Abstract
Lipids from the extremely halophilic Archaea, Haloarcula marismortui, contain abundant phytanyl diether phospholipids, namely archaetidic acid (AA), archaetidylglycerol (AG), archaetidylglycerosulfate (AGS), with mainly archaetidylglycerophosphate methyl ester (AGP-Me). These were accompanied by a triglycosyl archaeol (TGA), lacking characteristic sulfate groups. Tandem-mass spectrometry was employed to provide fingerprints for identifying these known lipids, as well as small amounts of unsaturated phospholipids. These contained 3 and 6 double bonds in their archaeol moiety, suggested by negative tandem-MS of intact phospholipids, as indicated by differences between their pseudo-molecular ion and specific fragment ions designated as π2. The core ether lipids were confirmed by electrospray ionization mass spectrometry (ESI-MS) as 2,3-di-O-phytanyl-sn-glycerol (C20, C20), which gave rise to a precursor-ion at m/z 660 [M+Li]+, and its fragment ion at m/z 379 [M+Li]+, consistent with mono-O-phytanyl-glycerol. Furthermore, lithiated ions at m/z 654 (MS1), 379 (MS2) and m/z 648 (MS1), 373 (MS2), combined with 1H/13C NMR chemical shifts at δ 5.31-121.6 (C2/2′-H2/2′), 5.08-124.9 (C6/6′-H6/6′) and 5.10-126.0 (10/10′-H10/10′) confirmed the presence of unsaturated homologs of archaeol. We carried out a comprehensive study on the lipids present in cells of H. marismortui. We used positive and negative ESI-MS with tandem-MS, which served as a fingerprint analysis for identifying the majority of component lipids.
Keywords: fingerprint analysis, glycolipid, 1H/13C NMR, Haloarcula marismortui, phospholipids, tandem-mass spectrometry
The halophilic archaea are usually found in salines, considered to be extremely hostile environments (1). In these, few microorganisms have adapted, notably those of the archaea family, Halobacteriaceae. Isolated from the hypersaline Dead Sea was Haloarcula marismortui, which required high salt concentrations for optimum growth (1).
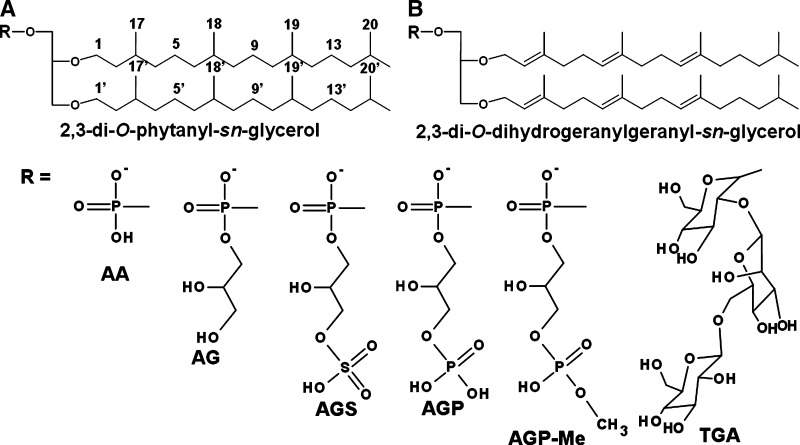
The archaeal lipids are particularly interesting, distinguished from lipids of other organisms by the presence of high amounts of ether lipids, which confer high resistance to support the osmotic stress of their environment. A common structure, designated as archaeol, is composed of two C-20 chains linked to glycerol via ether linkages (Fig. 1A). However, some archaea can also contain lipids with shorter and larger carbon chains, such as C-15 or C-25 linked to glycerol (2, 3), or a largest carbon framework (C-40), linked via four ether linkages to two glycerol units, such as in caldarchaeol structures (4, 5). Double bonds also occur in the carbon chain of archaeol (6, 7) (Fig. 1B).
Fig. 1.
Common structures of archaeal lipids: (A) saturated archaeol (B) hexa-unsaturated archaeol. Both structures could be linked to polar head group to give AA (archaetidic acid), AG (archaetidylglycerol), AGS (archaetidylglycerosulfate), AGP (archaetidylglycerophosphate), AGP-Me (archaetidylglycerophosphte methyl ester), and TGA (triglycosyl archaeol).
The archaeal lipids have been widely studied, and several structures have been characterized, such as glycolipids and phospholipids containing archaeol or caldarchaeol (8–10). The glycolipids from these extremophiles usually contain one or more sulfate groups, although it was previously found that H. marismortui (formerly Halobacterium marismortui) was capable of synthesizing a neutral glycolipid (11). In addition, a broad range of diphytanyl-O-glycero-phospholipids, such as archaetidylglycerol (AG), archaetidylglycerophosphate (AGP) and archaetidylglycerosulfate (AGS) have been characterized (Fig. 1A, B). Evans et al. (11) also found many pigments in H. marismortui, such as β-carotene, lycopene, and bacterioruberin, among others (11), although unsaturated archaeol lipids were not then detected.
However, on further investigation, a broad range of these compounds was found in H. marismortui when gas chromatography-mass spectrometry of their trimethysilyl derivatives detected from one to six double bonds (6).
Today, more improvements in analytical techniques allow detection of structures that would otherwise pass unobserved. One of the most powerful is mass spectrometry (MS), in which tandem-MS configuration has been successfully employed to characterize the structures of several lipids, such as glycolipids, phosphonolipids, sphingolipids, and archaeal lipids (4, 12–18). Furthermore, recent improvements in the preparation of lipid derivatives allow the employment of gas liquid chromatography-mass spectrometry (GC-MS) for a broad range of analyses (19).
In view of the recent interest in application of archaeal lipids in the liposomes due to their high stability against osmotic stress and potential resistance to degradation by esterases (20–24), we have carried out a comprehensive study on the lipids present in cells of H. marismortui. Employed were positive and negative electrospray ionization mass spectrometry (ESI-MS) with tandem-MS, which served as a fingerprint analysis for identifying the majority of component lipids.
MATERIALS AND METHODS
Haloarcula marismortui cultivation
Cells of H. marismortui were a kind gift from Professor Francisco Rodríguez-Valera (Universidad Miguel Hernández, Alicante, Spain). They were cultured in the medium used for growth of Halobaculum gomorrense, which consists of (g/L): NaCl 125 g; MgCl2.6H2O 160 g; K2SO4 5 g; CaCl2.2H2O 0.1 g; yeast extract 1 g; casamino acids 1 g, and starch 2 g (25). The pH was adjusted to 7.0 prior to autoclaving. Liquid cultures were grown using an orbital shaker (160 rpm) at 37°C. After 5 days, the culture was harvested by centrifugation (10,000 rpm), and the resulting pellet was washed twice with 2.5 mol/L NaCl, and then freeze dried.
Lipid extraction and fractionation
Lipids were obtained by extraction of cells (1.6 g) with 60 ml CHCl3-MeOH- (2:1 v/v), under reflux, for 2 h, which was repeated twice. The combined extract was evaporated under reduced pressure, and the residue dissolved in 2 ml CHCl3-MeOH (2:1, v/v), and polar lipids were precipitated by adding 20 vols of cold acetone to the solution (21), followed by centrifugation at 10,000 rpm. The resulting pellet was isolated from the soluble layer and dried. Total polar lipids were analyzed using silica gel 60G (Merck) thin layer chromatography (TLC). The separation was developed with CHCl3-MeOH-H2O (65:25:4, v/v), and the plates stained with orcinol-H2SO4 at 100°C for carbohydrate (26, 27), and the molybdate reagent for phosphorus detection (28).
The glycolipid present in the extract was purified by fractionation on a column (30 × 2 cm) containing silica gel 60G (Merck), by elution with CHCl3 with increasing proportions of MeOH (from 5% to 60%, v/v). Fractions of 5 ml were collected and analyzed by TLC and stained by orcinol-H2SO4, and molybdate reagent, as above. The purified glycolipid was analyzed by NMR and ESI-MS to indicate its structural components. Glycosidic linkages were characterized by methylation-GC-MS analysis (29).
NMR analysis of archaeal lipids
The total polar lipid fraction was solubilized in 400 μl of MeOD-CDCl3 (2:1, v/v) at a concentration of 100 mg/ml and analyzed using a Bruker Avance DRX-400 spectrometer with a 5 mm inverse probe. The spectra were obtained at 30°C, and the 13C chemical shifts measured in relation to Me4Si (δ = 0).
Sample preparation and ESI-MS setup
Positive and negative ESI-MS was carried out by atmospheric pressure ionization (API) with a tandem triple quadrupole, Quattro LC spectrometer (Waters) using nitrogen as nebulizer and desolvation gas. The samples (∼100 μg/ml) were prepared in MeOH-CHCl3-H2O (7:2:1, v/v), or deuterium exchanged by two successive solubilization/evaporation steps in MeOD-CHCl3-D2O (7:2:1, v/v), the same deuterated solvent being used for sample injection. The phospholipids were also analyzed as sodium salts in the positive ion mode. To induce deprotonation resulting in phospholipids as sodium salts, the same solvent above was used, but replacing water by aq. 0.1 M NaOH to pH 10.
The samples were directly injected into the ESI source with a syringe-pump (KDScientific) at a constant flow rate of 10 μl.min-1. The energy setup was: positive analysis, cone 110 V and the capillary 3.6 kV; negative analysis, cone 90 V and capillary 3 kV. The samples were acquired in the total ion current (TIC) mode. Tandem-MS was performed by collision induced dissociation mass spectrometry (CID-MS), using argon as collision gas. The energies used in CID-MS ranged between 80 and 90 V for positive and between 50 and 60 V for negative modes for intact lipid precursors, whereas those for the lithyated core ether lipids were 65 V for saturated, 50 V for tri-unsaturated, and 40 V for hexa-unsaturated chains.
Isolation and characterization of the core ether lipids
Core ether lipids (i.e., archaeol and unsaturated homologs) were obtained from the total polar lipid fraction by releasing them from the polar head group by acid and alkaline hydrolysis. The acid hydrolysis was performed with 1 mg of extract in 1 M MeOH-HCl (500 μl) in a sealed tube, at 100°C for 14 h. The archaeol was extracted by addition of CHCl3 (1.0 ml) plus H2O (0.5 ml). The CHCl3 layer was removed, evaporated under a nitrogen stream, and the residue was dissolved in MeOH-CHCl3-H2O (6:3:1, v/v), supplemented with 5 mM LiCl, for positive ESI-MS analysis.
To analyze the unsaturated archaeol, the polar lipid sample (1 mg) was dissolved in 3 ml of methanol. 1 ml 10% KOH (w/v) was added, and the mixture was maintained at 100°C for 3 h (7). After cooling to room temperature, the reaction mixture was extracted with CHCl3 to isolate the core ether lipids. The solvent was removed under a N2 stream, and the sample was dissolved in MeOH-CHCl3-H2O (6:3:1, v/v) supplemented with 5 mM LiCl for positive tandem-MS analysis.
RESULTS
Positive and negative ESI-MS scan of polar lipids
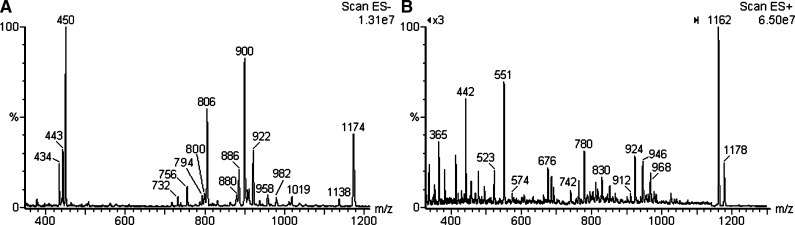
The polar lipid fraction, on precipitation with cold acetone, yielded 80 mg, in which the presence of phospholipids was suggested by blue TLC spots, on molybdate development (28), and one with orcinol-H2SO4 suggesting the presence of a glycolipid. Their characterization was based on their mass spectra in the positive and negative ESI mode. In the latter, many compounds were detected, and the ions were identified as AA at m/z 732 [M-H]-; AG at m/z 806 [M-H]-; AGS/AGP at m/z 443 [M-2H]2- and 886 [M-H]-; and AGP-Me at m/z 450 [M-2H]2-, 900 [M-H]-, and 922 [M-2H+Na]-. Two ions were consistent with the structure of a neutral glycolipid, with m/z 1138 [M-H]- and 1174 [M+Cl]-. Three ions were from phospholipids containing unsaturated archaeol, giving m/z 794, 800 (AG), and m/z 880 (AGS/AGP) (Fig. 2A).
Fig. 2.
Negative (A) and positive (B) scan ESI-MS of total polar lipid fraction from H. marismortui extract.
Positive ESI ionization resulted in a spectrum with a main ion at m/z 1162, consistent with sodiated TGA. The phospholipids gave rise to minor ions with m/z 756 (AA); 830 and 852 (AG); 954 (AGS/AGP); and 924, 946, and 968 (AGP-Me) (Fig. 2B). The structure of these lipids was confirmed by NMR and positive, negative tandem-MS. These results showed a lipid composition similar to those of previous investigations (7, 11, 12).
NMR analysis of total polar lipids
The 13C and 13C-DEPT-NMR spectra contained a mixture of signals with chemical shifts arising from glyco- and phospholipids. Those arising from phytanyl moiety were identified as follows: CH3 signals at δ 20.3 its inner chain, and δ 23.1 from its terminal chain, tertiary carbons (CH) were between δ 28.78 and 33.64, which confirmed the nature of the branched chains from arqueol lipids as these shifts are absent in linear carbon chains. DEPT-NMR served to locate the chemical shifts from CH2, with inverted signals at δ 25.2–25.6 and δ 37.6–40.2, and those of CH2-O from the ether linkages at δ 68.1–71.0 (see supplementary Fig. I). These results agree with previous findings (9, 30) and are consistent with the structure of archaeol.
In the 13C-NMR spectrum of the lipid extract, minor abundant signals at δ 120–140 can be attributed to double bounds. Moreover, its 1H spectrum contained two small signals at δ 5.10 and 5.30, consistent with olefinic protons (see supplementary Fig. II). These were better observed in a 2D 1H-13C HMQC spectrum, which showed connectivities between these carbons (δ 120–140) and olefinic protons as follows: δ 5.31/121.6 (H2,2′/C2,2′), 5.08/124.9 (H6,6′/C6,6′) and 5.10/126.0 (H10,10′/C-10,10′).
In its saturated chains, the 13C signals from CH3 ranged between δ 20 and 23, and 1H between δ 0.8 and 1.0. However, as found on HMQC NMR, the presence of double bounds shifted the CH3 signals to δ 1.57–16.45 (H18,18′-19,19′/C H18,18′-19,19′) and 1.65–16.9 (H17,17′/C17,17′). Connectivities between these methylic protons with those of olefines were provided on 1H/1H COSY, the signals occurring at δ 1.57–5.08/5.10 (H6,6′/H18,18′ and H10,10′/H19,19′) and 1.65–5.31 (H2,2′/H17,17′) (see supplementary Fig. III). These results confirmed the presence of unsaturated archaeol, consistent with the previously studied structure of 2,3-di-O-dihydrogeranylgeranyl glycerol diether (31).
The purified triglycosyl archaeol was also submitted to NMR spectroscopy. The anomeric region of carbohydrates was consistent with the presence of units of β-Glc at δ 104.25 (JC-1/H-1 = 158.5 Hz), α-Glc at δ 99.27 (JC-1/H-1 = 168.8 Hz), and α-Man at δ 97.39 (JC-1/H-1 = 168.1 Hz). Other carbohydrate signals including glycerol were distributed between δ 62.0 and 78.7. O-Linked glycosidic signals were at δ 78.66 and 68.18, consistent with O-substituted C-2 of Glcp and O-substituted C-6 of Manp units, respectively (see supplementary Fig. IV).
Analysis of the core ether lipid
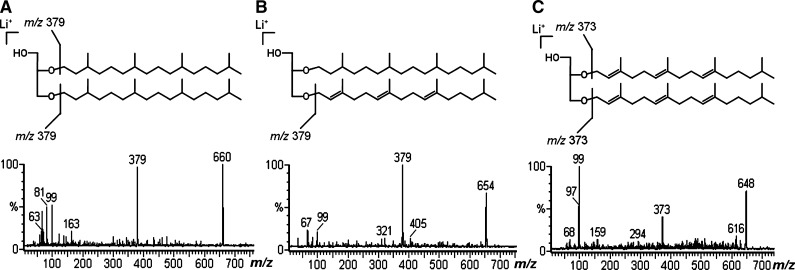
Considering that variations in the carbon chain-length of archaeol may occur, as with C-15, C-20, and C-25 (3), the core-ether lipid was isolated from PHG using two different hydrolytic methods to identify if the carbon chains lengths were equivalent. First, the archaeol moiety was obtained by acid hydrolysis (MeOH-HCl), the core lipid being analyzed by ESI-MS as Li+ adducts, which gave a main ion with m/z 660, consistent with archaeol, and a small one with m/z 379, identified as phytanyl linked to glycerol. Since the allyl ether linkage is more labile than the saturated ether, the ion at m/z 379 was probably formed by degradation of the allyl ether linkage present in the unsaturated archaeol chain during hydrolysis.
Because the 1H/13C NMR spectra revealed the presence of unsaturated lipid chains, which are labile by acidic hydrolysis, an alkaline hydrolysis was carried out to maintain their stability. The core lipid was first analyzed by ESI-MS as Na+ adducts, giving rise to ions at m/z 676, 670, and 664 consistent with saturated, tri-unsaturated, and hexa-unsaturated archaeol, which produced poor fragment-ions in collision induced dissociation-mass spectrometry (CID-MS). Thus, it was analyzed as Li+ adducts, giving a main ion at m/z 660 and minor ones at m/z 654 and 648, which were interpreted as arising from saturated, tri-unsaturated, and hexa-unsaturated archaeol, respectively. Furthermore, tandem-MS of the ion at m/z 660 gave a main fragment at m/z 379 (Fig. 3A). A similar result was obtained from an ion at m/z 654, which also gave rise to a main one with m/z 379 (Fig. 3B), as would be expected from a mono-phytanyl-O-glycerol. In the case of the ion with m/z 654, the formation of fragment at m/z 379 can be attributed to more labile allyl ether linkage, which was first removed by CID-MS, releasing the lithiated mono-phytanyl-O-glycerol. This is in agreement with fragmentation of the m/z 648, when less CID energy was necessary, and even so the main fragment was that at m/z 99, consistent with lithiated glycerol. However, an ion at m/z 373 was also present (Fig. 3C), indicating the presence of a lithiated dihydrogeranylgeranyl (tri-unsaturated phytanyl) linked to glycerol. The ion at m/z 99 (glycerol + Li+) was present in all CID-MS spectra obtained from the core ether lipid (Fig. 3A–C), indicating that the three ionic species were in lithiated form [M+Li]+, avoiding misinterpretation that could be caused by an incomplete lithiation with consequent protonation of the archaeol species.
Fig. 3.
Positive CID-MS from lithiated core ether lipids obtained by base hydrolysis, showing (A) saturated chain, (B) tri-unsaturated chain, and (C) hexa-unsaturated chain.
CID-MS fingerprints of intact archaeal phospholipids
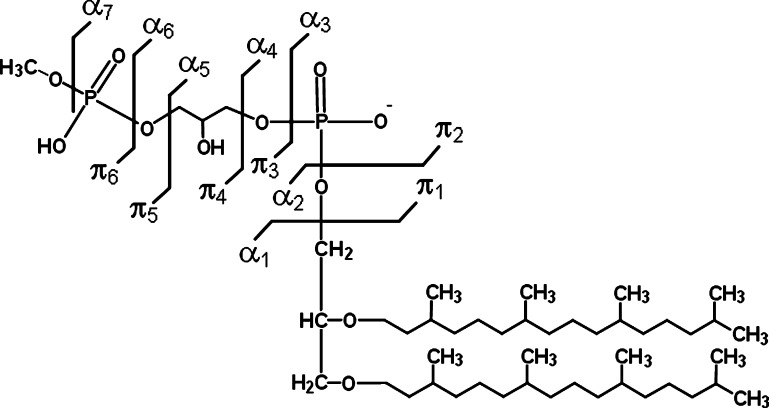
To visualize better correlation between the phospholipid structures and the fragmentation fingerprints, the fragment-ions obtained by tandem-MS were designated as αx, counting from the archaeol moiety, and πx counting from the phosphate group linked to archaeol. The subscript x indicates the breakdown position (e.g., α4, π2) (Fig. 4).
Fig. 4.
Schematic representation of breakdown sites on AGP-Me and nomenclature used to designate each observed fragment-ion.
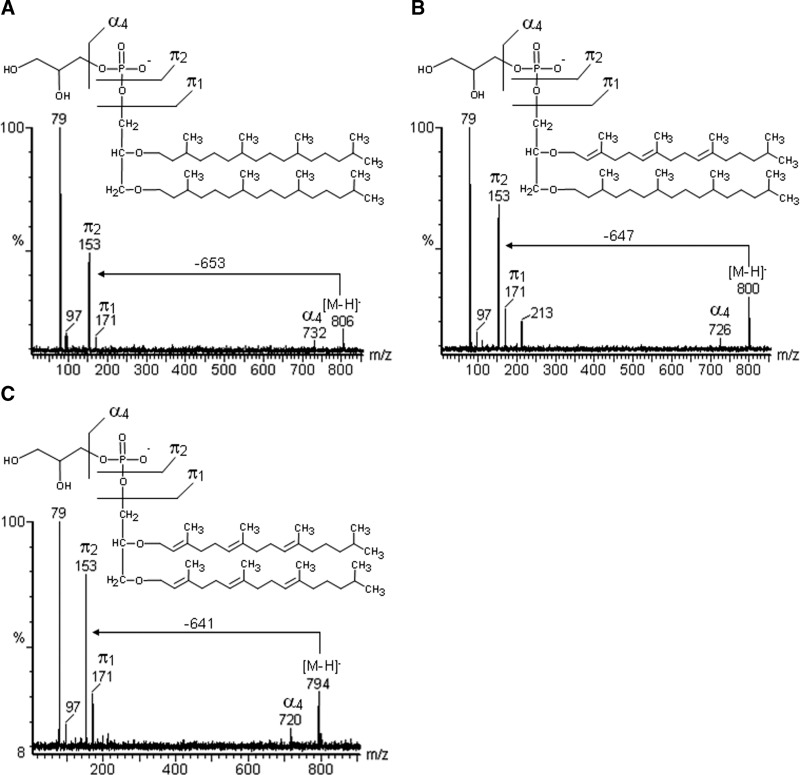
The simplest phospholipid detected in the negative mode was AA, at m/z 732 [M-H]-, which gave fragments at m/z 97 and 79, arising from its phosphate group (Table 1). Archaetidylglycerol (AG) at m/z 806 [M-H]- and its unsaturated analogs at m/z 800 [M-H]- and 794 [M-H]- yielded characteristic CID-MS fragments at m/z 171, 153, 97, and 79, corresponding to PHG (Fig. 5A–C). The ion at m/z 800 was consistent with an AG containing a tri-unsaturated archaeol, which yielded a neutral loss (PMI–π2) of 647 m.u., this corresponding to dihydrogeranylgeranyl-phytanyl-O-glycerol, whereas the compound with m/z 794 was identified as being hexa-unsaturated, giving rise to NL of 641 m.u., from di-O-dihydrogeranylgeranyl glycerol (Fig. 5B, C).
TABLE 1.
Positive and negative tandem-MS profiles of phospholipids
| Positive CID-MS fragmentation |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phospholipid | PMI | NL | π1 | π2 | π3 | π4 | π5 | π6 | α2 | α4 | α5 | α6 | α7 |
| AA [M+Na+]+a | 756 | — | 121 | — | — | — | — | — | — | — | — | — | — |
| AG [M+Na+]+a | 830 | — | 195 | 177 | — | — | — | — | 676 | 756 | — | — | — |
| AG [M-H++2Na+]+a | 852 | — | — | 199 | — | — | — | — | 676 | 778 | 834 | — | — |
| AG [M+Na+]+c | 818 | — | 195 | 177 | — | — | — | — | — | — | — | — | — |
| AGS [M-2H++3Na+]+a | 954 | — | 301 | — | 199 | 165 | 125 | — | 800 | 835 | — | — | |
| AGP-Me [M+Na+]+a | 924 | — | 289 | 271 | 209 | 191 | 135 | — | 676 | 756 | — | 830 | 893 |
| AGP-Me [M-H++2Na+]+a | 946 | — | 311 | 293 | 231 | 199d | 157 | 125d | 676 | 778 | 834 | 852 | 913 |
| AGP-Me [M-2H++3Na+]+a | 968 | — | 333 | 315 | — | 199d | 178 | 125d | 676 | 800 | 834 | 852 | 937 |
| Negative CID-MS fragmentation |
|||||||||||||
| AA [M-H+]-a | 732 | 653 | 97 | 79 | — | — | — | — | — | — | — | — | — |
| AG [M-H+]-a | 806 | 653 | 171 | 153 | — | — | — | — | — | 732 | — | — | — |
| AG [M-H+]-b | 800 | 647 | 171 | 153 | — | — | — | — | — | 726 | — | — | — |
| AG [M-H+]-c | 794 | 641 | 171 | 153 | — | — | — | — | — | 720 | — | — | — |
| AGS [M-H+]-a | 886 | 653 | — | 233 | 171 | 153 | 97 | 79 | — | 732 | — | 806 | — |
| AGS [M-2H++1Na+]-a | 908 | 653 | 273 | 255 | 171 | 153 | 97 | 79 | — | — | — | — | — |
| AGS [M-H+]-b | 880 | 647 | — | 233 | 171 | 153 | 97 | 79 | — | 726 | — | 800 | — |
| AGP-Me [M-H+]-a | 900 | 653 | — | 247 | 185 | — | 111 | 95 | 732 | 788 | 806 | 868 | |
| AGP-Me [M-2H++1Na+]-a | 922 | 653 | 287 | 269 | — | — | — | — | — | — | 788 | 806 | 890 |
CID-MS, collision induced dissociation-mass spectrometry; PMI, pseudo-moleuclar ion; NL, neutral loss (PMI-π2) in negative tandem-MS.
Saturated.
Tri-unsaturated.
Hexa-unsaturated.
Fragments from AGP-Me formed with loss of methyl group.
Fig. 5.
Negative tandem-MS fingerprints of AG containing saturated archaeol, indicated by the neutral loss (NL) of –653 m.u. (A), tri-unsaturated archaeol NL of –647 m.u. (B), and hexa-unsaturated archaeol of –641 m.u. (C).
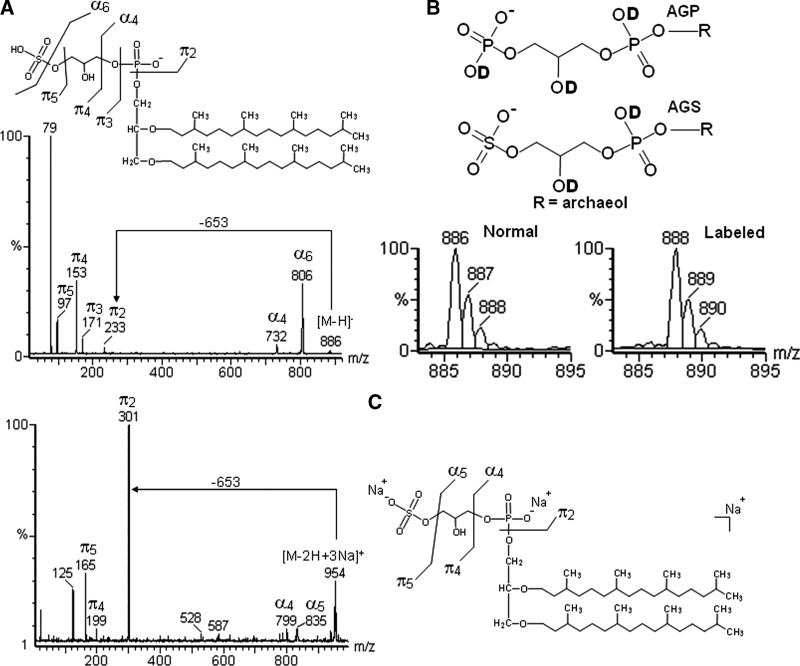
Two isobaric lipids have been reported in archaea, namely archaetidylglycerophosphate (AGP) and archaetidylglycerosulfate (AGS), each one with a nominal mass of 886 Da. To distinguish between them, a high-resolution mass spectrometer would be required. However, based on chemical differences between sulfate and phosphate groups, it was possible to infer the type of lipid present in the extract, even with a low resolution MS instrument.
To differentiate between the isobars, the sample was therefore hydrogen/deuterium (H/D) exchanged, considering that the negative ion of AGS should shift from m/z 886 (unlabeled) to 888 (labeled), whereas that of AGP would go from m/z 886 to m/z 889. Found in a nonlabeled mass spectrum, naturally occurring elemental isotopes appeared at m/z 886, 887, and 888 in a ratio of 1:0.5:0.07 and the labeled lipid gave rise to ions at m/z 888, 889, and 890, with same ratio (Fig. 6A, B). As the presence of labeled AGP should increase the intensity of the ion 889, these results mean that AGP was absent in the lipid extract. A tri-unsaturated AGS was also detected, with m/z 880, 881, 882 (unlabeled) and 882, 883, 884 (labeled), in a ratio of 1:0.5:0.07. The unsaturation was confirmed by their characteristic NL (647 m.u.), and other fragments formed on positive tandem-MS (Table 1).
Fig. 6.
(A) Negative tandem-MS fingerprints of AGS. (B) Sites for labeling in AGP and AGS, and ESI-MS of normal and deuterium labeled AGS. If present, AGP should increase the intensity of ion 889. (C) Positive tandem-MS fingerprints of tri-sodiated AGS (B) obtained with aq. NaOH.
To confirm the absence of AGP, the extract was dissolved in MeOH-CHCl3 containing aq. NaOH (pH 10.0), and positive ESI-MS was performed, substituting H+ with Na+ ions. Once more, the ion from AGP-3H++4Na+, expected at m/z 976, was not formed; instead, an ion at m/z 954 appeared, consistent with [AGS-2H++3Na]+ (Fig. 6C), providing supporting evidence of the absence of AGP.
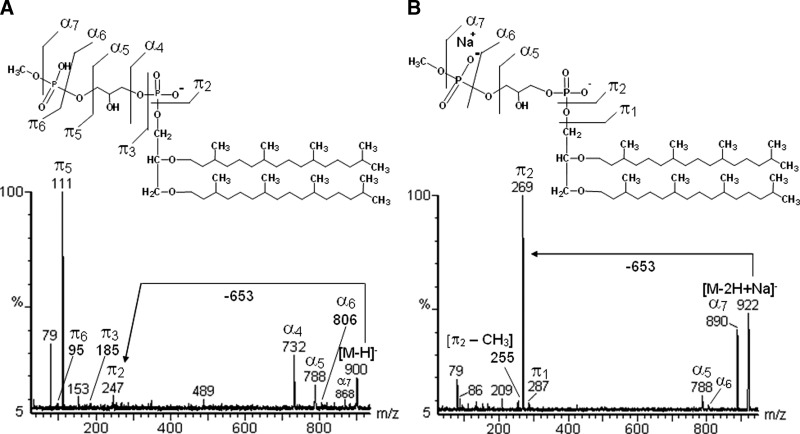
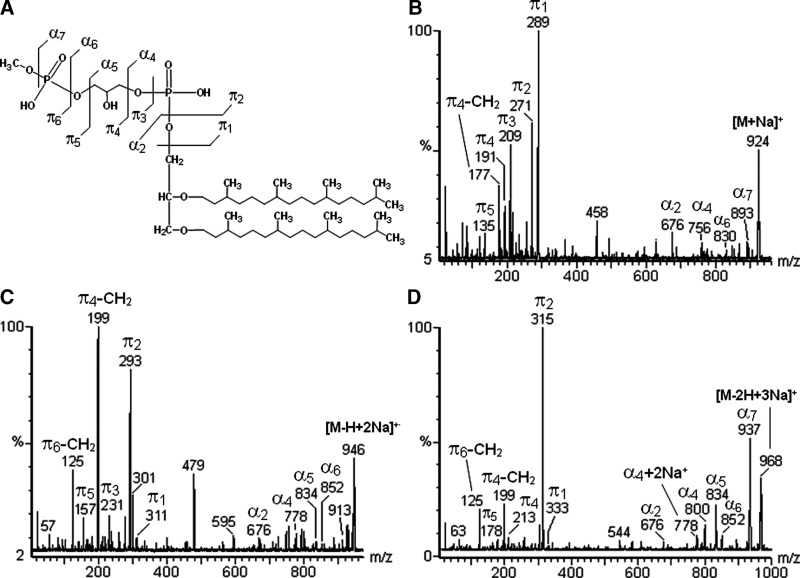
The negative ions from the lipid extract, at m/z 450 [M-2H]2-, 900 [M-H]-, and 922 [M-2H+Na+]-, as well as the positive ones at m/z 924 [M+Na]+, 946 [M-H+2Na]+, and 968 [M-2H+3Na]+ are consistent with an archaetidylglycerophosphate methyl ester (AGP-Me) (Fig. 2A, B). Furthermore, negative tandem-MS confirmed the presence of the phosphoglycerophosphate methyl ester group, with fragments at m/z 247 (π2) and 111 (π5) corresponding to PHG. In addition, negative fragments at m/z 788 (α5) and 732 (α4) corresponded to removal of phospho-methyl ester and glycerophospho-methyl ester groups, respectively (Fig. 7A). Positive tandem-MS confirmed the presence of the archaeol moiety, with a sodiated fragment at m/z 676. No unsaturated analog to AGP-Me has been found. Interestingly, the number of Na+ ions in the phospholipids alters its fragmentation partners, so that the resulting fragments corresponding to the same moiety in the molecule appeared in different intensities (Figs. 7A, 7B, 8A–D).
Fig. 7.
Negative tandem-MS fingerprints of AGP-Me, comparing the fragmentation partner of deprotonated (A) and mono-sodiated (B) ions.
Fig. 8.
(A) Positive tandem-MS fingerprints of AGP-Me, showing differences between mono- (B), di- (C), and tri-sodiated (D) ions.
ESI-MS fingerprints of triglycosyl archaeol
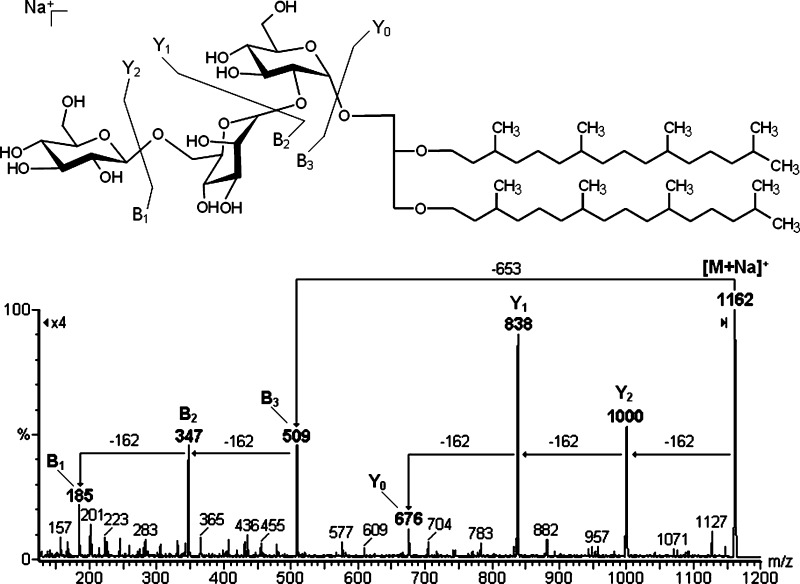
The tandem-MS analysis of TGA was then carried out, and the fragmentation profile was analyzed following the nomenclature proposed by Domon and Costelo (32) for glycosides. Accordingly, ions were designated Yj+, where Y indicates fragments containing the aglycone moiety (i.e., archaeol), and the subscript j the number of monosaccharide residues linked to the aglycone. Other ions were named Bi+, where B indicates fragments containing exclusively the carbohydrate moiety, and the subscript i the number of monosaccharide residues counting from the terminal unit. In both cases, the positive superscript symbol indicates the charge of the ion, in this case from Na+ adducts.
According to this nomenclature, the fragment-ions observed were Y2 at m/z 1000 [M-162]+, Y1 at m/z 838 [M-324]+, and Y0 at m/z 676 [M-486]+, which represent successive removal of hexose residues, and the fragments B3 at m/z 509, B2 at m/z 347, and B1 at m/z 185 only the saccharide moiety (Fig. 9).
Fig. 9.
Positive tandem-MS fingerprint of sodiated TGA. The fragment ions were named according to Domon and Costello's nomenclature (32). NL of –162 m.u. indicates the removal of hexosyl units.
The structure of each monosaccharide unit was confirmed by methylation analysis, with GC-MS of resulting partially methylated alditol acetates (PMAA) arising from each monosaccharide unit. Three peaks were identified based on our PMAA library (33), as 2,3,4,6-Me4-Glc; 3,4,6-Me3-Glc; and 2,3,4-Me3-Man, in a 1:1:1 molar ratio. This result is consistent with the results of Evans et al. (11) and our NMR data.
DISCUSSION
Identification of phospholipids
The first description of lipids from H. marismortui by Evans et al. (11) provided important information on the structure of its phospholipids. This was obtained mainly by TLC analysis and elemental composition, with the fine structure of a neutral triglycosyl archaeol being determined by methylation and NMR analysis. Klöppel and Fredrickson (12) later employed FAB-MS to provide more information, and also suggested the presence of unsaturated archaeol chains.
ESI-MS-MS is now used to provide more details on the structure of phospholipids and a neutral glycolipid from an extract of H. marismortui. The analytical results were similar to the first investigation (11), except for showing a high abundance of AGP-Me. Its presence appears to be important for maintaining cell integrity, since membranes from halophilic archaea must maintain their stability at high salt concentrations (3–5 M NaCl), whereas nonhalophilic membranes, lacking AGP-Me, are unstable and leak under such conditions (20).
We have also now proposed a simple and specific nomenclature for the phospholipid fragments, which facilitates their comparison, as well as their identification. As structural differences between lipids occur mainly in the polar head group (PHG), the key differences in the mass spectra were now found in fragments π1 and π2, which are characteristic of each phospholipid. The other fragments (π–type) must be evaluated cautiously, due to the formation of fragments with m/z 97 and 79, which are characteristic of phosphate and dehydrated phosphate groups respectively, which occurs throughout phospholipids.
Furthermore, it was possible to deduce the presence of saturated or unsaturated archaeol, by calculating the neutral losses by subtracting π2 from the pseudo-molecular ion (PMI) [M-H]-. Considering that a broad series of compounds could give an identical NL, the presence of double bonds was confirmed by 1H/13C-NMR spectroscopy, and the length of alkyl chains determined from the isolated core ether lipid, by CID-MS of lithiated molecules. Li+ was used since it produced more intense fragments than other common adducts, such as those of Na+ or K+. The differences among the ions arising from saturated and unsaturated archaeol species were 6 m.u., and similar differences should be present in case of incomplete lithiation, if protonated species were present. However, the presence of the ion at m/z 99 from glycerol [M+Li]+ in all CID-MS spectra confirmed the complete lithiation of archaeol species during ESI-MS analysis.
Thus in contrast with the findings of Klöppel and Fredrickson (12), who observed more intense ions in their mass spectra, which they attributed to unsaturated lipids, whereas we have now found less abundant ions, which arose from unsaturated phospholipids, appearing at m/z 800 and 794 (AG) and m/z 880 (AGS). Thus, by subtracting the fragment-ion π2 from PMI, we found an NL of 647 m.u. (from AG at m/z 800), consistent with tri-unsaturated archaeol (2,3-dihydrogeranylgeranyl-phytanyl glycerol) and one of 641 m.u. (from AG at m/z 794), consistent with hexa-unsaturated archaeol (2,3-dihydrogeranylgeranyl glycerol). An NL of 641 m.u. was also obtained from m/z 880, indicating the presence of tri-unsaturated AGS.
Differentiation between AGP and AGS
The isobaric AGP and AGS are barely distinguishable by mass spectrometry, except with high-resolution equipment. For them, a doubly-charged ion was observed at m/z 443. Although a triply-charged ion was absent, this could not represent any absence of AGP because the strong Coulombic repulsion should certainly prevent its formation. Consequently, isobaric differentiation was carried out using two strategies.
Strategy 1
Deuterium labeling should alter the ratio between the main isotopes found at m/z 888, 889, and 890, since AGP should provide three sites for labeling (i.e., one from free hydroxyl in glycerol, and two from phosphate groups), whereas AGS has only two sites (Fig. 6B). Consequently, the presence of AGP should increase the ratio of m/z 889 compared with 888, in comparison with the unlabeled ions at m/z 887 and 886. As the ratio was the same after deuterium exchange, AGP was absent.
Strategy 2
The same principle of deuterium labeling could be used to distinguish AGP from AGS using positive ESI-MS, by replacing H+ with Na+. The basis for such a distinction lies in the ability of AGS to form a tri-sodiated mono-charged ion with m/z 954 [M-2H++3Na+]+, whereas AGP should form a tetra-sodiated mono-charged ion at m/z 976, which did not appear in our analysis. Furthermore, tandem-MS of the precursor ion at m/z 954 gave rise to fragment at m/z 799, identified as arising from fragment α4, and being equivalent to a tri-sodiated AA moiety (Fig. 6C), confirming that the analytical conditions completely deprotonated the phosphate groups. This shows that the absence of an ion with m/z 976 is due to the absence of AGP and not to incomplete deprotonation, which would lead to an m/z 954 ion from AGP [M-2H++3Na+]+. Similarly, the ion at m/z 880 [M-H+]- from tri-unsaturated AGS, when labeled with deuterium, shifted to m/z 882, and in the presence of NaOH, formed sodium salts at m/z 948 [M-2H+ + 3Na+]+.
To ensure that pH 10.0 was sufficiently alkaline for complete deprotonation, we tested a standard of acyl phosphatidic acid (Sigma-Co.), which gave a main negative ion at m/z 674, consistent with PA containing oleic and palmitic acids. Thus, in positive ESI-MS the main ion was at m/z 741, arising from PA [M-2H++3Na+]+, but others from any mono- or di-sodiated ions were not present (data not shown).
Analysis of multi-sodiated phospholipids
The multi-charged and multi-sodiated ions formed on ESI-MS analysis could lead to overestimation and, consequently, misinterpretation of the results. Interestingly, the presence and the number of sodium ions present in each phospholipid modified its fragmentation pattern, another factor for misinterpreting the mass spectra. For example, AGP-Me in the negative ESI mode was found to form two ions at m/z 900 [M-H+]- and 922 [M-2H++Na+]-. This, at first glance at their fragmentation profile, suggested different compounds, since the base peak (i.e., the fragment reaching 100%) changed drastically from m/z 111 (π2), arising from the precursor ion at m/z 900, to 269 (π5) arising from the m/z 922 ion (Fig. 7A, B). With positive ESI detection, three pseudo-molecular ions for AGP-Me were formed at m/z 924 [M+Na]+, 946 [M-H+2Na]+, and 968 [M-2H+3Na]+, each one giving rise to specific fragmentation partners (Fig. 8A–D and Table 1). Similar results were obtained for the other phospholipids, such as AGS (Table 1). Thus supplementing the samples with an NaOH solution was an important technique to avoid formation of multi-sodiated ions, making the positive mass spectra simpler and easier to interpret.
Fingerprints of TGA
Evans et al. (11) commented on the presence of three spots of glycolipids on TLC analysis (positive with 0.5% α-naphthol/H2SO4). However, we found only one spot positive for carbohydrates with orcinol-H2SO4, which was isolated and gave rise to a sodiated ion at m/z 1162 in the positive ESI-MS mode. This was consistent with the structure of their neutral triglycosyl archaeol (11). Consequently, our sample was analyzed by tandem-MS, which confirmed the presence of a trisaccharide group composed of hexosyl units, which exhibited glycosidic cleavages similar to those found by Souza et al. (34), as occurred with flavonol glycosides. CID-MS led to formation of Yj fragments, resulting in the release of the adjacent monosaccharide residue, with a double bond formed by β-cleavage between C-1 and C-2 (32, 35), the fragments appearing as sodiated ions. The ions with m/z 1000 [M-162+Na]+, m/z 838 [M-324+Na]+, and m/z 676 [M-486+Na]+ represent fragments Y2+, Y1+, and Y0+ respectively. On the other hand, the masses lost in the Y-fragments were multiples of 162 m.u., indicating removal of mono-, di-, and trisacharide, respectively. In fact, these masses were not considered as neutral losses, as they appeared as sodiated B-fragments at m/z 509, 347, and 185 (i.e., B3+, B2+, and B1+ respectively), which correspond to the mono-, di-, and tri-hexosyl units. The sodiated fragment Y0+, appearing at m/z 676, confirms the complete saturation of the archaeol chain present in the glycolipid (Fig. 9).
To confirm its structure, triglycosyl archaeol was purified using column chromatography with silica-gel. Elution began with CHCl3, followed with increasing concentrations of MeOH. The glycolipid was eluted with CHCl3-MeOH (1:1, v/v), but TLC analysis indicated the presence of phospholipids, so anionic resin was added to remove these phospholipids. Thus the glycolipid could be identified as β-Glcp-(1→6)-α-Manp-(1→2)-α-Glcp-O-archaeol, according to methylation-GC-MS and NMR analyses (see supplementary data), being comparable with Evans et al. findings (11).
Supplementary Material
Acknowledgments
The authors wish to thank the Brazilian agencies Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq), Fundação Araucária, and PRONEX-Carboidratos for financial support, and Professor Francisco Rodríguez-Valera (Universidad Miguel Hernández, Alicante, Spain) for supplying the H. marismortui strain.
Abbreviations
AA, archaetidic acid
AG, archaetidylglycerol
AGP, archaetidylglycerophosphate
AGP-Me, archaetidylglycerophosphate methyl ester
AGS, archaetidylglycerosulfate
API, atmospheric pressure ionization
CID-MS, collision induced dissociation-mass spectrometry
ESI-MS, electrospray ionization-mass spectrometry
GC-MS, gas chromatography-mass spectrometry
HMQC, heteronuclear multiple quantum coherence
MS, mass spectrometry
m.u., mass units
m/z, mass to charge ratio
NL, neutral loss
NMR, nuclear magnetic resonance
PHG, polar head group
PMI, pseudo-molecular ion
TIC, total ion current
TGA, triglycosyl archaeol
TLC, thin layer chromatography
Published, JLR Papers in Press, March 2, 2009.
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
References
- 1.Oren A., M. Ginzburg, B. Z. Ginzburg, L. I. Hochstein, and B. E. Volcani. 1990. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int. J. Syst. Bacteriol. 40 209–210. [DOI] [PubMed] [Google Scholar]
- 2.Matsubara T., N. I. Tanaka, M. Kamekura, N. Moldoveanu, I. Ishizuka, H. Onishi, A. Hayashi, and M. Kates. 1994. Polar lipids of a non-alkaliphilic extremely halophilic archaebacterium strain 172: a novel bis-sulfated glycolipid. Biochim. Biophys. Acta. 1214 97–108. [DOI] [PubMed] [Google Scholar]
- 3.Mancuso C. A., G. Odham, G. Westerdahl, J. N. Reeve, and D. C. White. 1985. C15, C20, and C25 isoprenoid homologues in glycerol diether phospholipids of methanogenic archaebacteria. J. Lipid Res. 26 1120–1125. [PubMed] [Google Scholar]
- 4.Macalady J. L., M. M. Vestling, D. Baumler, N. Boekelheide, C. W. Kaspar, and J. F. Banfield. 2004. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles. 8 411–419. [DOI] [PubMed] [Google Scholar]
- 5.Fahy E., S. Subramaniam, H. A. Brown, C. K. Glass, A. H. Merrill, Jr., R. C. Murphy, C. R. H. Raetz, D. W. Russell, Y. Seyama, W. Shaw, et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46 839–861. [DOI] [PubMed] [Google Scholar]
- 6.Stiehl T., J. Rullkötter, and A. Nissenbaum. 2005. Molecular and isotopic characterization of lipids in cultured halophilic microorganisms from the Dead Sea and comparison with the sediment record of this hypersaline lake. Org. Geochem. 36 1242–1251. [Google Scholar]
- 7.Gibson J. A. E., M. R. Miller, N. W. Davies, G. P. Neill, D. S. Nichols, and J. K. Volkman. 2005. Unsaturated diether lipids in the psychrotrophic archaeon Halorubrum lacusprofundi. Syst. Appl. Microbiol. 28 19–26. [DOI] [PubMed] [Google Scholar]
- 8.Uda I., A. Sugai, K. Kon, S. Ando, Y. H. Itoh, and T. Itoh. 1999. Isolation and characterization of novel neutral glycolipids from Thermoplasma acidophilum. Biochim. Biophys. Acta. 1439 363–370. [DOI] [PubMed] [Google Scholar]
- 9.Ferrante G., J. R. Brisson, G. B. Patel, I. Ekiel, and G. D. Sprott. 1989. Structures of minor ether lipids isolated from the aceticlastic methanogen, Methanothrix concilii GP6l. J. Lipid Res. 30 1601–1609. [PubMed] [Google Scholar]
- 10.Smallbone B. W., and M. Kates. 1981. Structural identification of minor glycolipids in Halobacterium cutirubrum. Biochim. Biophys. Acta. 665 551–558. [Google Scholar]
- 11.Evans R. W., S. C. Kushwaha, and M. Kates. 1980. The lipids of Halobacterium marismortui, an extremely halophilic bacterium in the Dead Sea. Biochim. Biophys. Acta. 619 533–544. [DOI] [PubMed] [Google Scholar]
- 12.Klöppel K. D., and H. L. Fredrickson. 1991. Fast atom bombardment mass spectrometry as a rapid means of screening mixtures of ether-linked polar lipids from extremely halophilic archaebacteria for the presence of novel chemical structures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 562 369–376. [DOI] [PubMed] [Google Scholar]
- 13.Souza L. M., M. Iacomini, P. A. J. Gorin, R. S. Sari, M. A. Haddad, and G. L. Sassaki. 2007. Glyco– and sphingophosphonolipids from the medusa Phyllorhiza punctata: NMR and ESI-MS/MS fingerprints. Chem. Phys. Lipids. 145 85–96. [DOI] [PubMed] [Google Scholar]
- 14.Sassaki G. L., P. A. J. Gorin, C. A. Tischer, and M. Iacomini. 2001. Sulfonoglycolipids from the lichenized basidiomycete Dictyonema glabratum: isolation, NMR, and ESI-MS approaches. Glycobiology. 11 345–351. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro S. A., G. L. Sassaki, L. M. Souza, J. A. Meira, J. M. Araújo, D. A. Mitchell, L. P. Ramos, and N. Krieger. 2007. Molecular and structural characterization of the biosurfactant produced by Pseudomonas aeruginosa DAUPE 614. Chem. Phys. Lipids. 147 1–13. [DOI] [PubMed] [Google Scholar]
- 16.Sassaki G. L., P. A. J. Gorin, and M. Iacomini. 2001. Characterization of lyso-galactolipids, C-2 and C-3 O-acyl trigalactosylglycerol isomers, obtained from the lichenized fungus Dictyonema glabratum. FEMS Microbiol. Lett. 194 155–158. [DOI] [PubMed] [Google Scholar]
- 17.Toledo M. S., S. B. Levery, B. Bennion, L. L. Guimarães, S. A. Castle, R. Lindsey, M. Momany, C. Park, A. H. Straus, and H. K. Takahashi. 2007. Analysis of glycosylinositol phosphorylceramides expressed by the opportunistic mycopathogen Aspergillus fumigatus. J. Lipid Res. 48 1801–1824. [DOI] [PubMed] [Google Scholar]
- 18.Weik M., H. Patzelt, G. Zaccai, and D. Oesterhelt. 1998. Localization of glycolipids in membranes by in vivo labeling and neutron diffraction. Mol. Cell. 1 411–419. [DOI] [PubMed] [Google Scholar]
- 19.Sassaki G. L., L. M. Souza, R. V. Serrato, T. R. Cipriani, P. A. J. Gorin, and M. Iacomini. 2008. Application of acetate derivatives for gas chromatography-mass spectrometry: novel approaches on carbohydrates, lipids, amino acids analysis. J. Chromatogr. A. 1208 215–222. [DOI] [PubMed] [Google Scholar]
- 20.Tenchov B., E. M. Vescio, G. D. Sprott, M. L. Zeidel, and J. C. Mathai. 2006. Salt tolerance of archaeal extremely halophilic lipid membranes. J. Biol. Chem. 281 10016–10023. [DOI] [PubMed] [Google Scholar]
- 21.Choquet C. G., G. B. Patel, T. J. Beveridge, and G. D. Sprott. 1992. Formation of unilamellar liposomes from total polar lipid extracts of methanogens. Appl. Environ. Microbiol. 58 2894–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choquet C. G., G. B. Patel, T. J. Beveridge, and G. D. Sprott. 1994. Stability of pressure-extruded liposomes made from archaeobacterial ether lipids. Appl. Microbiol. Biotechnol. 42 375–384. [DOI] [PubMed] [Google Scholar]
- 23.Sprott G. D., J. Brisson, C. J. Dicaire, A. K. Pelletier, L. A. Deschatelets, L. Krishnan, G. B. Patel. 1999. A structural comparison of the total polar lipids from the human archaea Methanobrevibacter smithii and Methanosphaera stadtmanae and its relevance to the adjuvant activities of their liposomes. Biochim. Biophys. Acta. 1440 275–288. [DOI] [PubMed] [Google Scholar]
- 24.Sprott G. D., D. L. Tolson, G. B. Patel. 1997. Archaeosomes as novel antigen delivery systems. FEMS Microbiol. Lett. 154 17–22. [DOI] [PubMed] [Google Scholar]
- 25.Oren A., P. Gurevich, R. T. Gemmell, and A. Teske. 1995. Halobaculum gomorrense gen. nov., sp. nov., a novel extremely halophilic archaeon from the Dead Sea. Int. J. Syst. Bacteriol. 45 747–754. [DOI] [PubMed] [Google Scholar]
- 26.Skispki V. P. 1975. Thin layer chromatography of neutral glycolipids. Methods Enzymol. 35 396–425. [DOI] [PubMed] [Google Scholar]
- 27.Sassaki, G. L., L. M. Souza, T. R.; Cipriani, and M. Iacomini. 2008. TLC of Carbohydrates. In Thin Layer Chromatography in Phytochemistry. M. Waksmundzka-Hajnos, J. Sherma, and T. Kowalska, editors. CRC Press, Boca Raton, FL. 255–271.
- 28.Dittmer J. C., and R. L. Lester. 1964. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 15 126–127. [PubMed] [Google Scholar]
- 29.Ciucanu I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131 209–217. [Google Scholar]
- 30.Kates M., N. Moldoveanu, and L. C. Stewart. 1993. On the revised structure of the major phospholipid of Halobaterium salinarum. Biochim. Biophys. Acta. 1169 46–53. [DOI] [PubMed] [Google Scholar]
- 31.Gonthier I., M-N. Rager, P. Metzger, J. Guezennec, and C. Largeau. 2001. A di-O-dihydrogeranylgeranyl glycerol from Thermococcus S 557, a novel ether lipid, and likely intermediate in the biosynthesis of diethers in Archæa. Tetrahedron Lett. 42 2795–2797. [Google Scholar]
- 32.Domon B., and C. E. Costello. 1988. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5 397–409. [Google Scholar]
- 33.Sassaki G. L., P. A. J. Gorin, L. M. Souza, P. A. Czelusniak, and M. Iacomini. 2005. Rapid synthesis of partially O-methylated alditol acetate standards for GC-MS: some relative activities of hydroxyl groups of methyl glycopyranosides on Purdie methylation. Carbohydr. Res. 340 731–739. [DOI] [PubMed] [Google Scholar]
- 34.Souza L. M., T. R. Cipriani, R. V. Serrato, D. E. Costa, M. Iacomini, P. A. J. Gorin, and G. L. Sassaki. 2008. Analysis of flavonol glycoside isomers from leaves of Maytenus ilicifolia by offline and online high performance liquid chromatography-electrospray mass spectrometry. J. Chromatogr. A. 1207 101–109. [DOI] [PubMed] [Google Scholar]
- 35.Dell A. 1987. F.A.B.-Mass spectrometry of carbohydrates. Adv. Carbohydr. Chem. Biochem. 45 19–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.