in gelatin. Polyvinylpyrrolidone/sucrose-infiltrated specimens were sectioned at −110°C and picked up in methyl cellulose-sucrose. The sections were treated with 1% fish skin gelatin (Sigma-Aldrich) and 1% BSA and incubated with the ORP2 antibody (1:70) for 1 h. Secondary antibody (Protein A-10 nm gold; University of Utrecht, The Netherlands) was applied for 1 h, and sections were embedded in uranyl acetate-methyl cellulose and examined with a Jeol 1200 EX II electron microscope.
Protein purification
GST fusion proteins of ORP2 and MLN64 (Metastatic lymph node 64) START (Steroidogenic acute regulatory protein-related lipid transfer) domain were produced in E. coli BL21(DE3) and purified on glutathione sepharose 4B (GE Healthcare) according to the manufacturer's instructions. Protein concentrations of the specimens were determined by the DC assay (Bio-Rad, Hercules, CA). Before oxysterol binding experiments, the protein preparations were resolved on Laemmli gels, which were stained with Coomassie blue, scanned, and analyzed using Scion Image software (http://www.scioncorp.com/). According to this, adjustment of the protein amounts added was performed to ensure that the desired concentrations of the full-length fusion proteins were reached.
Charcoal-dextran oxysterol binding assay
Binding of [3H] labeled 7-ketocholesterol (65 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO), 25-hydroxycholesterol (20 Ci/mmol), 22(R)hydroxycholesterol (20 Ci/mmol; American Radiolabeled Chemicals), or 27-hydroxycholesterol (45 Ci/mmol; a gift from Prof. Ingemar Björkhem, Karolinska Institute, Huddinge, Sweden) to the purified GST-ORP2 was assayed as described previously (29). Briefly, proteins (0.25 or 1 μM) were incubated overnight at +4°C with 5, 10, 20, 40, and 80 nM (in some cases up to 160 nM) [3H]oxysterol in the absence or presence of a 40-fold excess of the corresponding unlabeled oxysterol (purchased from Sigma-Aldrich; except 27-hydroxycholesterol, which was from I. Björkhem). The free sterol was thereafter removed with charcoal-dextran, and the protein-bound [3H]sterol remaining in the supernatant was analyzed by liquid scintillation counting. Kd values were determined by Scatchard analysis. The data were normalized for specific radioactivity of the different labeled oxysterols.
Cholesterol binding assay
To be able to study the interaction of ORP2 and cholesterol in solution, cholesterol was complexed with methyl-β-cyclodextrin (mβCD; Sigma-Aldrich) according to (30). Briefly, 1 mg of mβCD was dissolved in 1 ml of binding buffer (10 mM HEPES, pH 7.4, 50 mM KCl, and 0.02% NP-40) and added on the top of the film of cholesterol and [3H]cholesterol (44 Ci/mmol; GE-Healthcare-Amersham) in a glass tube. The mixture was then probe sonicated 3 × 5 min and microcentrifuged at full speed for 15 min. For the cholesterol binding assay, 50 μg of GST (negative control), GST-ORP2, or GST-MLN64 START (positive control) were bound to glutathione sepharose beads, which were then incubated with cholesterol-mβCD (330 ng cholesterol per assay) for 30 min at room temperature. Competition of the cholesterol binding was tested using a 40-fold molar excess of 22(R)-hydroxycholesterol [22(R)OHC]. After the binding, the beads were washed and the radioactivity of the supernatant, the washes, and the pellet was measured by liquid scintillation counting.
Analysis of TG synthesis and breakdown
A431 cells on six-well plates were treated with siRNAs as described above. In the case of TG synthesis assay, 350 μM oleic acid-BSA complexes containing [3H]oleic acid (5 μCi per dish; 7 Ci/mmol; GE Healthcare-Amersham) were added to the cells 40 h after the transfection. The cells were harvested at different time points in 2% NaCl. Lipids were extracted using the Bligh-Dyer protocol, and the extracts were separated by TLC using hexane/diethyl ether/acetic acid/water (65:15:1:0.25). The TLC plates were stained with iodine, the TG spots identified by comigration with a triolein standard were scraped off, and the radioactivity was measured with a liquid scintillation counter. Protein concentrations of the cell specimens were measured with the DC Protein Assay (Bio-Rad), and the results were corrected with the protein amounts. In the case of TG breakdown assay, similar [3H]oleic acid-oleic acid-BSA complexes were added to cells after 24 h of transfection. After 18 h, the cells were washed three times with PBS, and the loading/labeling medium was replaced with chase medium (DMEM, 5% delipidated serum). The cells were harvested at several time points and treated further as in the TG synthesis assay.
Assay for [14C]cholesterol esterification
A431 cells on 35 mm dishes were treated with siRNAs as described above. After 6 h [14C]cholesterol (0.2 μCi/well; 58 mCi/mmol; GE Healthcare-Amersham) was added to the cells for 40 h. To analyze the distribution of the [14C]label between free cholesterol and CEs, the labeled cells were harvested in 2% NaCl and the lipids extracted using the Bligh-Dyer protocol. Aliquots of the cells were used for protein determination (see above). The neutral lipids were separated by TLC using petrolether/diethyl ether/acetic acid (60:40:1). The TLC plates were stained with iodine, the cholesterol and CE spots identified based on comigration with unlabeled standards were scraped off, and the radioactivity was measured with a liquid scintillation counter.
RESULTS
ORP2 binds several oxysterols with different affinities
We recently reported that ORP2 binds 25OHC with micromolar affinity (31). We now wanted to study the oxysterol binding properties of ORP2 more extensively. For this, we employed a charcoal-dextran pull-down assay of [3H]oxysterols with GST-tagged ORP2 produced in E. coli as previously. To determine the specific binding, we performed the assays in the absence or presence of 40-fold excess of unlabeled oxysterol, and the nonspecific binding was subtracted from the total binding (Fig. 1A). In these experiments 22(R)OHC was bound by ORP2 with the highest affinity (Kd = 1.4 × 10−8 M), followed by 7-ketocholesterol (Kd = 1.6 × 10−7 M) and 25OHC, which displayed weak but reproducible binding to the protein. However, no specific binding of 27-hydroxycholesterol was detected. We also tested the oxysterol binding properties of a mutant ORP2 (ORP2 I249W) previously shown to be defective in 25OHC binding (31). This mutant also failed to bind 22(R)OHC (Fig. 1A). From these assays, we conclude that ORP2 binds several oxysterols with different affinities, the highest affinity ligand so far being 22(R)OHC.
Fig. 1.
Binding of oxysterols and cholesterol by ORP2. A: Specific oxysterol binding. Different concentrations of [3H]oxysterols were incubated overnight with 1 μM GST-wild-type (wt) ORP2 or the mutant ORP2 I249W (indicated on the top). Unbound oxysterols were pulled down with charcoal-dextran, and the radioactivity in the supernatant was determined. Specific binding was determined by subtracting the values obtained in the presence of 40-fold excess of unlabeled oxysterol from those obtained with the [3H]sterol only. The data represent mean ± SEM (n = 4, except for 27OHC, n = 2). B: Cholesterol binding by ORP2. Fifty micrograms of GST-ORP2, GST (negative control), or the GST-tagged START domain of MLN64 (positive control) was bound to glutathione-sepharose beads and incubated with [3H]cholesterol-methyl-β-cyclodextrin complexes, after which the beads were pulled down. The bars represent the percentage of the radioactivity associated with the beads (mean ± SEM; n = 6; ***, P < 0.001, t-test). Competition of cholesterol binding by 22(R)OHC was determined by including a 40-fold excess of unlabeled 22(R)OHC in the assay.
ORP2 binds cholesterol
In a previous study, we presented data suggesting that overexpressed ORP2 enhances the intracellular transport of cholesterol (21). Therefore, we wanted to study whether ORP2 might, in addition to oxysterols, bind cholesterol. Since cholesterol is markedly more hydrophobic than oxysterols, we designed a cholesterol binding assay in which [3H]cholesterol is solubilized using methyl-β-cyclodextrin and subjected to pull down with GST-tagged recombinant proteins. The START domain of the cholesterol binding protein MLN64 was used as a positive (32) and GST as a negative control. In this assay, GST-ORP2 showed significant cholesterol binding over the background signal of GST. However, the binding was modest compared with that of the GST-MLN64 START domain (Fig. 1B). The specificity of the binding was tested by adding a 40-fold excess of unlabeled 22(R)OHC in the assay. The cholesterol binding was efficiently competed by the oxysterol, suggesting that the binding is specific and occurs in the same pocket as that of the oxysterols, as shown for the homologous yeast protein Osh4p (33).
ORP2 localizes to the surface of intracellular LDs
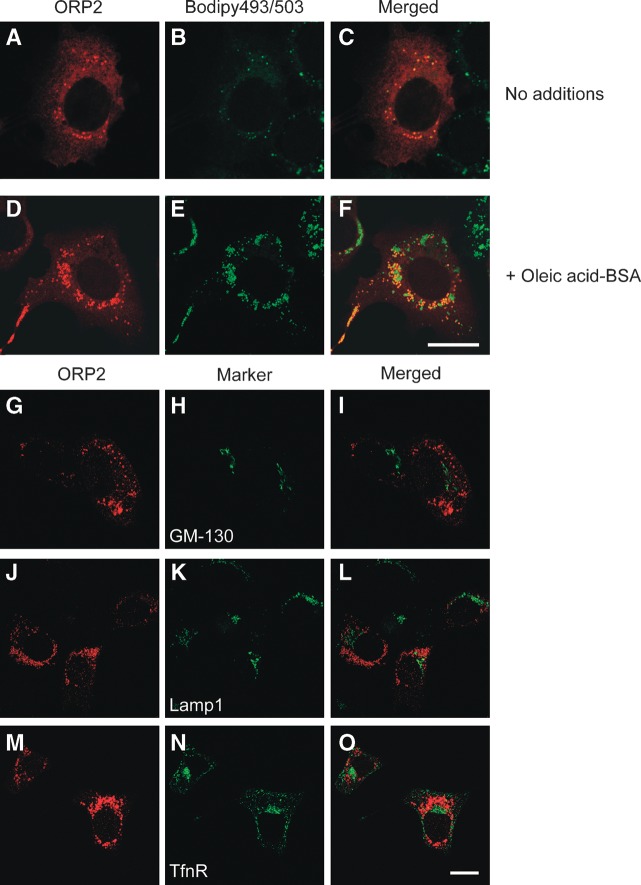
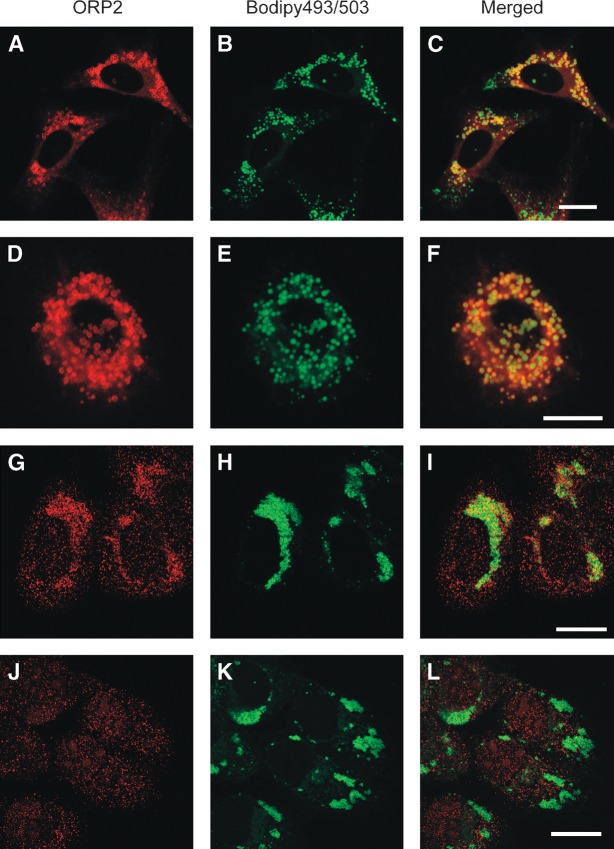
The subcellular localization of ORP2 has so far been unclear. The data of Laitinen et al. (26) suggested partial Golgi localization of human ORP2 in CHO cells, but when overexpressed in several human cell lines under standard culture conditions, the human ORP2 was diffusely distributed throughout the cytoplasm with no sign of Golgi association (Fig. 2A–C; data not shown). In this study, Xpress epitope-tagged ORP2 was expressed in the human epithelial carcinoma cell line A431 with abundant endogenous ORP2 expression. In the normal growth medium with 10% FBS ORP2 displayed a relatively even staining of the cytoplasmic compartment with additional small dot- or ring-like ORP2-positive structures. These structures were also found to be stained with Bodipy 493/503, a neutral lipid stain, suggesting that they may represent cytoplasmic LDs (Fig. 2A–C). When the cells were treated overnight with 350 μM oleic acid-BSA to enhance lipogenesis and LD formation, prominent ORP2 localization on Bodipy 493/503-positive spherical structures was evident (Fig. 2D–F). The spherical structures were not stained with antibodies recognizing the Golgi (GM130; Fig. 2G–I), lysosomes (Lamp1; Fig. 2J–L), or early/recycling endosomes (transferrin receptor; Fig. 2M–O). We thus concluded that ORP2 localizes on the surface of neutral LDs. The LD association was not unique for A431 cells: Overexpressed ORP2 displayed LD association also in other cell lines, such as HeLa, HepG2, and Cos7 (data not shown). ORP2 was also seen on the LD of stably transfected HeLa TRex cells induced to overexpress untagged ORP2 (21) (Fig. 3A–C), demonstrating that the localization is not specific for the Xpress epitope-tagged protein. Furthermore, ORP2 with the fluorescent HcRed protein fused at its C terminus displayed a similar LD localization, showing that the LD distribution is not a result from an artifactual association that could arise during the immunofluorescence staining protocol (Fig. 3D–F). This observation also demonstrates that modification of the C terminus of ORP2 does not interfere with the LD localization.
Fig. 2.
ORP2 localizes to the surface of intracellular LDs. Xpress-tagged ORP2 was expressed in A431 cells grown in the absence (A–C) or presence (D–F) of 350 μM oleic acid-BSA complexes. ORP2 (shown in red in the left-most panels) was stained with rabbit polyclonal ORP2 antibodies and the LDs with Bodipy 493/503 (in green in the center panels). The staining of ORP2 does not colocalize with other organelle markers (G–O). ORP2 was expressed in A431 cells grown in the presence of 350 μM oleic acid-BSA. ORP2 was stained with a rabbit antibody (in red in the left-most panels), the Golgi apparatus with GM-130 antibody (H), lysosomes with Lamp1 antibody (J), and early/recycling endosomes with transferrin receptor (TfnR) antibody (N). Merged images are shown on the right. Bars = 10 μM.
Fig. 3.
Specificity of ORP2 LD localization. A–C: Doxycyclin-induced TREx HeLa cells expressing untagged ORP2 were grown in the presence of 350 μM oleic acid-BSA complexes. ORP2 was stained with polyclonal ORP2 antibodies (A) and LD visualized with Bodipy 493/503 (B). D–F: HcRed-tagged ORP2 (D) was expressed in A431 cells treated with 350 μM oleic acid-BSA, and the LDs were stained with Bodipy 493/503 (E) in the absence of detergent permeabilization of the cells. G–L: The endogenous ORP2 in A431 cells treated with 350 μM oleic acid-BSA was stained with polyclonal rabbit ORP2 antibodies (G) or with the same antibodies preincubated with GST-ORP2 to inhibit specific immunoreactivity (J). LDs were stained with Bodipy 493/503 (H and K). Merged images are shown on the right. Bars = 10 μM.
Importantly, a faint staining of endogenous ORP2 with the rabbit antibody could be seen in the area of LD accumulation in nontransfected, oleic acid-loaded A431 cells, providing evidence that the endogenous ORP2 also associates with the LD (Fig. 3G–I). The endogenous staining appeared punctuate, being thus somewhat different from the ring-like LD staining observed for the overexpressed ORP2. This staining vanished when the ORP2 antibody was preincubated with purified GST-ORP2 (Fig. 3J–L), confirming the specificity of the immunoreactivity.
To test the specificity of ORP2 LD association, ORP1L and ORP3 were transfected in A431 cells loaded with oleic acid. As previously reported for other cell lines (14, 15), ORP1L localized to late endo/lysosomal structures (see supplementary Fig. IA–C) and ORP3 to the ER and the cytosol (see supplementary Fig. 1D–F). No colocalization of these proteins with Bodipy 493/503 was detected, suggesting that the LD association is specific for ORP2.
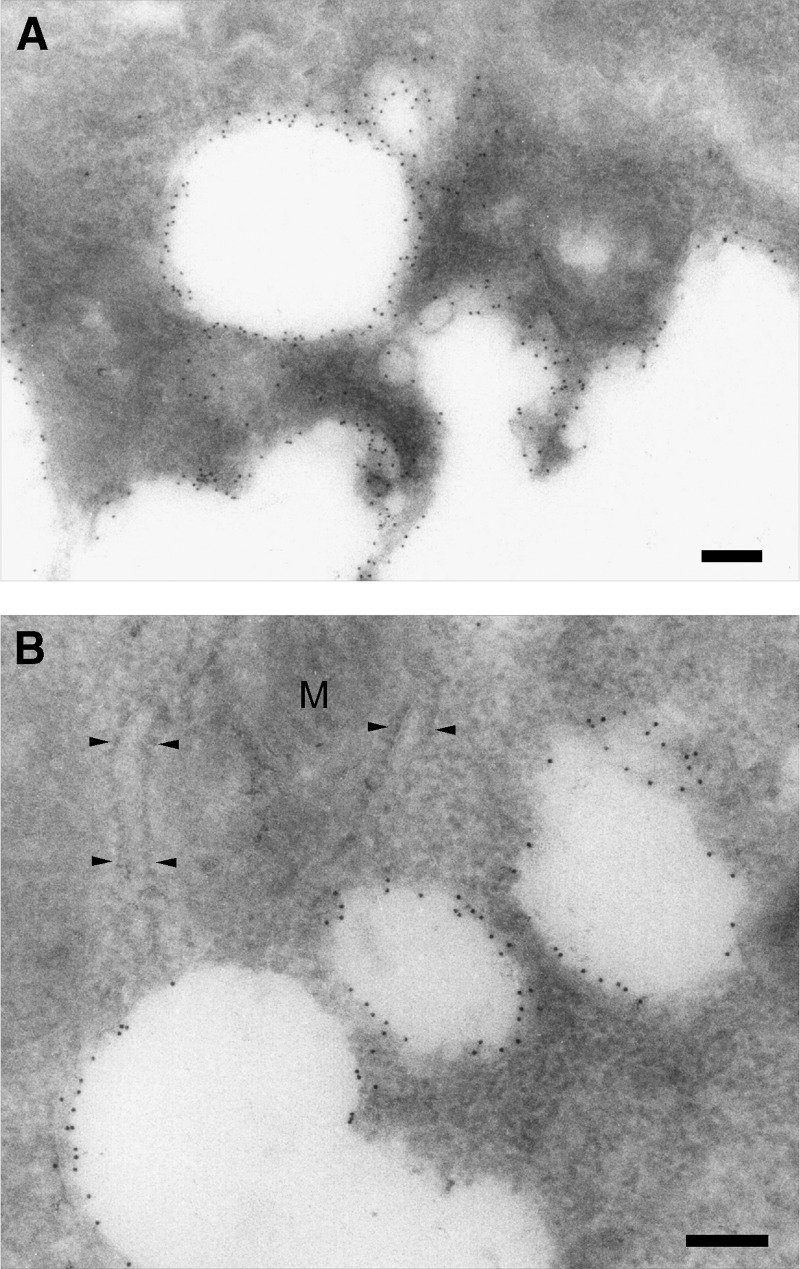
Immunoelectron microscopic localization of ORP2
To investigate the ORP2 localization at the ultrastructural level, we carried out cryosection immunoelectron microscopy of transfected A431 cells using the polyclonal rabbit antibody. Immunogold was detected lining LDs, while specific staining of other membrane structures was not detectable (Fig. 4). Even though the FFAT motif of ORP2 could specify ER targeting (22), no significant labeling of ER elements was observed. Furthermore, immunolabeling was prominent on LD surfaces at which no closely associated ER membrane was evident. This observation is consistent with the fact that no significant ER association of ORP2 was detected in immunofluorescence experiments (data not shown).
Fig. 4.
Immunoelectron microscopy confirms ORP2 localization on the surface of LDs. ORP2 was expressed in A431 cells treated with 350 μM oleic acid-BSA, and the cells were processed for cryo-immunoelectron microscopy using ORP2 antibodies. Abundant immunogold labeling of LD surfaces (white areas) was observed (A). No label was seen over the ER (arrowheads in B); only occasional individual gold particles were found associated with ER membranes in close vicinity of LDs. M, mitochondrion. Bars = 200 nm.
ORP2 overexpression causes dispersal of LDs
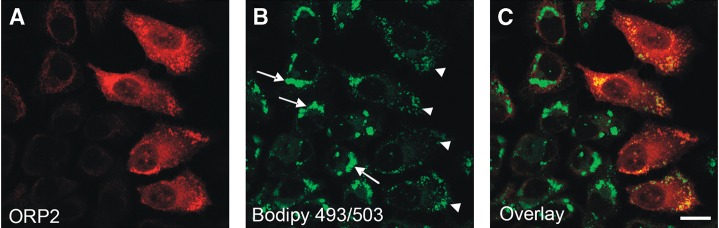
The LD in A431 cells are typically clustered at a juxtanuclear location. Fluorescence microscopy observation of oleic acid-loaded cells revealed that the distribution of the LD in cells overexpressing ORP2 was different from the neighboring untransfected cells. The droplets were no longer clustered but were dispersed throughout the cytoplasm (Fig. 5). Even though the present observation was made under overexpression conditions, it suggests that ORP2 is capable of modulating the LD organization, which could impact the metabolism of neutral lipids in these organelles.
Fig. 5.
ORP2 overexpression causes a dispersal of LDs. A431 cells transfected with the ORP2 cDNA were treated overnight with 350 μM oleic acid-BSA, fixed, and processed for immunofluorescence microscopy using rabbit ORP2 antibodies. LD clusters in untransfected cells are indicated with arrows and dispersed LD in ORP2 overexpressing cells with arrowheads. A: Staining for ORP2. B: LDs visualized with Bodipy 493/503. C: Overlay of the channels. Bar = 10 μm.
22(R)hydroxycholesterol inhibits the LD association of ORP2
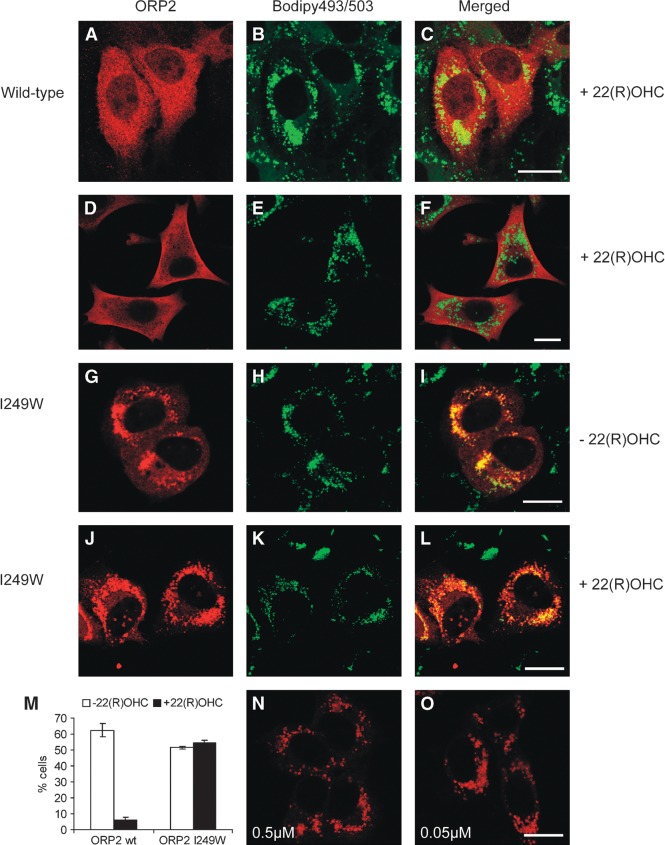
The addition of oxysterols to cells has been shown to affect the localization of OSBP (17) and an ORP3:OSBP chimera (15). We wanted to test if the same applies to ORP2. A431 cells overexpressing ORP2 were treated overnight with 350 μM oleic acid-BSA to induce LD formation in the presence of 5 μM 22(R)OHC. The association of ORP2 with LDs was clearly inhibited in the oxysterol-treated cells. On the vehicle (ethanol) treated control cover slips, ORP2 was associated with LDs in 63% of the transfected cells, while only 6% of the cells on 22(R)OHC-treated cover slips displayed LD association, the ORP2 showing a diffuse, often dotty cytoplasmic distribution (Fig. 6A–C; quantification in 6M). The same effect was detected in stably transfected HeLa cells induced to express ORP2: The 22(R)OHC treatment abolished the LD association and led to an altered distribution of ORP2 with a prominent plasma membrane aspect (Fig. 6D–F). To investigate whether this effect is dependent on the oxysterol binding by ORP2, we expressed the oxysterol binding defective mutant I249W in A431 cells subjected to the same treatments as above. In the absence of oxysterol, the mutant associated with the LD similar to the wild type protein (Fig. 6G–I, M). However, in contrast to wild-type ORP2, addition of 22(R)OHC did not affect the LD association of ORP2 I249W (54% of the cells; Fig. 6J–L, M).
Fig. 6.
22(R)OHC inhibits the LD localization of ORP2. A–C: A431 cells were transfected with Xpress-tagged ORP2 and treated with 350 μM oleic acid-BSA and 5 μM 22(R)OHC overnight. ORP2 was stained with ORP2 antibody (A) and LDs with Bodipy 493/503 (B). D–F: Doxycyclin-induced TREx HeLa cells expressing ORP2 were treated with oleic acid-BSA and 22(R)OHC as above. G–L: Oxysterol binding deficient mutant ORP2 I249W was expressed in A431 cells that were treated with oleic acid-BSA in the absence (G–I) or presence (J–L) of 5 μM 22(R)OHC and stained as above. M: Quantification of the localization data. Wild-type ORP2 (wt) or ORP2 I249W were expressed in A431 cells grown in the presence of 350 μM oleic acid-BSA. The cells were treated with the vehicle (ethanol; white bars) or 5 μM 22(R)OHC (black bars) overnight and stained as above. The LD association of ORP2 (three cover slips analyzed) is expressed as a proportion of cells displaying clear ORP2 LD localization (mean ± SEM, 100 transfected cells counted from each cover slip). N, O: ORP2 expressing oleic acid-BSA treated cells were incubated with 0.5 μM [N] or 0.05 μM (O) 22(R)OHC. Only the ORP2 channel (red) is shown. Bars = 10 μm.
When testing the concentration dependency of the 22(R)OHC effect on ORP2 LD localization, we found that 0.05 μM 22(R)OHC was not able to inhibit the LD association, and 0.5 μM oxysterol had a mild effect detectable in some cells (Fig. 6N, O). It is hard to estimate the effective intracellular oxysterol concentration reached; the 0.05 μM and possibly also the 0.5 μM concentration may be insufficient to reach full ligand occupancy in live cells. As expected, the other oxysterols displaying lower or no detectable affinity for ORP2, 7-ketocholesterol, 25OHC, and 27OHC were unable to inhibit the ORP2 LD association at 5 μM concentration (data not shown). The data suggest that ligand binding to ORP2 is capable of inducing a conformational change that inhibits the association of the protein with LD surface.
ORP2 silencing slows down the TG breakdown
To silence ORP2 expression in A431 cells, four different siRNAs were designed. Of these, ORP2 siRNAs 1, 2, and 4 showed the highest silencing efficiency compared with a control siRNA (Fig. 7A) and were used for further studies. The effect of ORP2 silencing on LD morphology was assessed. A431 cells were treated with siRNAs for 72 h in 10% fetal calf serum (FCS) and oleic acid-containing medium, after which the LD were stained with Bodipy 493/503. ORP2 silencing appeared to result in clearly increased intensity of perinuclear LD staining as compared with control siRNA-treated cells (Fig. 7B, left panels). When the cells were first treated for 48 h with siRNAs in 10% FCS and oleic acid containing medium and then chased for 24 h in medium containing 5% delipidated FCS to induce TG breakdown, the effect of ORP2 silencing became clear. Cells treated with control siRNA lost nearly all the LDs as judged by Bodipy 493/503 staining, but the ORP2 siRNA-treated cells still had clearly visible LDs (Fig. 7B, right panels), suggesting that the silencing of ORP2 stabilizes the LD and slows down the mobilization of neutral lipids.
Fig. 7.
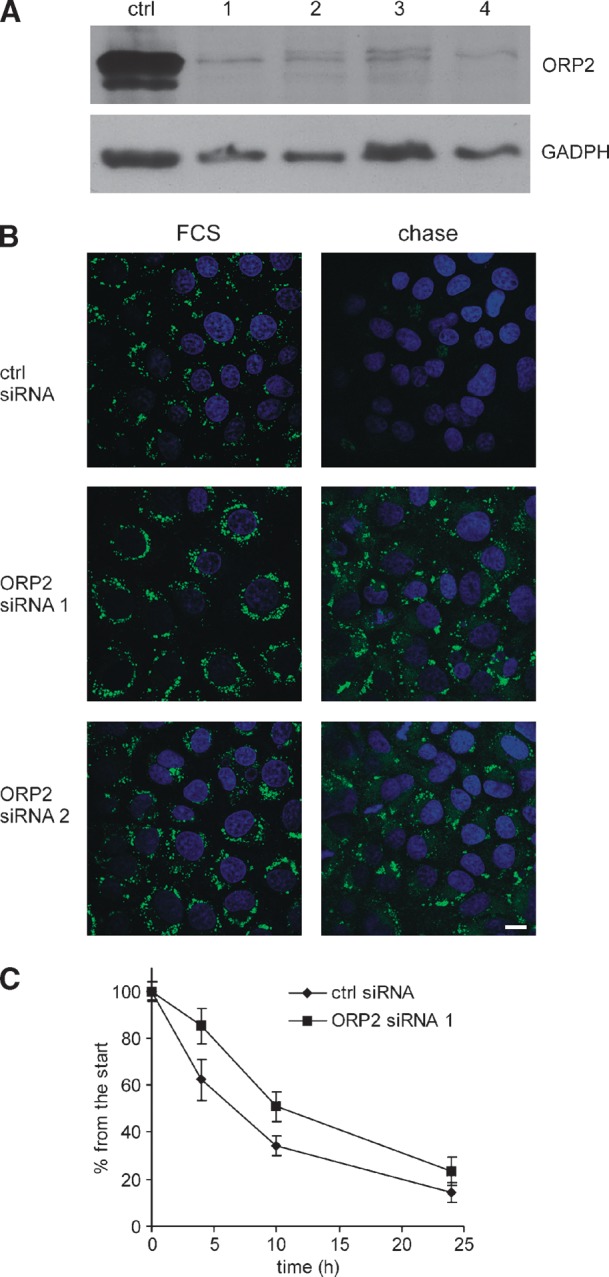
Silencing of ORP2 stabilizes cellular TGs. A: Lysates (20 μg of total protein) of A431 cells treated with a control and four ORP2 specific siRNAs (1–4, indicated on the top) were analyzed by Western blotting. ORP2 was detected with polyclonal rabbit antibodies, and GADPH antibody was used to control for equal protein loading. B: A431 cells were either treated with siRNAs for 72 h in 10% FCS and oleic acid-containing medium (panels labeled “FCS”) or treated for 48 h with siRNAs in 10% FCS and oleic acid-containing medium and then chased for 24 h in medium containing 5% delipidated FCS (panels labeled “chase”). The cells were fixed and stained with Bodipy 493/503 and 4′,6-diamidino-2-phenylindole to visualize LDs and nuclei, respectively. Bar = 10 μm. C: A431 cells were treated with control siRNA or ORP2 siRNA 1 and subjected to a 16-h treatment with 350 μM [3H]oleic acid-BSA, after which they were chased in medium containing 5% delipidated serum and harvested at different time points. The amount of [3H]TG was determined and corrected for the amount of total cell protein. The figure shows the percentage of [3H]TG of that at the zero time point and represents the mean ± SEM of three independent experiments.
To investigate the phenomenon using a biochemical approach, A431 cells were treated with control or ORP2 siRNAs and labeled for 16 h with a mixture of unlabeled and [3H]oleic acid bound to BSA. The cells were then chased in a medium containing delipidated serum and analyzed at different time points. The [3H]TGs were quantified and the results normalized for cell protein. After the labeling period (the zero time point) the amount of [3H]TG was 29 ± 15% higher in the cells subjected to ORP2 silencing. During the chase, the amount of [3H]TG declined more slowly in ORP2 silenced cells than in the controls (presented as percentage of the DPM/mg protein at the start of chase for ORP2 siRNA 1 in Fig. 7C). Similar results were obtained with the other ORP2 siRNAs tested (siRNAs 2 and 4; see supplementary Fig. II). ORP2 silencing had no significant effect on [3H]oleic acid incorporation into TG during a 3-h time course (data not shown). The results suggest that the endogenous ORP2 facilitates the breakdown of TG.
ORP2 silencing increases CEs in oleic acid-loaded cells
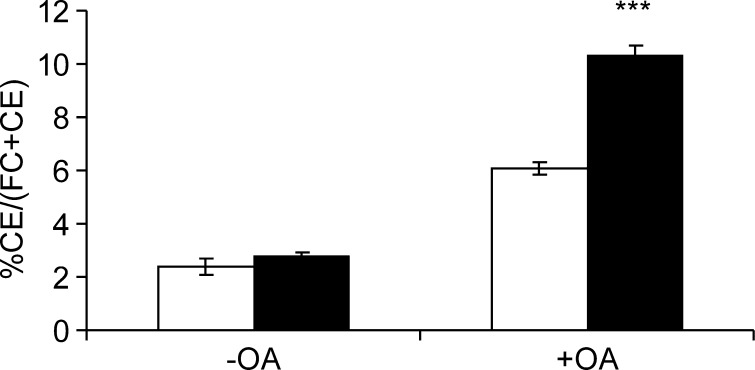
In addition to TGs, LDs store cholesterol in the form of CEs. Since there are suggested connections between the metabolism of TG and CE in LDs (34, 35), we wanted to study the effect of ORP2 silencing on cholesterol esterification in A431 cells in normal growth medium or under oleic acid loading conditions. The cells were treated for 48 h with ORP2 or control siRNAs and concomitantly labeled for 40 h with [14C]cholesterol. Part of the cell specimens were treated overnight with 350 μM oleic acid-BSA. The [14C]CE radioactivity as percentage of total cellular [14C]cholesterol was determined. In the normal growth conditions (DMEM, 10% FBS), ORP2 silencing had no effect on the quantity of [14C]CE (Fig. 8). When TG synthesis and LD formation were enhanced by overnight treatment with oleic acid-BSA, a difference between cells treated with ORP2 and control siRNAs became evident. The amount of [14C]CE in oleic acid-loaded cells subjected to ORP2 silencing was 87% higher than in the controls. The results were the same irrespective of whether they were calculated as percentage [14C]CE of total [14C]cholesterol or CE radioactivity per mg cell protein (data not shown). Notably, the amount of [14C]CE was also increased in control siRNA treated cells upon oleic acid-BSA treatment, probably due the fact that oleic acid is a good substrate for ACAT (36, 37).
Fig. 8.
Silencing of ORP2 increases the amount of CEs in the presence of elevated TG levels. Control siRNA (shown in white) or ORP2 siRNA 1 (in black) treated A431 cells were labeled with [14C]cholesterol for 40 h and grown for 16 h in the presence (+OA) or absence (−OA) of 350 μM oleic acid-BSA. Lipids were extracted and resolved by TLC, and the distribution of [14C]cholesterol between free cholesterol and CEs was determined. The results show the percentage of [14C]CEs of the total [14C]cholesterol and represent a mean (± SEM) of three experiments (n = 3; ***, P < 0.001, t-test).
DISCUSSION
The starting point of this study is the novel finding that ORP2 localizes on intracellular LDs. The LD localization was prominent when the cells were loaded with oleic acid-BSA complexes, but ORP2 could be detected on LDs also in unloaded cells. The localization was detected in a number of human cell lines, and its specificity was controlled for in several ways. Although ORP2 localizes to the LD in every cell model we have studied, the protein has not been identified as a LD component in the studies identifying the mammalian LD proteome (38–41). Consistently, we were unable to detect endogenous ORP2 in LD fractions of A431 cells using the sucrose gradient isolation procedure described in (42) (data not shown). We find it likely that ORP2 readily dissociates from the LD during the gradient centrifugation procedures used for LD isolation.
We identified 22(R)OHC as the highest affinity ligand of ORP2 among the oxysterols tested, the Kd 1.4 × 10−8 M being similar to that of OSBP for 25OHC, 8–10 × 10−9 M (43, 44). Addition of 22(R)OHC to cells inhibited the LD association of ORP2; on the other hand, a mutant ORP2 (I249W) defective in sterol binding was constitutively LD bound even in the presence of 22(R)OHC. Although the present findings do not allow us to draw firm conclusions of how ligand occupancy modulates ORP2 function, they demonstrate that ligand binding by ORP2 has the capacity to regulate the localization of the protein. A shift of subcellular localization has also been described for OSBP, which resides in the cytosol or cytoplasmic vesicles and in the ER in the absence of oxysterol ligand and translocates to Golgi membranes when 25OHC is added (17). Thereby ligand binding activates OSBP function in ceramide transport from the ER to the Golgi complex, resulting in enhanced sphingomyelin synthesis (45). One interpretation of our data is that ORP2 differs from OSBP in its functional regulation by ligand. The effects of ORP2 silencing on neutral lipid metabolism would be consistent with a function at the LD surface, and ligand binding could in this case remove the protein from its site of function. However, we do not know whether ORP2 on the LD surface is unliganded or occupied by an unknown endogenous cellular ligand that might be displaced by the added 22(R)OHC. The fact that the mutant ORP2 I249W localizes constitutively on the LD indicates that unliganded ORP2 may be capable of LD association. Liganding of OSBP by cholesterol or 25OHC has distinct effects on OSBP function in modulating the phosphorylation and activity of extracellular signal-regulated kinases: Cholesterol-OSBP facilitates dephosphorylation of the extracellular signal-regulated kinase, while the oxysterol-bound OSBP fails to do this (46). The present data leave open the interesting question whether interaction of ORP2 with cholesterol or oxysterols might similarly result in different functional outcomes. Detailed characterization of the functional role of ORP2 ligand occupancy remains a topic for future studies.
A common property of a number of LD proteins is the ability to shift dynamically between LD and the ER (47, 48) or the plasma membrane (49, 50) induced by changes in cellular fatty acid or cholesterol supply. Our data suggest that ORP2 behaves in a similar fashion. Known LD proteins do not seem to have any common LD targeting motif. Small Rab GTPases associate with LD via a C-terminal hydrophobic isoprenoid moiety. Caveolin-1, NADH sterol dehydrogenase-like protein, and diacylglycerol acyl transferase 2 are inserted into the LD surface and core lipid via a long hydrophobic segment most likely forming a hairpin-like structure [reviewed in (25)]. The exact determinants for LD targeting of the perilipin-adipophilin-TIP47 family proteins, major LD surface components, are even today somewhat controversial. However, these proteins appear to use amphipatic α-helices for binding to the LD surface phospholipid monolayer (25, 51). Based on the present data, the molecular model of ORP2 structure (31), and the high-resolution structure of Osh4p (33), we envision that the ORP2 lid region and N terminus regulate the LD association in a ligand-dependent manner. Whether the ability of ORP2 to bind acidic phospholipids (21) plays a role in the LD targeting is an interesting issue to be addressed in future work.
What is the function of ORP2? The present finding that ORP2 binds cholesterol suggests that the previously observed enhanced cholesterol transport in cell overexpressing ORP2 (21, 26) may be due to an excess of exogenous cytosolic ORP2 capable of transferring cholesterol between membranes. We now believe that the major function of ORP2 occurs on LD and involves the metabolism of TGs and CEs. The silencing of ORP2 resulted in a decreased rate of TG hydrolysis under lipid depletion conditions, suggesting that the endogenous ORP2 facilitates the breakdown of TG in this situation. Furthermore, in oleic acid-loaded cells, ORP2 knockdown resulted in increased cellular [14C]CE in radiolabeling experiments. These findings are in accordance with previous results demonstrating a decrease in both cellular TG and CE in CHO cells overexpressing ORP2 (27). The mechanism by which ORP2 could contribute to lipolysis is at present not known; it could plausibly act to facilitate the hydrolytic activity of the major lipases acting sequentially on the LD TGs, adipocyte triglyceride lipase, hormone-sensitive lipase, and monoacylglycerol lipase (52). We made attempts to elucidate by coimmunoprecipitation whether ORP2 might interact physically with adipocyte triglyceride lipase present abundantly in A431 cells, but the results were not conclusive (data not shown).
This study forms a starting point for investigation of the detailed mechanisms by which ORP2 impacts on neutral lipids. Our results identify ORP2 as a sterol receptor present on LD and provide evidence for its role in the regulation of neutral lipid metabolism, putatively as a factor that integrates the cellular metabolism of TGs with that of cholesterol.
Supplementary Material
Acknowledgments
The authors are grateful to Anna Molenda-Radzikowska, Seija Puomilahti, and Pirjo Ranta for expert technical assistance, to Prof. Elina Ikonen for the GST-MLN64 START domain expression construct, to Prof. Ingemar Björkhem for kindly providing the [3H] and unlabeled 27-hydroxycholesterol, and to Prof. Neale Ridgway (Dalhousie University, Halifax, Canada) for help in setting up the oxysterol binding assay. The authors thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki for providing laboratory facilities.
Abbreviations
22(R)OHC, 22(R)-hydroxycholesterol
25OHC, 25-hydroxycholesterol
CE, cholesteryl ester
CHO, Chinese hamster ovary
ER, endoplasmic reticulum
FCS, fetal calf serum
FFAT, two phenylalanines in an acidic tract
GST, glutathione S-transferase
LD, lipid droplet
mab, monoclonal mouse antibody
mβCD, methyl-β-cyclodextrin
ORD, oxysterol binding protein-related domain
ORP, oxysterol binding protein-related protein
OSBP, oxysterol binding protein
siRNA, short interfering RNA
SREBP, sterol regulatory element binding protein
TG, triglyceride
This study was supported by the Academy of Finland (Grants 113013, 118720, and 121457 to V.M.O.), the Sigrid Juselius Foundation, the Finnish Foundation for Cardiovascular Research, the Magnus Ehrnrooth Foundation, the Finnish Cultural Foundation (V.M.O.), the Biomedicum Helsinki Foundation (R.H.), the Perklén Foundation, and the Liv och Hälsa Foundation (N.B). Moreover, the work was partially funded by the European Union program LIPIDOMICNET (Grant 202272). R.H. is a member of the Helsinki Biomedical Graduate School, the support of which is acknowledged.
Published, JLR Papers in Press, February 17, 2009.
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
References
- 1.Björkhem I., and U. Diczfalusy. 2002. Oxysterols: friends, foes, or just fellow passengers? Arterioscler. Thromb. Vasc. Biol. 22 734–742. [DOI] [PubMed] [Google Scholar]
- 2.Gill S., R. Chow, and A. J. Brown. 2008. Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog. Lipid Res. 47 391–404. [DOI] [PubMed] [Google Scholar]
- 3.Russell D. W. 2000. Oxysterol biosynthetic enzymes. Biochim. Biophys. Acta. 1529 126–135. [DOI] [PubMed] [Google Scholar]
- 4.Schroepfer G. J., Jr. 2000. Oxysterols: modulators of cholesterol metabolism and other processes. Physiol. Rev. 80 361–554. [DOI] [PubMed] [Google Scholar]
- 5.Zelcer N., and P. Tontonoz. 2006. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishnan A., Y. Ikeda, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor F. R., and A. A. Kandutsch. 1985. Oxysterol binding protein. Chem. Phys. Lipids. 38 187–194. [DOI] [PubMed] [Google Scholar]
- 8.Yan D., and V. M. Olkkonen. 2008. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265 253–285. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri S., Y. J. Im, J. H. Hurley, and W. A. Prinz. 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairn G. D., and C. R. McMaster. 2008. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell. Mol. Life Sci. 65 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang M., B. G. Kearns, A. Gedvilaite, S. Kagiwada, M. Kearns, M. K. Fung, and V. A. Bankaitis. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15 6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 12.Saito K., K. Fujimura-Kamada, H. Hanamatsu, U. Kato, M. Umeda, K. G. Kozminski, and K. Tanaka. 2007. Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev. Cell. 13 743–751. [DOI] [PubMed] [Google Scholar]
- 13.Schulz T. A., and W. A. Prinz. 2007. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim. Biophys. Acta. 1771 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson M., V. Bocher, M. Lehto, G. Chinetti, E. Kuismanen, C. Ehnholm, B. Staels, and V. M. Olkkonen. 2003. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol. Biol. Cell. 14 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehto M., R. Hynynen, K. Karjalainen, E. Kuismanen, K. Hyvärinen, and V. M. Olkkonen. 2005. Targeting of OSBP-related protein 3 (ORP3) to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp. Cell Res. 310 445–462. [DOI] [PubMed] [Google Scholar]
- 16.Levine T. P., and S. Munro. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8 729–739. [DOI] [PubMed] [Google Scholar]
- 17.Ridgway N. D., P. A. Dawson, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1992. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyles J. P., and N. D. Ridgway. 2004. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp. Cell Res. 297 533–547. [DOI] [PubMed] [Google Scholar]
- 19.Fairn G. D., and C. R. McMaster. 2005. The roles of the human lipid-binding proteins ORP9S and ORP10S in vesicular transport. Biochem. Cell Biol. 83 631–636. [DOI] [PubMed] [Google Scholar]
- 20.Fairn G. D., and C. R. McMaster. 2005. Identification and assessment of the role of a nominal phospholipid binding region of ORP1S (oxysterol-binding-protein-related protein 1 short) in the regulation of vesicular transport. Biochem. J. 387 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynynen R., S. Laitinen, R. Käkelä, K. Tanhuanpää, S. Lusa, C. Ehnholm, P. Somerharju, E. Ikonen, and V. M. Olkkonen. 2005. Overexpression of OSBP-related protein 2 (ORP2) induces changes in cellular cholesterol metabolism and enhances endocytosis. Biochem. J. 390 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewen C. J., A. Roy, and T. P. Levine. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto T., Y. Ohsaki, J. Cheng, M. Suzuki, and Y. Shinohara. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tauchi-Sato K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277 44507–44512. [DOI] [PubMed] [Google Scholar]
- 25.Thiele C., and J. Spandl. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20 378–385. [DOI] [PubMed] [Google Scholar]
- 26.Laitinen S., M. Lehto, S. Lehtonen, K. Hyvarinen, S. Heino, E. Lehtonen, C. Ehnholm, E. Ikonen, and V. M. Olkkonen. 2002. ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J. Lipid Res. 43 245–255. [PubMed] [Google Scholar]
- 27.Käkelä R., K. Tanhuanpää, S. Laitinen, P. Somerharju, and V. M. Olkkonen. 2005. Overexpression of OSBP-related protein 2 (ORP2) in CHO cells induces alterations of phospholipid species composition. Biochem. Cell Biol. 83 677–683. [DOI] [PubMed] [Google Scholar]
- 28.Spector A. A. 1986. Structure and lipid binding properties of serum albumin. Methods Enzymol. 128 320–339. [DOI] [PubMed] [Google Scholar]
- 29.Taylor F. R., and A. A. Kandutsch. 1985. Use of oxygenated sterols to probe the regulation of 3-hydroxy-3-methylglutaryl-CoA reductase and sterologenesis. Methods Enzymol. 110 9–19. [DOI] [PubMed] [Google Scholar]
- 30.Leppimäki P., J. Mattinen, and J. P. Slotte. 2000. Sterol-induced upregulation of phosphatidylcholine synthesis in cultured fibroblasts is affected by the double-bond position in the sterol tetracyclic ring structure. Eur. J. Biochem. 267 6385–6394. [DOI] [PubMed] [Google Scholar]
- 31.Suchanek M., R. Hynynen, G. Wohlfahrt, M. Lehto, M. Johansson, H. Saarinen, A. Radzikowska, C. Thiele, and V. M. Olkkonen. 2007. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 405 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujishita Y., and J. H. Hurley. 2000. Structure and lipid transport mechanism of a StAR-related domain. Nat. Struct. Biol. 7 408–414. [DOI] [PubMed] [Google Scholar]
- 33.Im Y. J., S. Raychaudhuri, W. A. Prinz, and J. H. Hurley. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lada A. T., M. C. Willingham, and R. W. St Clair. 2002. Triglyceride depletion in THP-1 cells alters cholesteryl ester physical state and cholesterol efflux. J. Lipid Res. 43 618–628. [PubMed] [Google Scholar]
- 35.Zhao B., B. J. Fisher, R. W. St Clair, L. L. Rudel, and S. Ghosh. 2005. Redistribution of macrophage cholesteryl ester hydrolase from cytoplasm to lipid droplets upon lipid loading. J. Lipid Res. 46 2114–2121. [DOI] [PubMed] [Google Scholar]
- 36.Chang T. Y., C. C. Chang, N. Ohgami, and Y. Yamauchi. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22 129–157. [DOI] [PubMed] [Google Scholar]
- 37.Seo T., P. M. Oelkers, M. R. Giattina, T. S. Worgall, S. L. Sturley, and R. J. Deckelbaum. 2001. Differential modulation of ACAT1 and ACAT2 transcription and activity by long chain free fatty acids in cultured cells. Biochemistry. 40 4756–4762. [DOI] [PubMed] [Google Scholar]
- 38.Brasaemle D. L., G. Dolios, L. Shapiro, and R. Wang. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279 46835–46842. [DOI] [PubMed] [Google Scholar]
- 39.Fujimoto Y., H. Itabe, J. Sakai, M. Makita, J. Noda, M. Mori, Y. Higashi, S. Kojima, and T. Takano. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644 47–59. [DOI] [PubMed] [Google Scholar]
- 40.Liu P., Y. Ying, Y. Zhao, D. I. Mundy, M. Zhu, and R. G. Anderson. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279 3787–3792. [DOI] [PubMed] [Google Scholar]
- 41.Umlauf E., E. Csaszar, M. Moertelmaier, G. J. Schuetz, R. G. Parton, and R. Prohaska. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279 23699–23709. [DOI] [PubMed] [Google Scholar]
- 42.Brasaemle, D. L., and N. E. Wolins. 2006. Isolation of lipid droplets from cells by density gradient centrifugation. Curr. Protoc. Cell Biol. Chapter 3: Unit 3.15. [DOI] [PubMed]
- 43.Dawson P. A., N. D. Ridgway, C. A. Slaughter, M. S. Brown, and J. L. Goldstein. 1989. cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J. Biol. Chem. 264 16798–16803. [PubMed] [Google Scholar]
- 44.Dawson P. A., D. R. Van der Westhuyzen, J. L. Goldstein, and M. S. Brown. 1989. Purification of oxysterol binding protein from hamster liver cytosol. J. Biol. Chem. 264 9046–9052. [PubMed] [Google Scholar]
- 45.Perry R. J., and N. D. Ridgway. 2006. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell. 17 2604–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P. Y., J. Weng, and R. G. Anderson. 2005. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 307 1472–1476. [DOI] [PubMed] [Google Scholar]
- 47.Kuerschner L., C. Moessinger, and C. Thiele. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9 338–352. [DOI] [PubMed] [Google Scholar]
- 48.Ohashi M., N. Mizushima, Y. Kabeya, and T. Yoshimori. 2003. Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J. Biol. Chem. 278 36819–36829. [DOI] [PubMed] [Google Scholar]
- 49.Le Lay S., E. Hajduch, M. R. Lindsay, X. Le Liepvre, C. Thiele, P. Ferre, R. G. Parton, T. Kurzchalia, K. Simons, and I. Dugail. 2006. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 7 549–561. [DOI] [PubMed] [Google Scholar]
- 50.Pol A., S. Martin, M. A. Fernandez, C. Ferguson, A. Carozzi, R. Luetterforst, C. Enrich, and R. G. Parton. 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 15 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48 2547–2559. [DOI] [PubMed] [Google Scholar]
- 52.Zechner R., P. C. Kienesberger, G. Haemmerle, R. Zimmermann, and A. Lass. 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 50 3–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.