Abstract
We employed a novel approach to identify the key loci that harbor genes influencing lipoprotein metabolism in approximately 2,000 pedigreed baboons fed various diets differing in levels of fat and cholesterol. In this study, 126 overlapping traits related to both LDL and HDL metabolism were normalized and subjected to genome-wide linkage screening. As was expected, the traits were highly, but not completely, correlated. We exploited the information in these correlated traits by focusing on those genomic regions harboring quantitative trait loci (QTL) for multiple traits, reasoning that the more influential genes would impact a larger number of traits. This study identified five major QTL clusters (each with at least two significant logarithm of the odds scores >4.7), two of which had not been previously reported in baboons. One of these mapped to the baboon ortholog of human chromosome 1p32-p34 and influenced concentrations of LDL-cholesterol on Basal and high-fat, low-cholesterol diets. The other novel QTL cluster mapped to the baboon ortholog of human chromosome 12q13.13-q14.1 and influenced LDL size properties on high-fat, low-cholesterol and high-fat, high-cholesterol, but not Basal, diets. Confirming the value of this approach, three of the QTL clusters replicated published linkage findings for the same or similar traits.
Keywords: linkage analysis, quantitative trait locus, genetic, primate
Atherosclerosis is a complex disease with etiologies attributable to a variety of environmental, metabolic, and genetic factors. Perhaps the strongest predictor of atherosclerosis is circulating levels of lipoprotein lipids. However, lipoproteins are themselves quite heterogeneous in terms of composition, both lipid and protein, and in terms of regulatory metabolic pathways. It is not surprising, therefore, that a large number of different factors have been identified in various studies to exert significant influence on lipoprotein phenotypes and associated risk of atherosclerosis.
Our group has studied genetic factors that influence lipoprotein phenotypes in pedigreed baboons as a closely related nonhuman primate model of human lipoprotein metabolism. While it is unlikely that the same functional polymorphisms will be found in both humans and baboons, our overall hypothesis has been that genes important to baboon lipoprotein metabolism will also be important to human lipoprotein metabolism.
Of course, a gene can be identified as having great impact in baboons but appear to be unimportant in parallel human studies due to little or no functional variation. Even in this case, however, identification of the gene may help explain rare lipoprotein disorders in humans or may be useful in developing therapeutic drug regimens.
In the course of our studies, we have measured lipoprotein phenotypes in approximately 2,000 pedigreed and genotyped baboons while they were feeding on various diets differing in levels of fat and cholesterol. In this study, we pooled these data to conduct linkage analyses on a large number of interrelated lipoprotein phenotypes. As a first-step attempt at prioritization, we focus here on pleiotropic loci harboring genes that influence clusters of intercorrelated traits and that therefore are likely to play a fundamental role in lipoprotein metabolism.
METHODS
Pedigreed baboons
The baboons from which data were obtained in this study were organized into a single, unbroken six-generation pedigree configuration with 2,426 individuals. This pedigree, with approximately 384 founders, has resulted from managed breeding of olive baboons (Papio hamadryas anubis), yellow baboons (P. h. cynocephalus), and their hybrid progeny over the past three decades. Statistical power to detect, localize, and characterize genetic effects in a pedigree derives largely from the information contained in the relative pair classes contained therein. In this complex pedigree, full sibships ranged in size from 2 (n = 372) to 12 (n = 10), with the median = 5, and there were 50 additional classes of relative pairs in this extended pedigree. Examples of these pair classes included parent-offspring (n = 350), half sibling (n = 6855), half avuncular (n = 6414), and double half first cousin (n = 754) pairs.
Diet protocol
Nearly 2,000 of these pedigreed baboons were sampled while feeding on one or more diets differing in level of fat and cholesterol (i.e., Basal diet low in fat and cholesterol, HFHC diet high in fat and cholesterol, and HFLC diet high in fat but low in cholesterol); the diets and the diet experiment have been described previously (1). Animals were fasted overnight, immobilized with ketamine, and bled from the femoral artery. Serum was frozen at −80°C in single-use aliquots, protected from desiccation and oxidation (2).
Animal care and maintenance were performed by the veterinary resources staff at Southwest Foundation for Biomedical Research, a facility certified by the Association for Assessment and Accreditation of Laboratory Animal Care International. All procedures were approved by the Institutional Animal Care and Use Committee and conformed to the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Measurement of lipid and lipoprotein traits
Methods for measuring lipid and lipoprotein traits have been reported previously; below are given brief descriptions and references for the methods employed in measuring traits used in this study. Cholesterol and triglyceride (TG) concentrations were determined enzymatically using commercial reagents in a clinical chemistry analyzer (3). HDL-cholesterol (HDL-C) was measured in the supernatant after heparin-Mn+2 precipitation (4), and nonHDL-C was calculated as the difference between total and HDL-C. Concentrations of apolipoprotein AI (apoA1), apoB, and apoE were determined using an immunoturbidometric approach with commercial reagents in a clinical chemistry analyzer (5, 6). Lp(a) concentrations were measured using a sandwich-style ELISA (7).
We used composite gradient gels to resolve LDLs and HDLs on the basis of size (8) and Sudan black B to quantify lipoprotein cholesterol size distributions. Fractional cholesterol absorbance among apoB-containing lipoproteins was estimated for VLDL1 (V1-C; 36–33 nm), VLDL2 (V2-C; 33–30 nm), LDL1 (L1-C; 30–28 nm), LDL2 (L2-C; 28–27 nm), LDL3 (L3-C; 27–26 nm), and LDL4 (L4-C; 26–24 nm). Fractional cholesterol absorbance among HDLs was estimated for HDL1A (H1A-C; 24–20 nm), HDL1B (H1B-C; 20–13 nm), HDL2 (H2-C; 13–9.9 nm), and HDL3 (H3-C; 9.9–8.2 nm). In addition, serum lipoproteins also were resolved in HDL gels (9), and apoE size distributions were quantified by immunoblotting procedures as described (10, 11); fractional apoE absorbance among lipoproteins was estimated for apoB-containing lipoproteins (VL-E; 36–25 nm), HDL1A (H1A-E; 25–21 nm), HDL1B (H1B-E; 21–13 nm), HDL2 (H2-E; 13–9.9 nm), and HDL3 (H3-E; 9.9–7.2 nm). An estimate of serum concentration for each fraction was obtained by multiplication with nonHDL or HDL-C or apoE concentrations as appropriate. For cholesterol distributions only, we estimated median diameters for apoB-containing lipoproteins (Lmed-C; 24–36 nm) and HDLs (Hmed-C; 8.2–24 nm), which were defined as the diameter where half the absorbance was on larger, and half was on smaller, particles. LDL peak particle diameter was also estimated at the same time (8).
Serum activity levels of two lipoprotein-associated enzymes, lipoprotein-associated phospholipase A2 (Lp-PLA2), and paraoxonase (PON) were also measured as described elsewhere. Lp-PLA2 was assayed using 2-thio-PAF as substrate in a commercial microplate-based colorimetric assay (12). PON was assayed using paraoxon as substrate in a microplate-based colorimetric assay (13).
Genotyping and construction of the map
Quantitative trait locus (QTL) localization analyses reported in this article used an updated version of the second-generation baboon whole-genome genetic linkage map that is based largely on human microsatellite markers that also are polymorphic in baboons (14, 15). This enables extrapolation of results to the orthologous human genomic locations; in this text, therefore, map locations will refer to the human chromosomal regions. Based on genotype data from 2,044 baboons, the version of the map used for this study included 287 markers mapped at 1,000:1 odds, at an average of 7.7 cM across the 20 baboon autosomes. Links to information regarding the current whole-genome linkage map for this species (including marker characteristics and genotyping conditions) can be found at http://baboon.sfbrgenetics.org/. Genotype data from all 2,044 animals on which the map is based were also used to estimate multipoint identity-by-descent (IBD) coefficients for this study.
Statistical genetic analyses
Data for 42 traits on each of three diets were available for these analyses. Because the data spanned several different experiments with differing objectives, there was a variable number of animals with data for any one trait. Each trait was preadjusted for a standard set of covariates (i.e., age, sex, and weight) that have proven to be significant covariates in many previous studies, even if one (or more) covariate was not significant for the trait. On average, these covariates accounted for 5% of total phenotypic variance (range was 0 to 32%), so the bulk of raw trait variance was retained in the residual. The residual variables were then normalized by fitting to a Gaussian distribution and standardized such that mean and SD were 0 and 1, respectively. These transformations were done in the software package SOLAR (16), and the normalized traits were used for the analyses described below.
Simple Pearson correlation analyses were performed on the transformed adjusted traits using a commercial statistics package (Stata 9.2; College Station, TX). We estimated the genetic effect, or heritability (h2), as the proportion of the residual variance in each trait that was attributable to the additive effects of genes (i.e., σ2G/σ2P). Genome-wide multipoint linkage screening of these traits was done using maximum-likelihood-based variance decomposition routines implemented in SOLAR. Briefly, we estimated the amount of variance attributable to a QTL (σ2Q) by specifying the expected genetic covariances between arbitrary relative pairs as a function of the IBD relationships at a given marker locus assumed to be tightly linked to a locus that influences the trait (16). Markov Chain Monte Carlo routines, implemented in Loki (17), were used to estimate multipoint IBD probabilities for all relative pairs in the single pedigree. We conducted likelihood ratio tests for linkage at 1-cM intervals across all autosomes. Evidence for linkage was expressed in terms of logarithm of the odds (LOD) scores, obtained as the quotient of the likelihood ratio test statistic divided by the value loge10 (18).
Genome-wide probability values incorporated information on marker density and on the structure of the pedigree and they were calculated as suggested by Feingold, Brown, and Siegmund (19); by this calculation, a LOD = 2.75 was associated with genome-wide significance level of 0.05, a conventional criterion for QTL significance. However, based on our consideration of 126 traits, and employing a Bonferroni approach, we defined as significant in this study a QTL with a LOD score >4.7, which is the value associated with a genome-wide significance level of 0.00040 (i.e., 0.05/126) in this pedigree. We then defined as suggestive a QTL satisfying the conventional criterion of genome-wide significance (i.e., a LOD score >2.75). The purpose of imposing this relatively stringent criterion was to focus attention on the most important and reliable QTLs.
Bivariate analyses were employed to test the contrasting hypotheses of pleiotropy versus coincident linkage to explain the QTL clusters. These tests included the trait having the strongest evidence of a QTL paired with each of the other traits having a significant QTL. Implemented in SOLAR, bivariate multipoint linkage analyses identified the genomic location returning the highest joint (bivariate) LOD score within the cluster interval (20). At that point, the model partitioned the genetic variance for each trait by estimating the QTL heritability (h2Q), defined as the proportion of total phenotypic variance explained by the joint QTL (σ2Q/σ2P), and the residual heritability, defined as (σ2G/σ2P)−h2Q. In addition, the additive genetic correlation (ρQ) between traits attributable to the QTL was calculated in the model based on estimated IBD allele sharing. Hypotheses of QTL pleiotropy versus coincident linkage were tested by comparing the likelihoods of models in which ρQ was estimated to those of models in which it was fixed at zero; a ρQ significantly >0 implied pleiotropy. Complete QTL pleiotropy was tested by comparing models in which ρQ was estimated to models in which it was fixed at 1 (absolute value); a ρQ not significantly different from 1 was interpreted as evidence for complete QTL pleiotropy (21). However, in none of these tests did a pleiotropic QTL account for all the additive genetic variance.
RESULTS
Lipid and lipoprotein traits
For the purposes of multiple studies, we obtained measures of 42 lipoprotein traits for animals fed a series of three defined diets differing in levels of fat and cholesterol. The traits included various measures of lipoprotein properties, including concentrations and size distribution characteristics of several lipids and apolipoproteins. Half the traits were categorized as LDL related and half as HDL related. Each of the traits was significantly heritable at the P < 0.05 level, and heritabilities ranged from 6% to 71% for the 126 traits (Table 1).
TABLE 1.
Residual heritability ± SEM (%) for 21 LDL-related and 21 HDL-related traits measured in baboons fed three diets
| Basal |
HFLC |
HFHC |
||||||
|---|---|---|---|---|---|---|---|---|
| Group | Traita | n | h2 | n | h2 | n | h2 | |
| LDL | Frac | V1-C | 1,707 | 10.2 ± 3.6 | 1,298 | 27.6 ± 5.1 | 1,721 | 25.3 ± 4.4 |
| LDL | Frac | V2-C | 1,707 | 37.3 ± 4.3 | 1,298 | 45.7 ± 4.6 | 1,721 | 44.5 ± 4.6 |
| LDL | Frac | L1-C | 1,707 | 31.4 ± 4.9 | 1,298 | 30.1 ± 5.1 | 1,721 | 42.5 ± 5.2 |
| LDL | Frac | L2-C | 1,707 | 15.8 ± 4.1 | 1,298 | 23.6 ± 5.8 | 1,721 | 28.0 ± 5.0 |
| LDL | Frac | L3-C | 1,707 | 24.9 ± 4.6 | 1,298 | 38.6 ± 5.8 | 1,721 | 44.4 ± 5.1 |
| LDL | Frac | L4-C | 1,707 | 21.1 ± 4.0 | 1,298 | 34.0 ± 5.5 | 1,721 | 36.1 ± 4.6 |
| LDL | Frac | VL-E | 690 | 16.0 ± 6.6 | 690 | 26.7 ± 7.2 | 689 | 42.3 ± 7.7 |
| LDL | Diam | Lmed-C | 1,648 | 30.1 ± 4.6 | 1,258 | 39.0 ± 5.6 | 1,658 | 48.6 ± 5.1 |
| LDL | Diam | LDLPPD | 1,648 | 25.0 ± 4.3 | 1,258 | 29.4 ± 5.3 | 1,658 | 45.3 ± 5.1 |
| LDL | Conc | NonHDL-C | 1,937 | 53.2 ± 3.8 | 1,299 | 61.1 ± 4.6 | 1,956 | 58.3 ± 3.9 |
| LDL | Conc | TG | 1,085 | 22.9 ± 5.3 | 1,212 | 31.4 ± 5.5 | 1,126 | 29.0 ± 6.3 |
| LDL | Conc | Lp(a) | 681 | 70.5 ± 6.0 | 689 | 66.1 ± 6.2 | 686 | 71.2 ± 5.7 |
| LDL | Conc | ApoB | 1,229 | 38.8 ± 5.3 | 1,269 | 52.1 ± 5.2 | 1,256 | 46.4 ± 5.2 |
| LDL | Conc | V1-C | 1,648 | 21.4 ± 3.9 | 1,258 | 40.8 ± 5.1 | 1,658 | 41.0 ± 4.5 |
| LDL | Conc | V2-C | 1,648 | 38.8 ± 4.1 | 1,258 | 49.5 ± 4.4 | 1,658 | 53.2 ± 4.7 |
| LDL | Conc | L1-C | 1,648 | 38.5 ± 4.5 | 1,258 | 44.9 ± 4.9 | 1,658 | 58.0 ± 4.6 |
| LDL | Conc | L2-C | 1,648 | 37.3 ± 4.6 | 1,258 | 51.6 ± 5.6 | 1,658 | 30.4 ± 5.5 |
| LDL | Conc | L3-C | 1,648 | 25.5 ± 4.5 | 1,258 | 28.7 ± 5.7 | 1,658 | 35.0 ± 5.4 |
| LDL | Conc | L4-C | 1,648 | 23.7 ± 4.6 | 1,258 | 36.7 ± 6.2 | 1,658 | 27.8 ± 4.9 |
| LDL | Conc | VL-E | 590 | 12.0 ± 4.7 | 660 | 23.5 ± 6.3 | 628 | 47.3 ± 8.0 |
| LDL | Act | Lp-PLA2 | 713 | 69.3 ± 7.2 | 706 | 55.7 ± 8.4 | 716 | 66.8 ± 6.8 |
| HDL | Frac | H1A-C | 1,707 | 24.6 ± 4.0 | 1,298 | 21.4 ± 4.6 | 1,721 | 22.6 ± 4.2 |
| HDL | Frac | H1B-C | 1,707 | 29.4 ± 4.3 | 1,298 | 35.4 ± 5.1 | 1,721 | 35.4 ± 4.6 |
| HDL | Frac | H2-C | 1,707 | 17.8 ± 4.1 | 1,298 | 33.6 ± 5.2 | 1,721 | 28.8 ± 4.0 |
| HDL | Frac | H3-C | 1,707 | 37.3 ± 4.2 | 1,298 | 32.6 ± 5.2 | 1,721 | 35.0 ± 4.4 |
| HDL | Frac | H1A-E | 690 | 23.5 ± 6.1 | 690 | 31.2 ± 7.2 | 689 | 37.1 ± 7.3 |
| HDL | Frac | H1B-E | 690 | 12.6 ± 7.1 | 690 | 10.9 ± 6.7 | 689 | 25.9 ± 7.5 |
| HDL | Frac | H2-E | 690 | 20.0 ± 5.2 | 690 | 32.8 ± 6.8 | 689 | 43.4 ± 7.3 |
| HDL | Frac | H3-E | 690 | 9.7 ± 5.8 | 690 | 5.8 ± 3.7 | 689 | 19.1 ± 6.2 |
| HDL | Diam | Hmed-C | 1,648 | 38.5 ± 4.6 | 1,258 | 40.4 ± 5.8 | 1,658 | 36.0 ± 4.7 |
| HDL | Conc | HDLC | 1,937 | 55.2 ± 4.0 | 1,299 | 61.8 ± 5.0 | 1,956 | 49.1 ± 4.2 |
| HDL | Conc | ApoA1 | 1,234 | 45.1 ± 4.9 | 1,270 | 44.7 ± 4.7 | 1,257 | 39.0 ± 4.9 |
| HDL | Conc | ApoE | 1,197 | 34.7 ± 5.5 | 1,270 | 36.6 ± 5.4 | 1,239 | 37.5 ± 5.7 |
| HDL | Conc | H1A-C | 1,648 | 25.4 ± 3.9 | 1,258 | 31.7 ± 4.9 | 1,658 | 24.3 ± 4.3 |
| HDL | Conc | H1B-C | 1,648 | 40.7 ± 4.8 | 1,258 | 39.0 ± 5.2 | 1,658 | 41.8 ± 4.9 |
| HDL | Conc | H2-C | 1,648 | 43.9 ± 4.4 | 1,258 | 40.1 ± 5.3 | 1,658 | 34.5 ± 4.6 |
| HDL | Conc | H3-C | 1,648 | 32.2 ± 4.3 | 1,258 | 17.3 ± 4.0 | 1,658 | 30.2 ± 4.4 |
| HDL | Conc | H1A-E | 590 | 35.5 ± 7.8 | 660 | 38.6 ± 7.7 | 628 | 45.9 ± 8.1 |
| HDL | Conc | H1B-E | 590 | 26.8 ± 7.6 | 660 | 32.9 ± 8.0 | 628 | 33.8 ± 7.3 |
| HDL | Conc | H2-E | 590 | 13.0 ± 5.7 | 660 | 15.1 ± 6.0 | 628 | 11.1 ± 6.3 |
| HDL | Conc | H3-E | 590 | 19.1 ± 7.6 | 660 | 9.9 ± 5.3 | 628 | 14.4 ± 7.3 |
| HDL | Act | PON | 1,178 | 54.2 ± 5.4 | 1,155 | 67.7 ± 5.1 | 1,185 | 59.1 ± 5.4 |
LDLPPD, LDL peak particle diameter; Lp-PLA2, lipoprotein-associated phospholipase A2.
Trait names are defined in Methods; preceding the name is the type of measure: Frac, fraction; Diam, diameter; Conc, concentration; Act, enzyme activity.
These traits were by no means independent. Correlation analyses of the transformed traits confirmed the expected high degree of intercorrelation among the traits. For example, the average Pearson correlation coefficient (absolute value) was 0.202, and 78% of all possible pairwise correlations were significant at the P < 0.05 level. Thus, we anticipated developing substantial evidence for pleiotropy among these traits.
Screen for QTLs influencing lipid and lipoprotein traits
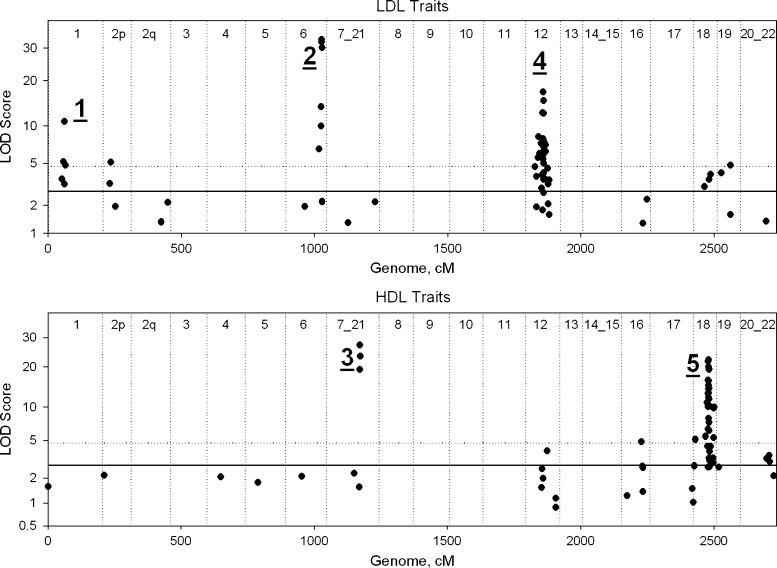
Each trait was individually preadjusted for sex, age, and weight, transformed, and then subjected to a genome-wide linkage screen. Figure 1 plots, for LDL (upper panel) and HDL (lower panel) traits, the genomic locations of the primary QTL for each trait. For convenience, we have sorted and labeled the baboon genome using the homologous human designations. Of the 126 traits, 87 (69%; 45 LDL-related and 42 HDL-related traits) returned LOD scores >2.75 (i.e., genome-wide P < 0.05) and 57 of those (45%; 28 LDL-related and 29 HDL-related traits) were significant after adjustment for multiple testing (i.e., LOD >4.7; genome-wide P < 0.00040). The location and peak LOD score of the primary QTL for each of the 126 traits in this study are given in supplementary Table I (http://baboon.sfbrgenetics.org/Bab_SupplementalData/Rainwater2009.php?pageNum_rsDRSuppDataList=0&totalRows_rsDRSuppDataList=126). This table also provides links to the multipoint LOD score plots for all autosomes for each of the 126 traits.
Fig. 1.
Chromsomal locations of peak LOD score for the primary QTL for each of 63 LDL-related (upper panel) and 63 HDL-related (lower panel) traits. Baboon chromosomal designations are indicated as the corresponding human chromosome orthologs. The horizontal lines indicate LOD scores defined as suggestive (2.75; solid) and significant (4.7; dotted). The five major clusters are indicated by the underlined numerals.
A major QTL cluster was defined as two or more significant QTLs whose 1-LOD support intervals enclosed the peak location of the predominant QTL. Under this definition, it would be possible for the cluster to result from the actions of a single pleiotropic gene. We hypothesized that such a pleiotropic gene, influencing multiple traits, might play a key role in lipoprotein metabolism. In this study, we identified five clusters of significant QTLs that satisfied this criterion, and Table 2 lists the QTLs, their locations, and peak LOD scores. It is noteworthy that each of these clusters is anchored by a predominant QTL with LOD score exceeding 10.
TABLE 2.
Locations of peak LOD scores for traits comprising five major QTL clusters
| Cluster | Chromosome | cM | LOD | Trait | Diet | |
|---|---|---|---|---|---|---|
| 1 | 1 | 55 | 5.16 | Conc | NonHDL-C | HFLC |
| 1 | 1 | 60 | 10.76 | Conc | NonHDL-C | Basal |
| 1 | 1 | 64 | 4.81 | Conc | L3-C | Basal |
| 2 | 6 | 124 | 6.63 | Frac | V2-C | HFLC |
| 2 | 6 | 132 | 13.63 | Frac | V2-C | Basal |
| 2 | 6 | 132 | 10.01 | Conc | V2-C | Basal |
| 2 | 6 | 134 | 32.98 | Conc | Lp(a) | HFLC |
| 2 | 6 | 134 | 32.09 | Conc | Lp(a) | HFHC |
| 2 | 6 | 135 | 30.09 | Conc | Lp(a) | Basal |
| 3 | 7_21 | 124 | 19.21 | Act | PON | Basal |
| 3 | 7_21 | 124 | 27.25 | Act | PON | HFLC |
| 3 | 7_21 | 126 | 23.37 | Act | PON | HFHC |
| 4 | 12 | 45 | 5.59 | Frac | L3-C | HFLC |
| 4 | 12 | 51 | 6.04 | Conc | L2-C | HFHC |
| 4 | 12 | 63 | 12.37 | Diam | LDLPPD | HFHC |
| 4 | 12 | 64 | 5.43 | Frac | V1-C | HFHC |
| 4 | 12 | 64 | 17.11 | Diam | Lmed-C | HFHC |
| 4 | 12 | 64 | 5.87 | Conc | V1-C | HFHC |
| 4 | 12 | 65 | 7.96 | Frac | L1-C | HFLC |
| 4 | 12 | 65 | 6.23 | Diam | Lmed-C | HFLC |
| 4 | 12 | 66 | 5.01 | Conc | L1-C | HFLC |
| 4 | 12 | 67 | 15.02 | Frac | V2-C | HFHC |
| 4 | 12 | 67 | 6.86 | Frac | L3-C | HFHC |
| 4 | 12 | 67 | 12.26 | Conc | V2-C | HFHC |
| 4 | 12 | 69 | 7.53 | Conc | L3-C | HFHC |
| 5 | 18 | 46 | 5.46 | Frac | H3-C | HFLC |
| 5 | 18 | 51 | 10.92 | Frac | H1A-C | HFHC |
| 5 | 18 | 54 | 16.11 | Conc | HDLC | Basal |
| 5 | 18 | 54 | 10.13 | Conc | H1B-E | HFLC |
| 5 | 18 | 54 | 12.88 | Conc | H1A-C | HFHC |
| 5 | 18 | 55 | 6.39 | Frac | H1A-C | HFLC |
| 5 | 18 | 55 | 21.71 | Conc | HDLC | HFLC |
| 5 | 18 | 56 | 19.92 | Diam | Hmed-C | HFLC |
| 5 | 18 | 56 | 22.18 | Conc | H1B-C | HFLC |
| 5 | 18 | 56 | 13.99 | Diam | Hmed-C | HFHC |
| 5 | 18 | 57 | 13.04 | Frac | H1B-C | Basal |
| 5 | 18 | 57 | 11.93 | Diam | Hmed-C | Basal |
| 5 | 18 | 57 | 16.03 | Conc | H1B-C | Basal |
| 5 | 18 | 57 | 14.85 | Frac | H1B-C | HFLC |
| 5 | 18 | 57 | 8.04 | Conc | H1A-C | HFLC |
| 5 | 18 | 58 | 19.34 | Frac | H2-C | HFLC |
| 5 | 18 | 58 | 19.19 | Conc | ApoE | HFLC |
| 5 | 18 | 58 | 6.21 | Conc | H1A-E | HFLC |
| 5 | 18 | 58 | 14.10 | Frac | H2-C | HFHC |
| 5 | 18 | 58 | 11.63 | Conc | ApoE | HFHC |
| 5 | 18 | 59 | 7.37 | Conc | ApoE | Basal |
| 5 | 18 | 60 | 10.15 | Frac | H2-C | Basal |
Chromosome numbering is based on that of the homologous human chromosomes. Traits indicated in bold exhibited the highest LOD score for a cluster.
Description of QTL clusters
Cluster 1 comprised three QTLs for concentrations of LDL and LDL subfractions on Basal and HFLC, but not HFHC, diets; two suggestive QTLs in the vicinity helped confirm this cluster. Human microsatellite markers mapping near this cluster included D1S2130, D1S515, and D1S192 (which were located at 48.2, 62.3, and 68.4 cM, respectively; all locations represent map distance in cM from the pter-most marker locus on the chromosome). Cluster 2 comprised six QTLs for intermediate-size lipoproteins, such as Lp(a), on all diets. Nearby markers included D6S404, D6S1698, and D6S503 (124.5, 127.2, and 146.9 cM). Cluster 3 comprised QTLs for paraoxonase activity levels on each of the three diets. Nearby microsatellite markers included D7S821 and D7S2204 (123.4 and 132.4 cM). Cluster 4 comprised 13 QTLs on chromosome 12; the QTLs are for LDL-related traits on the two high-fat diets, including various estimates of size distribution and concentration of apoB-containing lipoprotein cholesterol; four suggestive QTLs helped confirm this cluster. Four LOD scores >10 in this cluster were flanked by markers D12S297 and D12S75 (62.6 and 72.6 cM). Finally, Cluster 5 comprised 22 QTLs for a variety of HDL-related traits, including size distributions and concentrations of HDL cholesterol, apoE, and apoA1. Seventeen LOD scores >10 were flanked by human microsatellite markers D18S475, D18S72, and D18S1156 (41.8, 51.9, and 61.5 cM). The baboon microsatellite marker D18Spha1 (57.4 cM) is located within exon 9 of the structural locus for endothelial lipase (LIPG) (22).
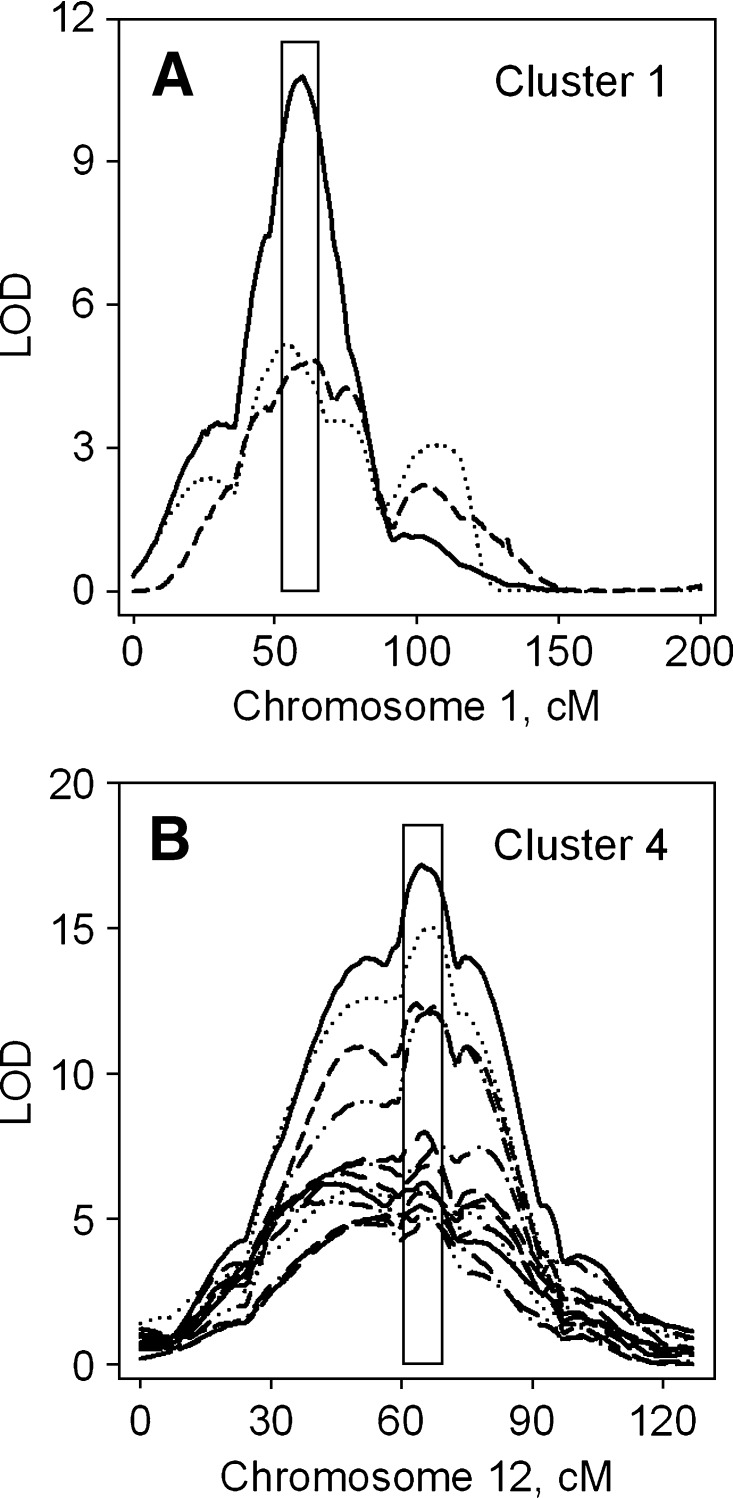
Of these five major QTL clusters, two, clusters 1 and 4, had not been reported previously in baboons. Figure 2 graphs the multipoint LOD score plots for the three traits comprising Cluster 1 on chromosome 1 (panel A) and for the 13 traits comprising Cluster 4 on chromosome 12 (panel B). Below, we further characterize the effects of genes that might be responsible for clusters 1 and 4.
Fig. 2.
Multipoint LOD score plots for the traits comprising QTL Clusters 1 (A) and 4 (B) on the baboon orthologs of human chromosomes 1 and 12, respectively. The boxes indicate the 1-LOD drop intervals for the predominant QTL.
Further characterization of Cluster 1 QTLs on the baboon ortholog of human chromosome 1
The gene(s) responsible for Cluster 1 obviously influences LDL metabolism, but in this case the specific effects are on cholesterol concentrations and in diets with low cholesterol levels. The trait with the highest LOD score peak for this cluster was NonHDL-C on Basal diet (LOD = 10.76). To test the hypotheses of QTL pleiotropy versus coincident linkage, we conducted bivariate linkage analyses that paired this trait with each of the other traits with significant QTLs (Table 3). First, it should be noted that on average the QTL explained less than half the additive genetic variance for these traits. For the pair nonHDL-C on Basal and HFLC diets, the QTL correlation estimated to be 1, whereas for nonHDL-C and concentration of LDL1-C on Basal diet the correlation was high (0.79) but significantly different from 1. Still, the QTL correlations are all significantly greater than zero, suggesting that a single pleiotropic gene is largely responsible for this cluster.
TABLE 3.
Results from bivariate linkage analyses
| Joint |
Trait |
1 |
Trait |
2 |
P |
|||
|---|---|---|---|---|---|---|---|---|
| Trait 1 (Diet) | Trait 2 (Diet) | LOD | h2R | h2Q | h2R | h2Q | ρQ | (|ρQ| = 1) |
| Chromosome 1 | ||||||||
| Conc NonHDL-C (Basal) | Conc NonHDL-C (HFLC) | 11.2 | 26.8 | 22.8 | 43.1 | 14.5 | 1.000 | 1.000 |
| Conc NonHDL-C (Basal) | Conc L3-C (Basal) | 13.2 | 28.0 | 23.9 | 11.9 | 13.6 | 0.795 | 0.001 |
| Chromosome 12 | ||||||||
| Diam Lmed-C (HFHC) | Frac L3-C (HFLC) | 17.8 | 29.2 | 16.9 | 29.3 | 6.5 | −1.000 | 1.000 |
| Diam Lmed-C (HFHC) | Conc L2-C (HFHC) | 18.0 | 30.0 | 15.7 | 24.0 | 7.4 | −0.472 | 0.008 |
| Diam Lmed-C (HFHC) | Diam LDLPPD (HFHC) | 16.2 | 30.2 | 16.5 | 29.0 | 13.9 | 0.983 | 0.100 |
| Diam Lmed-C (HFHC) | Frac V1-C (HFHC) | 15.5 | 29.8 | 16.1 | 18.8 | 5.7 | 0.880 | 0.301 |
| Diam Lmed-C (HFHC) | Conc V1-C (HFHC) | 16.2 | 29.4 | 17.9 | 36.1 | 6.4 | 0.932 | 0.362 |
| Diam Lmed-C (HFHC) | Diam Lmed-C (HFLC) | 17.5 | 28.5 | 16.3 | 26.8 | 7.6 | 0.999 | 0.993 |
| Diam Lmed-C (HFHC) | Frac L1-C (HFLC) | 19.2 | 29.9 | 15.7 | 18.5 | 9.6 | 1.000 | 1.000 |
| Diam Lmed-C (HFHC) | Conc L1-C (HFLC) | 16.6 | 29.9 | 15.4 | 36.8 | 6.7 | 0.991 | 0.918 |
| Diam Lmed-C (HFHC) | Frac L3-C (HFHC) | 19.2 | 33.0 | 14.3 | 33.1 | 8.0 | −1.000 | 1.000 |
| Diam Lmed-C (HFHC) | Conc V2-C (HFHC) | 16.4 | 29.9 | 17.1 | 42.4 | 11.4 | 0.957 | 0.165 |
| Diam Lmed-C (HFHC) | Frac V2-C (HFHC) | 17.0 | 29.5 | 15.9 | 31.8 | 13.1 | 0.987 | 0.431 |
| Diam Lmed-C (HFHC) | Conc L3-C (HFHC) | 15.9 | 29.6 | 16.4 | 24.3 | 9.4 | −0.979 | 0.493 |
The models included the indicated trait pairs from Cluster 1 on chromosome 1 and Cluster 4 on chromosome 12; the table gives joint LOD score, proportion (%) of trait variances attributable to the residual (h2R) and QTL (h2Q) genetic components, the QTL correlation (ρQ), and the P value for the test of complete QTL pleiotropy. ρQ is the QTL-specific additive genetic correlation; the P value is from the loge-likelihood ratio test of the hypothesis that ∣ρQ∣ = 1.
To help narrow the region in which to search for candidate genes, we identified the 1-LOD support interval for nonHDL-C on Basal diet to be 53–65 cM on the baboon map. Selection of this region was confirmed by the observation that the peak LOD scores for the other two traits with significant LODs also fell within this interval. Under the reasonable assumption of cross-species conservation in orthologous syntenic regions known to exhibit conserved microsatellite marker order (15), we interpolated QTL locations on the human physical map based on genetic distances between the QTL and flanking markers. By this process, this QTL support interval maps to 1p32-p34, a region that is 50.2–67.6 mb from pter [physical locations were taken from National Center for Biotechnology Information build 36.1 of March 2006, by use of the University of California, Santa Cruz Human Genome Browser (23)].
Further characterization of Cluster 4 QTLs on the baboon ortholog of human chromosome 12
The gene(s) responsible for Cluster 4 obviously influences LDL metabolism and, more specifically, it appears to influence LDL size distributions on the high-fat diet. This conclusion is based on the observation that most of the significant QTLs were for fractions or diameter data, including the three highest LODs in the cluster. The trait with the highest LOD score peak was Lmed-C on HFHC diet (LOD = 17.11). To test the hypotheses of QTL pleiotropy versus coincident linkage, we conducted bivariate linkage analyses that paired this trait with each of the 12 other traits exhibiting significant LOD scores (Table 3). In this cluster, the QTL explained about a quarter of the additive genetic variance in each trait. All of the 12 QTL correlations were significantly different from zero at the P < 0.05 level, and all but one of the QTL correlations was indistinguishable from unity. These observations suggest that a single pleiotropic gene is responsible for the major QTL cluster observed on the baboon ortholog of human chromosome 12.
To help narrow the region in which to search for candidate genes for this QTL, we determined the 1-LOD support interval for Lmed-C on HFHC to be 61–69 cM on the baboon map. Further corroborating the selection of this interval, the LOD score peaks for all but two of the clustered traits in Table 2 fell within this same interval. Interpolations based on nearby human microsatellite markers suggested this interval corresponds approximately to cytogenetic location 12q13.13-q14.1 and physical location 48.7–61.2 mb on the map of human chromosome 12.
DISCUSSION
In this study, we conducted genome screens for 126 lipid and lipoprotein traits that included size and concentration measures of protein and lipid components of LDLs and HDLs. A key focus of the experiments underlying this study was detection of diet effects on QTLs, namely the effects of increasing levels of dietary fat (comparing Basal and HFLC diets) and of increasing levels of dietary cholesterol in the high-fat environment (comparing HFLC and HFHC diets). In some cases, a QTL showed no evidence of diet effects; for example, PON and Lp(a) QTLs on each diet mapped to the same genomic regions. However, more intriguing was the detection of QTLs for a trait only on certain diets or even on different chromosomes depending on diet. For example, significant QTLs for nonHDL-C were detected on chromosomes 1 (Basal and HFLC diets) and 19 (HFHC diet), suggestive of diet-by-genotype interaction. There can be many reasons why QTLs influencing a trait might differ across diets, including the possibility that our criterion for declaring significance was too stringent (or not sufficiently stringent). While we cannot exclude the possibility that Type 1 and Type 2 errors were responsible for apparent diet differences, a likely interpretation is genotype-by-diet interaction, wherein the effects of different genes predominate depending on dietary environment. In fact, some of our previous baboon studies have reported evidence of diet-by-genotype interaction for HDL- and LDL-related traits (13, 24–26). However, additional explorations of novel diet-by-genotype interactions developed in this study await future analyses.
This study focused on characterizing loci that harbor genes important to lipoprotein metabolism. For nearly 70% of the 126 traits, we detected a QTL that was significant by conventional criteria (i.e., genome-wide P < 0.05). To minimize the possibility of Type 1 error due to the multiple tests, we elected to restrict the criterion for significance to be a genome-wide P value < 0.0004, which is equivalent to a LOD score of 4.7 in our baboon pedigree. Using this criterion, we detected significant QTLs for 57 (45%) of the traits. It should be noted that this criterion is probably overly conservative, given the observed degree of intercorrelation among traits that serves to reduce the effective number of independent traits. The significant QTLs were found on the baboon orthologs of human chromosomes 1, 2, 6, 7, 12, 16, 18, and 19.
To help prioritize for further characterization, we attempted to localize pleiotropic QTLs that influenced multiple lipoprotein traits under the assumption that the genes underlying them are likely to play fundamental roles in lipoprotein metabolism. Therefore, we defined a major QTL cluster as two or more QTLs whose 1-LOD support intervals enclosed the peak of the predominant QTL. In fact, 47 of the 57 significant QTLs in this study colocalized into one of five major QTL clusters under this definition.
Two of these major clusters (Cluster 2 and Cluster 3) mapped to chromosomal regions containing the appropriate structural locus. LPA is the structural locus on chromosome 6 for the Lp(a)-specific protein apo(a). Previous studies in baboons (7, 27, 28) and humans (29) have demonstrated genetic variation at this locus accounts for the majority of variation in Lp(a) levels. In this study, the locus also appears to influence lipoprotein particles in V2-C size range (30–33 nm), which is the size range that would contain baboon Lp(a) particles. PON1 is the structural locus on chromosome 7 for the PON protein found in serum. Our previous study in baboons detected a QTL in this region (13), and numerous studies in humans (30–33) have demonstrated significant effects of PON1 polymorphisms on PON activity.
Cluster 5 mapped to the baboon ortholog of human chromosome 18 and included 22 significant QTLs for HDL traits. The significant traits tended to be for larger HDLs that are enriched in apoE (10). This QTL cluster confirms our previous study involving many of the same traits and animals and in which we reported promoter variants in LIPG, the structural locus for endothelial lipase on chromosome 18, to regulate HDLC levels (22).
Finally, two of the QTL clusters, Cluster 1 and Cluster 4, identified chromosomal regions in which we have not previously localized QTLs for lipoprotein-related traits. Both these clusters had multiple significant QTLs and they reflected pleiotropic effects on various LDL traits. However, the two clusters differed markedly in the types of LDL measures that were affected. Below, we discuss the two clusters in greater detail.
Cluster 1 included three significant and two suggestive QTLs that represented measures of LDL-C and LDL subfraction concentrations on the two low cholesterol diets. The QTLs clustered in a genomic region corresponding to human chromosome 1p32-p34. Falling within the 1-LOD support interval for the largest signal are 105 genes (23, 34), including LRP8 (encoding LDL receptor-related protein 8, apoER2, which is an apoE receptor), SCP2 (encoding sterol carrier protein-2), and PCSK9 (encoding proprotein convertase subtilisin/kexin type 9). Although it binds apoE, apoER2 is found only in brain and testis and so is not likely to play a role in lipoprotein metabolism of the circulation (35). SCP-2 plays important roles in intracellular lipid metabolism and signaling (36) but does not appear to play a direct role in LDL metabolism. However, PCSK9 is a particularly strong candidate, based on an extensive list of polymorphisms that modulate LDL receptor levels, in both positive and negative directions, and thereby LDL-C levels [reviewed in (37)]. In fact, there was a strong correlation (r = 0.54, P = 0.001, n = 115) in diabetic humans between circulating levels of PCSK9 protein and LDL-C (38). PCSK9 hepatic transcript expression in mice is substantially reduced by dietary cholesterol (39), which could help explain why we have detected significant QTLs only on low cholesterol diets in baboons.
Genome-wide single-nucleotide polymorphism association studies have been reported recently for various populations, and several have investigated effects on LDL-related measures (40–44). These studies have identified significant associations of LDL-C with polymorphisms in various genes. With respect to chromosome 1, several genes have been implicated for LDL-C [CELSR2/PSRC1/SORT1 (40, 42, 43) and PCSK9 (40, 43, 44)] and for TG [ANGPTL3 (40, 43) and GALNT2 (43)]. Of these implicated genes, the closest to our QTL cluster is PCSK9. Although PCSK9 is an excellent candidate for the QTL cluster, our bivariate analyses do not exclude entirely the possibility that several loci on chromosome 1 independently influence LDL-C concentrations. Future research will focus on identifying functional polymorphisms associated with variation in baboon LDL-C.
Cluster 4 included 13 significant and four suggestive QTLs that represented measures of LDL size and concentration properties on the two high-fat diets. Given the number of significant signals and the stringent criteria we imposed for declaring significance, there can be little doubt this cluster reflects a key locus influencing LDL metabolism. The peaks of all but two of the significant QTLs in this cluster mapped within the 9-cM 1-LOD drop interval of the predominant QTL for Lmed-C on HFHC diet. Falling within this interval on the human physical map are 212 genes (23, 34). There are several among these genes that play key roles in lipoprotein metabolism: SOAT2 (encoding SOAT2, steroyl O-acyltransferase 2, which has long been known as acyl-CoA:cholesterol acyltransferase 2 or ACAT2), APOF (encoding apoF or lipid transfer inhibitor protein), and LRP1 (encoding low density lipoprotein-related protein 1 or α2-macroglobulin receptor). LRP1 is ubiquitously expressed and functions in endocytosis and signal transduction (35, 45). In binding and internalizing a number of lipoprotein ligands, LRP1 plays a key role in atherosclerosis (46). ApoF is a lipoprotein constituent that is an inhibitor of cholesteryl ester transfer protein; its presence on lipoproteins modulates cholesteryl ester transfer protein-mediated remodeling between VLDL and both LDL and HDL particles (47, 48). SOAT2 expression is specifically localized to hepatocytes and enterocytes; the enzyme produces cholesteryl esters that are incorporated into apoB-containing lipoproteins secreted by these cell types (49). Studies in humans and other primates suggest that variation in SOAT2 is associated with response to dietary cholesterol and plasma lipid levels (50, 51).
Each of these genes is a plausible biological candidate for QTL Cluster 4. Given that all but one of the 12 traits tested had genetic correlations with Lmed-C on HFHC that were high and not different from one, the data suggest there is only one pleiotropic gene responsible for this cluster. Future research will be directed toward identifying that gene. We also will want to determine whether specific polymorphisms at this locus underlie variation in human LDL phenotypes. However, it may be difficult to extrapolate the present findings to free-living human populations, given that our ability to detect this gene in baboons was no doubt due, at least in part, to our ability to precisely control diet.
In summary, we have adopted a novel approach to identify highly influential QTLs for lipoprotein metabolism using data from a large baboon pedigree. Our results identified five major QTL clusters in the baboon genome. Three of the clusters confirmed previously reported signals, but two of the QTL clusters were novel and had not been reported previously in baboons. The novel clusters reflected genes influencing various aspects of LDL metabolism and they localized to genomic regions in which plausible candidate genes for lipoprotein metabolism are found in humans. Future research will focus on identifying the gene(s) responsible for each QTL cluster and, ultimately, identifying the specific polymorphisms in baboons and humans that are responsible for LDL variation.
Acknowledgments
The authors thank Jim Bridges, Perry H. Moore, Deborah E. Newman, Wendy R. Shelledy, Jane F. VandeBerg, and Joel Whitehead for technical assistance.
Abbreviations
apo, apolipoprotein
HDL-C, HDL-cholesterol
HFHC, diet high in fat and cholesterol
HFLC, diet high in fat but low in cholesterol
IBD, identity-by-descent
LOD, logarithm of the odds
PON, paraoxonase
QTL, quantitative trait loci
TG, triglyceride
The research was supported by a grant (P01 HL-028972) from the National Institutes of Health; the primate resources were supported by National Institutes of Health Grant P51 RR-013986 to the Southwest National Primate Research Center; and the research was performed in facilities constructed with support from National Institutes of Health Grants C06 RR-13556, C06 RR-014578, C06 RR-15456, C06 RR-17332, and C06 RR-017515.
Published, JLR Papers in Press, March 8, 2009.
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a table.
References
- 1.Singh A. T. K., D. L. Rainwater, C. M. Kammerer, R. M. Sharp, M. Poushesh, W. R. Shelledy, and J. L. VandeBerg. 1996. Dietary and genetic effects on LDL size measures in baboons. Arterioscler. Thromb. Vasc. Biol. 16 1448–1453. [DOI] [PubMed] [Google Scholar]
- 2.Cheng M-L., S. C. Woodford, J. L. Hilburn, and J. L. VandeBerg. 1986. A novel system for storage of sera frozen in small aliquots. J. Biochem. Biophys. Methods. 13 47–51. [DOI] [PubMed] [Google Scholar]
- 3.Rainwater D. L., C. M. Kammerer, K. D. Carey, B. Dyke, J. F. VandeBerg, W. R. Shelledy, P. H. Moore, Jr., M. C. Mahaney, H. C. McGill, Jr., and J. L. VandeBerg. 2002. Genetic determination of HDL variation and response to diet in baboons. Atherosclerosis. 161 335–343. [DOI] [PubMed] [Google Scholar]
- 4.Lipid Research Clinics Program. 1974. Manual of Laboratory Operations. Vol. 1. Lipid and Lipoprotein Analysis. (DHEW Publ. No. (NIH) 75–628). US Government Printing Office, Washington D.C.
- 5.Rainwater D. L., C. M. Kammerer, L. A. Cox, J. Rogers, K. D. Carey, B. Dyke, M. C. Mahaney, H. C. McGill, Jr., and J. L. VandeBerg. 2002. A major gene influences variation in large HDL particles and their response to diet in baboons. Atherosclerosis. 163 241–248. [DOI] [PubMed] [Google Scholar]
- 6.Kammerer C. M., D. L. Rainwater, L. A. Cox, J. L. Schneider, M. C. Mahaney, J. Rogers, and J. L. VandeBerg. 2002. Locus controlling LDL cholesterol response to dietary cholesterol is on baboon homologue of human chromosome 6. Arterioscler. Thromb. Vasc. Biol. 22 1720–1725. [DOI] [PubMed] [Google Scholar]
- 7.Rainwater D. L., C. M. Kammerer, and J. L. VandeBerg. 1999. Evidence that multiple genes influence baseline concentrations and diet response of Lp(a) in baboons. Arterioscler. Thromb. Vasc. Biol. 19 2696–2700. [DOI] [PubMed] [Google Scholar]
- 8.Rainwater D. L., P. H. Moore, Jr., W. R. Shelledy, T. D. Dyer, and S. H. Slifer. 1997. Characterization of a composite gradient gel for the electrophoretic separation of lipoproteins. J. Lipid Res. 38 1261–1266. [PubMed] [Google Scholar]
- 9.Rainwater D. L., D. W. Andres, A. L. Ford, W. F. Lowe, P. J. Blanche, and R. M. Krauss. 1992. Production of polyacrylamide gradient gels for the electrophoretic resolution of lipoproteins. J. Lipid Res. 33 1876–1881. [PubMed] [Google Scholar]
- 10.Rainwater D. L., C. M. Kammerer, M-L. Cheng, M. L. Sparks, and J. L. VandeBerg. 1992. Distribution of specific apolipoproteins detected by immunoblotting of baboon lipoproteins resolved by polyacrylamide gradient gel electrophoresis. Biochem. Genet. 30 143–158. [DOI] [PubMed] [Google Scholar]
- 11.Hsueh W-C., B. D. Mitchell, J. E. Hixson, and D. L. Rainwater. 2000. Effects of the apoE polymorphism on plasma lipoproteins in Mexican Americans. Ann. Epidemiol. 10 524–531. [DOI] [PubMed] [Google Scholar]
- 12.Vinson A., M. C. Mahaney, L. A. Cox, J. Rogers, J. L. VandeBerg, and D. L. Rainwater. 2008. A pleiotropic QTL on 2p influences serum Lp-PLA2 activity and LDL cholesterol concentration in a baboon model for the genetics of atherosclerosis risk factors. Atherosclerosis. 196 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainwater D. L., M. C. Mahaney, X. L. Wang, J. Rogers, L. A. Cox, and J. L. VandeBerg. 2005. Determinants of variation in serum paraoxonase enzyme activity in baboons. J. Lipid Res. 46 1450–1456. [DOI] [PubMed] [Google Scholar]
- 14.Rogers J., M. C. Mahaney, S. M. Witte, S. Nair, D. Newman, S. Wedel, L. A. Rodriguez, K. S. Rice, S. H. Slifer, A. Perelygin, et al. 2000. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 67 237–247. [DOI] [PubMed] [Google Scholar]
- 15.Cox L. A., M. C. Mahaney, J. L. VandeBerg, and J. Rogers. 2006. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics. 88 274–281. [DOI] [PubMed] [Google Scholar]
- 16.Almasy L., and J. Blangero. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath S. C., G. L. Snow, E. A. Thompson, C. Tseng, and E. M. Wijsman. 1997. MCMC segregation and linkage analysis. Genet. Epidemiol. 14 1011–1016. [DOI] [PubMed] [Google Scholar]
- 18.Ott, J. 1998. Analysis of Human Genetic Linkage. The Johns Hopkins University Press, Baltimore, MD.
- 19.Feingold E., P. O. Brown, and D. Siegmund. 1993. Gaussian models for genetic linkage analysis using complete high- resolution maps of identity by descent. Am. J. Hum. Genet. 53 234–251. [PMC free article] [PubMed] [Google Scholar]
- 20.Williams J. T., E. P. Van, L. Almasy, and J. Blangero. 1999. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am. J. Hum. Genet. 65 1134–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almasy L., T. D. Dyer, and J. Blangero. 1997. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet. Epidemiol. 14 953–958. [DOI] [PubMed] [Google Scholar]
- 22.Cox L. A., S. Birnbaum, M. C. Mahaney, D. L. Rainwater, J. T. Williams, and J. L. VandeBerg. 2007. Identification of promoter variants in baboon endothelial lipase that regulate high-density lipoprotein cholesterol levels. Circulation. 116 1185–1195. [DOI] [PubMed] [Google Scholar]
- 23.Kent W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahaney M. C., J. Blangero, D. L. Rainwater, G. E. Mott, A. G. Comuzzie, J. W. MacCluer, and J. L. VandeBerg. 1999. Pleiotropy and genotype by diet interaction in a baboon model for atherosclerosis - A multivariate quantitative genetic analysis of HDL subfractions in two dietary environments. Arterioscler. Thromb. Vasc. Biol. 19 1134–1141. [DOI] [PubMed] [Google Scholar]
- 25.Rainwater D. L., C. M. Kammerer, M. C. Mahaney, J. Rogers, L. A. Cox, J. L. Schneider, and J. L. VandeBerg. 2003. Localization of genes that control LDL size fractions in baboons. Atherosclerosis. 168 15–22. [DOI] [PubMed] [Google Scholar]
- 26.Vinson A., M. C. Mahaney, V. P. Diego, L. A. Cox, J. Rogers, J. L. VandeBerg, and D. L. Rainwater. 2008. Genotype-by-diet effects on co-variation in Lp-PLA2 activity and LDL cholesterol concentration in baboons fed an atherogenic diet. J. Lipid Res. 49 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainwater D. L., G. S. Manis, and J. L. VandeBerg. 1989. Hereditary and dietary effects on apolipoprotein[a] isoforms and Lp[a] in baboons. J. Lipid Res. 30 549–558. [PubMed] [Google Scholar]
- 28.Hixson J. E., M. L. Britten, G. S. Manis, and D. L. Rainwater. 1989. Apolipoprotein(a) (apo(a)) glycoprotein isoforms result from size differences in apo(a) mRNA in baboons. J. Biol. Chem. 264 6013–6016. [PubMed] [Google Scholar]
- 29.Boerwinkle E., C. C. Leffert, J. Lin, C. Lackner, G. Chiesa, and H. H. Hobbs. 1992. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 90 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa L. G., T. B. Cole, G. P. Jarvik, and C. E. Furlong. 2003. Functional genomics of the paraoxonase (PON1) polymorphisms: effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annu. Rev. Med. 54 371–392. [DOI] [PubMed] [Google Scholar]
- 31.Deakin S. P., and R. W. James. 2004. Genetic and environmental factors modulating serum concentrations and activities of the antioxidant enzyme paraoxonase-1. Clin. Sci. (Lond.). 107 435–447. [DOI] [PubMed] [Google Scholar]
- 32.Li H. L., D. P. Liu, and C. C. Liang. 2003. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J. Mol. Med. 81 766–779. [DOI] [PubMed] [Google Scholar]
- 33.Rainwater D. L., S. Rutherford, T. D. Dyer, E. D. Rainwater, S. A. Cole, J. L. VandeBerg, L. Almasy, J. Blangero, J. W. MacCluer, and M. C. Mahaney. 2009. Determinants of variation in human serum paraoxonase activity. Heredity. 102 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karolchik D., A. S. Hinrichs, T. S. Furey, K. M. Roskin, C. W. Sugnet, D. Haussler, and W. J. Kent. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32 D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May P., J. Herz, and H. H. Bock. 2005. Molecular mechanisms of lipoprotein receptor signalling. Cell. Mol. Life Sci. 62 2325–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder F., B. P. Atshaves, A. L. McIntosh, A. M. Gallegos, S. M. Storey, R. D. Parr, J. R. Jefferson, J. M. Ball, and A. B. Kier. 2007. Sterol carrier protein-2: new roles in regulating lipid rafts and signaling. Biochim. Biophys. Acta. 1771 700–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert G. 2007. Unravelling the functional significance of PCSK9. Curr. Opin. Lipidol. 18 304–309. [DOI] [PubMed] [Google Scholar]
- 38.Lambert G., N. Ancellin, F. Charlton, D. Comas, J. Pilot, A. Keech, S. Patel, D. R. Sullivan, J. S. Cohn, K. A. Rye, et al. 2008. Plasma PCSK9 concentrations correlate with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment. Clin. Chem. 54 1038–1045. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell K. N., R. E. Soccio, E. M. Duncan, E. Sehayek, and J. L. Breslow. 2003. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 44 2109–2119. [DOI] [PubMed] [Google Scholar]
- 40.Willer C. J., S. Sanna, A. U. Jackson, A. Scuteri, L. L. Bonnycastle, R. Clarke, S. C. Heath, N. J. Timpson, S. S. Najjar, H. M. Stringham, et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kooner J. S., J. C. Chambers, C. A. Aguilar-Salinas, D. A. Hinds, C. L. Hyde, G. R. Warnes, F. J. Gomez Perez, K. A. Frazer, P. Elliott, J. Scott, et al. 2008. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 40 149–151. [DOI] [PubMed] [Google Scholar]
- 42.Sandhu M. S., D. M. Waterworth, S. L. Debenham, E. Wheeler, K. Papadakis, J. H. Zhao, K. Song, X. Yuan, T. Johnson, S. Ashford, et al. 2008. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 371 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kathiresan S., O. Melander, C. Guiducci, A. Surti, N. P. Burtt, M. J. Rieder, G. M. Cooper, C. Roos, B. F. Voight, A. S. Havulinna, et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kathiresan S., O. Melander, D. Anevski, C. Guiducci, N. P. Burtt, C. Roos, J. N. Hirschhorn, G. Berglund, B. Hedblad, L. Groop, et al. 2008. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 358 1240–1249. [DOI] [PubMed] [Google Scholar]
- 45.May P., E. Woldt, R. L. Matz, and P. Boucher. 2007. The LDL receptor-related protein (LRP) family: an old family of proteins with new physiological functions. Ann. Med. 39 219–228. [DOI] [PubMed] [Google Scholar]
- 46.Llorente-Cortes V., and L. Badimon. 2005. LDL receptor-related protein and the vascular wall: implications for atherothrombosis. Arterioscler. Thromb. Vasc. Biol. 25 497–504. [DOI] [PubMed] [Google Scholar]
- 47.Paromov V. M., and R. E. Morton. 2003. Lipid transfer inhibitor protein defines the participation of high density lipoprotein subfractions in lipid transfer reactions mediated by cholesterol ester transfer protein (CETP). J. Biol. Chem. 278 40859–40866. [DOI] [PubMed] [Google Scholar]
- 48.He Y., D. J. Greene, M. Kinter, and R. E. Morton. 2008. Control of cholesteryl ester transfer protein activity by sequestration of lipid transfer inhibitor protein in an inactive complex. J. Lipid Res. 49 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudel L. L., R. G. Lee, and P. Parini. 2005. ACAT2 is a target for treatment of coronary heart disease associated with hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 25 1112–1118. [DOI] [PubMed] [Google Scholar]
- 50.Rudel L. L., M. Davis, J. Sawyer, R. Shah, and J. Wallace. 2002. Primates highly responsive to dietary cholesterol up-regulate hepatic ACAT2, and less responsive primates do not. J. Biol. Chem. 277 31401–31406. [DOI] [PubMed] [Google Scholar]
- 51.He, X ., Y. Lu, N. Saha, H. Yang, and C. K. Heng. 2005. Acyl-CoA: cholesterol acyltransferase-2 gene polymorphisms and their association with plasma lipids and coronary artery disease risks. Hum. Genet. 118 393–403. [DOI] [PubMed] [Google Scholar]