Abstract
Apolipoproteins (apo) E and C-I are components of triglyceride (TG)-rich lipoproteins and impact their metabolism. Functional polymorphisms have been established in apoE but not in apoC-I. We studied the relationship between apoE and apoC-I gene polymorphisms and plasma lipoproteins and coronary artery disease (CAD) in 211 African Americans and 306 Caucasians. In African Americans but not in Caucasians, apoC-I H2-carriers had significantly lower total and LDL cholesterol and apoB levels, and higher glucose, insulin, and HOMA-IR levels compared with H1 homozygotes. Differences across CAD phenotypes were seen for the apoC-I polymorphism. African-American H2-carriers without CAD had significantly lower total cholesterol (P < 0.001), LDL cholesterol (P < 0.001), and apoB (P < 0.001) levels compared with H1 homozygotes, whereas no differences were found across apoC-I genotypes for African Americans with CAD. Among African-American apoC-I H1 homozygotes, subjects with CAD had a profile similar to the metabolic syndrome (i.e., higher triglyceride, glucose, and insulin) compared with subjects without CAD. For African-American H2-carriers, subjects with CAD had a pro-atherogenic lipid pattern (i.e., higher LDL cholesterol and apoB levels), compared with subjects without CAD. ApoC-I genotypes showed an ethnically distinct phenotype relationship with regard to CAD and CAD risk factors.
Keywords: apolipoprotein C-I, apolipoprotein E, coronary artery disease, ethnicity, polymorphism
Apolipoprotein (apo) C-I is a constituent of triglyceride (TG)-rich lipoproteins. ApoC-I is reported to inhibit hepatic lipase and to interfere with lipoprotein clearance by the LDL receptor, the LDL receptor-related protein (LRP) and the VLDL receptor (1–4). Overexpression of apoC-I in transgenic mice produces combined hyperlipidemia along with increased postprandial lipemia (5–7). Increased levels of plasma apoC-I and VLDL apoC-I in hypertriglyceridemic subjects are associated with an increased production of VLDL apoC-I (8). These studies have raised the possibility that apoC-I may play an important role in clinical dyslipidemia and coronary artery disease (CAD).
Genetic variability of apolipoprotein E (apoE) is a major determinant of plasma lipoprotein levels (9). Furthermore, apoE genotype frequencies differ between African Americans and Caucasians (10, 11). Variation of apoE genotypes has been strongly and consistently associated with plasma lipid levels and risk of CAD (12–16). In contrast to apoE, less information is available on apoC-I genetic variation in humans or on their possible contributions to clinically relevant lipoprotein phenotypes. A DNA polymorphism produced by a CGTT insertion 317-bp 5′ to the apoC-I transcription initiation site (apoC-I -317insCGTT) has been described and is at present the only known common apoC-I gene variant (17, 18). This polymorphism has been termed the HpaI RFLP, with “H2” designating the insertion allele (and consequent presence of the HpaI DNA restriction enzyme site) and “H1” designating the deletion allele (and absence of the HpaI site). The H1 and H2 alleles of apoC1 have been reported to show an ethnically distinct pattern of linkage disequilibrium with alleles of the adjacent apoE gene (18–20). In our prior study of an elderly, multiracial, community-based population, there was a strong association of apo ɛ3 with the apoC-I H1 allele and of apo ɛ4 with apoC-I H2 allele in non-Hispanic Caucasians. In contrast, in African-Americans, these associations were significantly weaker, while apo ɛ2 was closely associated with H2 in both groups (18). Similar ethnic differences in the genetic association of the two markers were found in a comparison of Portuguese with Africans from Sao Tome e Principe (19).
In contrast to the wealth of information on apoE genotypes, the relationship of apoC-I genotypes with CAD has not been addressed. In the current study, we investigated the association between genetic polymorphism of apoE and apoC-I and cardiovascular disease and risk factors across African-American and Caucasian ethnicity. This approach allowed us to assess differences across both CAD phenotypes and apoC-I and apoE genotypes for each ethnic group.
MATERIALS AND METHODS
Subjects
Subjects were recruited from a patient population scheduled for diagnostic coronary arteriography either at Harlem Hospital Center in New York City or at the Mary Imogene Bassett Hospital in Cooperstown, NY. The clinical characteristics of the study population and the study design, including inclusion and exclusion criteria, have been described previously, and notably, exclusion criteria included use of lipid-lowering drugs (16, 21). Briefly, a total of 648 patients, 401 men and 247 women, ethnically self-identified as Caucasian (n = 344), African American (n = 232), or Other (n = 72) were enrolled. The present report is based on the findings in 306 Caucasians (201 males, 105 females) and 211 African Americans (123 males, 88 females) in whom lipid levels and apoE and apoC-I genotypes were available. The study was approved by the Institutional Review Boards at Harlem Hospital, the Mary Imogene Bassett Hospital, Columbia University College of Physicians and Surgeons, and University of California Davis, and informed consent was obtained from all subjects.
Clinical and biochemical assessment
Fasting blood samples were drawn approximately 2–4 h before the catheterization procedure, and serum and plasma samples were stored at −80°C prior to analysis. Serum triglycerides, total and HDL cholesterol, and glucose were determined using standard enzymatic procedures, and LDL cholesterol levels were calculated as described (22–25). Homeostasis model assessment–insulin resistance (HOMA-IR) was calculated using the updated model available from the Oxford Centre for Endocrinology and Diabetes (26, 27).
Determination of apoE and apoC-I genotypes
ApoE genotypes were determined as described previously (16, 21, 28). Genotypic status for the HpaI apoC-I promoter polymorphism was determined as described by a two-step nested PCR followed by restriction digestion with HpaI and electrophoresis on 1.5% agarose (ultraPure Agarose, Life Technologies, Gaithersburg, MD) (18).
Coronary angiography
Two readers, who were blinded to patient identity, clinical diagnosis, lipoprotein, and genotype results, recorded the localization and extent of luminal narrowing for 15 segments of the major coronary arteries. Presence of CAD was defined as the presence of at least 50% stenosis in any 1 of 15 coronary artery segments. Of the patients without CAD, the majority (80.5%) had less than 25% stenosis, and of the patients with CAD, 81% had greater than 75% stenosis. A composite cardiovascular score (0–75) was calculated based on determination of presence of stenosis on a scale of 0–5 of 15 predetermined coronary artery segments.
Statistics
Analysis of data was done with SPSS statistical analysis software (SPSS Inc., Chicago, IL). Results were expressed as means ± SD. Triglyceride and insulin levels and cardiovascular composite score were logarithmically transformed to achieve normal distributions. Proportions were compared between groups using χ2 analysis, and Fisher exact test where appropriate. Group means for clinical parameters were compared using one-way ANOVA and post hoc analyses were performed by the Tukey-HSD test. Means for CAD groups were compared using Student's t-test. Multiple logistic regression analysis was applied to predict the variables that independently and significantly contributed to the dependent variable: the presence of cardiovascular disease. All analyses were two-tailed, and P-values less than 0.05 were considered statistically significant.
RESULTS
Effect of apoC-I polymorphism on clinical parameters
The effect of apoC-I allele status on lipoprotein, apolipoprotein, and glucose, insulin and HOMA-IR measurements is shown in Table 1. In Caucasians, no effect of apoC-I allele status on any of the clinical parameters was evident. On the other hand, statistically significant effects of the apoC-I polymorphism on clinical parameters were noted in African Americans, with a gradual decrease in total cholesterol levels with increasing number of apoC-I H2 alleles (209 ± 47, 190 ± 41, and 172 ± 39 mg/dl for H1/H1, H1/H2 and H2/H2, respectively; P < 0.001). As seen in the table, essentially the same pattern was found for LDL cholesterol and apoB levels. There was no difference in HDL cholesterol or apoA-I levels across apoC-I genotype. Glucose, insulin, and HOMA-IR levels were significantly higher among African-American apoC-I H2 homozygotes compared with H1 homozygotes or H1/H1 heterozygotes.
TABLE 1.
Clinical characteristics by apoC-I genotype across ethnicity
| African Americans |
Caucasians |
|||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | H1/H1 (n = 103) | H1/H2 (n = 88) | H2/H2 (n = 20) | P | H1/H1 (n = 191) | H1/H2 (n = 101) | H2/H2 (n = 14) | P |
| Total cholesterol (mg/dl) | 209 ± 47 | 190 ± 41a | 172 ± 39a | <0.001 | 193 ± 39 | 200 ± 40 | 191 ± 53 | NS |
| LDL cholesterol (mg/dl) | 136 ± 41 | 117 ± 40a | 101 ± 41a | <0.001 | 120 ± 32 | 122 ± 36 | 116 ± 43 | NS |
| HDL cholesterol (mg/dl) | 49 ± 17 | 50 ± 19 | 47 ± 16 | NS | 40 ± 12 | 41 ± 12 | 39 ± 14 | NS |
| Triglyceride (mg/dl) | 117 ± 47 | 116 ± 55 | 122 ± 72 | NS | 170 ± 90 | 196 ± 117 | 174 ± 74 | NS |
| ApoA-I (mg/dl) | 132 ± 31 | 129 ± 29 | 126 ± 27 | NS | 121 ± 22 | 124 ± 24 | 113 ± 20 | NS |
| ApoB (mg/dl) | 143 ± 42 | 126 ± 37a | 113 ± 40a | 0.001 | 133 ± 32 | 138 ± 40 | 130 ± 41 | NS |
| Glucose (mg/dl) | 122 ± 56 | 113 ± 42 | 146 ± 81b | 0.048 | 128 ± 61 | 138 ± 69 | 105 ± 23 | NS |
| Insulin (μU/ml) | 22 ± 27 | 18 ± 20 | 30 ± 26b | 0.035 | 26 ± 35 | 30 ± 48 | 20 ± 17 | NS |
| HOMA-IR | 2.6 ± 2.4 | 2.4 ± 2.5 | 4.2 ± 3.3a,b | 0.011 | 3.5 ± 4.3 | 3.5 ± 4.8 | 2.6 ± 2.1 | NS |
Data are means ± SD. Apo, apolipoprotein; HOMA-IR, homeostasis model assessment–insulin resistance; NS, not significant. P values were calculated using one-way ANOVA and post hoc analyses were performed with the Tukey-HSD test for two independent samples. Values for triglycerides, insulin and HOMA-IR were logarithmically transformed before analyses. Nontransformed values are shown in the table.
P < 0.05 compared with H1 homozygote subjects.
P < 0.05 compared with H1/H2 heterozygote subjects.
Clinical parameters by CAD across ApoC-I genotype
We next evaluated the impact of apoC-I genotype on the same clinical parameters in patients with two different clinical phenotypes, presence or absence of CAD (Table 2). In doing so, we were able to undertake two different comparisons. First, a comparison across apoC-I genotypes provided information for a given phenotype (presence or absence of CAD). Second, a comparison within a given genotype (H1 homozygotes or H2-carriers) allowed a comparison across phenotypes (e.g., presence or absence of CAD).
TABLE 2.
Clinical characteristics by CAD across ApoC-I genotype in Caucasians and African Americans
| Without CAD |
With CAD |
|||||
|---|---|---|---|---|---|---|
| H1/H1 | H1/H2, H2/H2 | P | H1/H1 | H1/H2, H2/H2 | P | |
| African Americans | n = 59 | n = 55 | n = 41 | n = 50 | ||
| Total cholesterol (mg/dl) | 206 ± 41 | 173 ± 38 | <0.001 | 215 ± 56 | 200 ± 40b | NS |
| LDL cholesterol (mg/dl) | 134 ± 35 | 102 ± 37 | <0.001 | 141 ± 49 | 126 ± 39b | NS |
| HDL cholesterol (mg/dl) | 50 ± 16 | 51 ± 19 | NS | 48 ± 17 | 48 ± 17 | NS |
| Triglyceride (mg/dl) | 109 ± 44 | 101 ± 39 | NS | 128 ± 51a | 130 ± 69b | NS |
| ApoA-I (mg/dl) | 136 ± 32 | 131 ± 30 | NS | 127 ± 27 | 125 ± 27 | NS |
| ApoB (mg/dl) | 138 ± 37 | 112 ± 34 | <0.001 | 152 ± 48 | 134 ± 36b | NS |
| Glucose (mg/dl) | 112 ± 36 | 111 ± 43 | NS | 136 ± 75a | 124 ± 47 | NS |
| Insulin (μU/ml) | 20 ± 31 | 18 ± 18 | NS | 25 ± 23 | 24 ± 26 | NS |
| HOMA-IR | 2.1 ± 1.5 | 2.4 ± 2.3 | NS | 3.4 ± 3.2a | 3.1 ± 3.1 | NS |
| Caucasians | n = 76 | n = 52 | n = 110 | n = 60 | ||
| Total cholesterol (mg/dl) | 187 ± 39 | 195 ± 41 | NS | 198 ± 40 | 203 ± 41 | NS |
| LDL cholesterol (mg/dl) | 114 ± 29 | 120 ± 34 | NS | 126 ± 35a | 125 ± 39 | NS |
| HDL cholesterol (mg/dl) | 42 ± 14 | 43 ± 12 | NS | 39 ± 11a | 40 ± 12 | NS |
| Triglyceride (mg/dl) | 157 ± 93 | 168 ± 90 | NS | 179 ± 88a | 218 ± 127b | 0.038 |
| ApoA-I (mg/dl) | 124 ± 24 | 125 ± 22 | NS | 119 ± 20 | 122 ± 26 | NS |
| ApoB (mg/dl) | 124 ± 29 | 136 ± 46 | NS | 141 ± 34a | 139 ± 35 | NS |
| Glucose (mg/dl) | 118 ± 64 | 130 ± 58 | NS | 134 ± 59 | 137 ± 74 | NS |
| Insulin (μU/ml) | 22 ± 27 | 34 ± 60 | NS | 29 ± 40 | 22 ± 19 | NS |
| HOMA-IR | 2.9 ± 3.5 | 3.2 ± 5.1 | NS | 3.8 ± 4.7 | 3.3 ± 3.7 | NS |
Apo, apolipoprotein; CAD, coronary artery disease; HOMA-IR, homeostasis model assessment–insulin resistance. Data are means ± SD. P values calculated using Student's t-test. Values for triglycerides, insulin and HOMA-IR were logarithmically transformed before analyses. Nontransformed values are shown.
P < 0.05 compared with H1/H1 across CAD status.
P < 0.05 compared with H1/H2, H2/H2 across CAD status.
In the first comparison (i.e., apoC-I genotypes in patients with or without CAD), African-American H2-carriers without CAD had significantly lower total cholesterol (P < 0.001), LDL cholesterol (P < 0.001), and apoB (P < 0.001) levels compared with H1 homozygotes. However, no significant difference were seen across apoC-I genotypes for African Americans with CAD. There was no significant difference between Caucasian apoC-I H1 homozygotes and H2-carriers without CAD, whereas triglyceride levels were higher in Caucasian H2-carriers with CAD (P < 0.05).
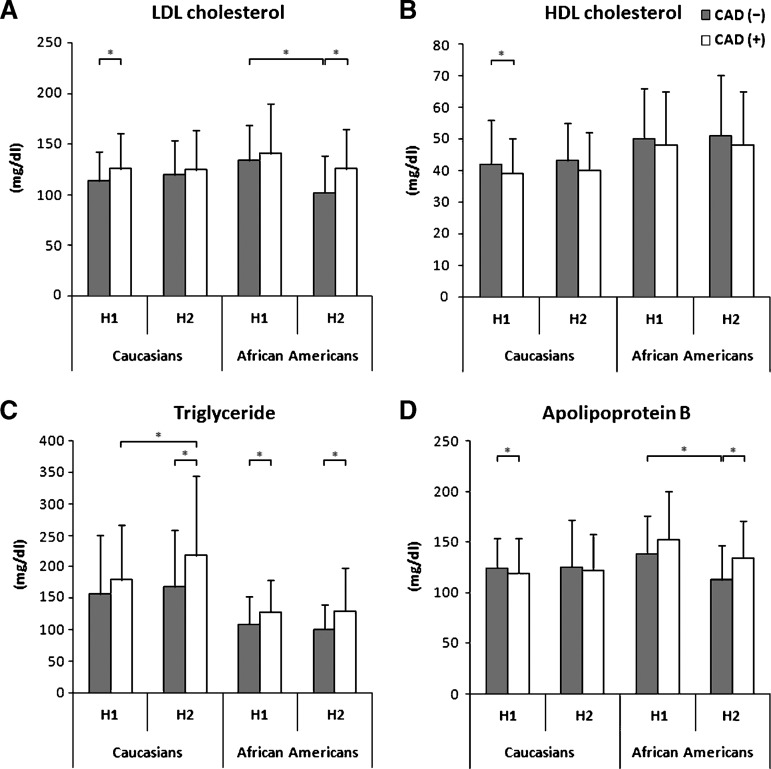
In the second comparison, we found significant differences for a number of parameters for a given apoC-I genotype (H1 homozygotes or H2-carriers) across CAD status among African Americans. As seen in Table 2 and highlighted for lipid components in Fig. 1, African-American apoC-I H1 homozygotes with CAD had significantly higher triglyceride (P < 0.05), glucose (P < 0.05), and HOMA-IR (P < 0.05) levels compared with patients with the same genotype without CAD. In contrast, African-American apoC-I H2-carriers with CAD had significantly higher total cholesterol (P = 0.001), LDL cholesterol (P < 0.05) and apoB (P = 0.001) levels compared with patients with the same genotype without CAD. These results suggest that parameters associated with the metabolic syndrome (triglyceride, insulin, and glucose) differed across CAD status for H1 homozygotes, whereas H2-carriers with CAD had a more pronounced pro-atherogenic lipid pattern (higher levels of total and LDL cholesterol, triglycerides, and apoB) compared with the same genotype without CAD. For Caucasians, the pattern was different. Caucasian apoC-I H1 homozygotes with CAD had a pro-atherogenic lipid phenotype (i.e., higher LDL cholesterol, triglyceride, and apoB levels, and lower HDL cholesterol levels) compared with subjects carrying the same genotype without CAD. The only significant difference among apoC-I H2-carriers was a higher triglyceride level for subjects with CAD.
Fig. 1.
Distribution of LDL cholesterol (A), HDL cholesterol (B), triglyceride (C) and apolipoprotein B (D) levels by CAD and apoC-I genotype in Caucasians and African Americans. * = P < 0.05.
Prevalence of apoC-I and apoE genotype
In our previous study in a different population, we observed linkage disequilibrium between apoE and apoC-I genotypes (18). In the present study, the prevalence of apoC-I genotype differed between Caucasians and African Americans. Caucasians had higher H1 and lower H2 allele frequency and, consequently, a higher frequency of H1/H1 and H1/H2 genotypes (P = 0.002 and P = 0.038, respectively), while African Americans had significantly higher prevalence of H2/H2 (P = 0.041). An association of apoC-I allele status with apoE allele status was observed for both ethnic groups. Apo ɛ3 homozygotes (genotype E3/E3), the major genotype group in both ethnicities, was strongly associated with apoC-I H1/H1 in Caucasians, while the association was less pronounced among African Americans (P < 0.001) (Table 3). Thus, only 6 of the 193 (3%) Caucasian apo ɛ3 homozygosity carried the apoC-I H2-allele, whereas 18 of 77 (23%) African-American apo ɛ3 homozygotes were apoC-I H2-carriers, all being H1/H2 heterozygotes. Conversely, only 2 of the 69 (3%) Caucasian apo ɛ4-carriers (apo E3/E4 and E4/E4) were apoC-I H1 homozygotes, while the corresponding number for African Americans was substantially higher [40 of 87 apo ɛ4-carriers (46%)].
TABLE 3.
ApoC-I genotype by apoE genotype
| ApoC-I genotype |
||||
|---|---|---|---|---|
| ApoE genotype | Ethnicity | H1/H1 | H1/H2 | H2/H2 |
| E2/E2 | African Americans | 0 | 1 (50%) | 1 (50%) |
| Caucasians | 0 | 0 | 3 (100%) | |
| E2/E4 | African Americans | 2 (13%) | 8 (53%) | 5 (34%) |
| Caucasians | 1 (14%) | 2 (29%) | 4 (57%) | |
| E2/E3 | African Americans | 2 (7%) | 23 (77%) | 5 (16%) |
| Caucasians | 1 (3%) | 33 (97%) | 0 | |
| E3/E3 | African Americans | 59 (77%) | 18 (23%) | 0 |
| Caucasians | 187 (97%) | 5 (3%) | 1 (0%) | |
| E3/E4 | African Americans | 35 (46%) | 32 (42%) | 9 (12%) |
| Caucasians | 2 (3%) | 61 (97%) | 0 | |
| E4/E4 | African Americans | 5 (45%) | 6 (55%) | 0 |
| Caucasians | 0 | 0 | 6 (100%) | |
| Total | African Americans | 103 (49%) | 88 (42%) | 20 (9%) |
| Caucasians | 191 (62%) | 101 (33%) | 14 (5%) | |
Apo, apolipoprotein. Values represent number of subjects; values in parentheses are percentages of each group.
Clinical parameters by CAD across ApoC-I genotype in African-American apo ɛ4-carriers
As the apoC-I genotype showed a less pronounced pattern of linkage disequilibrium with apoE genotypes in African Americans, we next compared the relation between apoC-I genotypes and clinical parameters across CAD in African-American apo ɛ4-carriers (Table 4). As for the entire group, African-American ɛ4-carriers without CAD had higher total and LDL cholesterol and apoB levels among apoC-I H1 homozygotes, while no significant differences were seen across apoC-I genotypes for subjects with CAD. When comparing intra-genotype differences across CAD phenotype, glucose and HOMA-IR was higher among apoC-I H1 homozygotes with CAD compared with subjects without CAD. In contrast, apoC-I H2-carriers with CAD had higher total and LDL cholesterol levels compared with the same genotypes without CAD (Table 4).
TABLE 4.
Clinical characteristics of African Americans across ApoE and ApoC-I genotypes
| Without CAD |
With CAD |
|||||
|---|---|---|---|---|---|---|
| African Americans apo ɛ4 | H1/H1 (n = 23) | H1/H2, H2/H2 (n = 24) | P | H1/H1 (n = 17) | H1/H2, H2/H2 (n = 23) | P |
| Total cholesterol (mg/dl) | 211 ± 39 | 180 ± 37 | 0.008 | 234 ± 65 | 209 ± 48b | NS |
| LDL-C (mg/dl) | 141 ± 36 | 111 ± 35 | 0.006 | 158 ± 55 | 137 ± 48b | NS |
| HDL-C (mg/dl) | 48 ± 10 | 49 ± 19 | NS | 49 ± 14 | 47 ± 18 | NS |
| TG (mg/dl) | 109 ± 43 | 100 ± 39 | NS | 134 ± 42 | 127 ± 69 | NS |
| ApoA-I (mg/dl) | 130 ± 23 | 123 ± 24 | NS | 132 ± 24 | 120 ± 28 | NS |
| ApoB (mg/dl) | 148 ± 39 | 122 ± 33 | 0.019 | 168 ± 52 | 144 ± 42 | NS |
| Glucose (mg/dl) | 112 ± 20 | 119 ± 54 | NS | 159 ± 96a | 121 ± 47 | NS |
| Insulin (μU/ml) | 26 ± 47 | 20 ± 24 | NS | 31 ± 27 | 19 ± 18 | NS |
| HOMA-IR | 2.2 ± 1.4 | 2.7 ± 3.2 | NS | 3.9 ± 3.5a | 2.5 ± 2.2 | NS |
Apo, apolipoprotein; CAD, coronary artery disease; HOMA-IR, homeostasis model assessment–insulin resistance. Data are means ± SD. P values were calculated using Student's t-test. Values for triglycerides, insulin and HOMA-IR were logarithmically transformed before analyses. Nontransformed values are shown.
P < 0.05 compared with H1/H1 across CAD status.
P < 0.05 compared with H1/H2, H2/H2 across CAD status.
Multiple logistic regression
We performed multiple logistic regression analyses to identify the variables that independently and significantly contributed to the presence of cardiovascular disease. When only apoE and apoC-I genotypes were considered, carrying apo ɛ2 was significantly associated with protection for CAD (odds ratio [OR] = 0.37, P = 0.043) in African Americans. After adjusting for LDL cholesterol, the association between apo ɛ2 and CAD was no longer significant (P = 0.147). However, inclusion of LDL cholesterol in the model uncovered a protective effect of the apoC-I H2 allele for CAD (OR = 0.45, P = 0.025) in African Americans but not in Caucasians. After further adjustment for other established risk factors, such as age, gender, smoking, BMI, HDL cholesterol and triglyceride, the association of apoC-I and apoE genotypes with CAD were no longer significant in either ethnic group.
DISCUSSION
We have previously reported on a protective effect of the apo ɛ2 allele on CAD in African Americans (16). In this study, we addressed a combined effect of two closely located genes (i.e., apoE and apoC-I) on lipoprotein pattern and presence of CAD. The main novel finding of our study was that the relationship of apoC-I and apoE genotype polymorphisms with plasma lipoproteins and cardiovascular disease phenotype differed between African Americans and Caucasians. African-American apoC-I H1 homozygotes with CAD had a clinical pattern similar to the metabolic syndrome (i.e., higher triglyceride, glucose, and insulin), whereas apo C-I H2-carriers with CAD had a more pronounced pro-atherogenic lipid pattern (i.e., higher LDL cholesterol, triglyceride, and apoB levels) in both cases compared with subjects with corresponding genotypes without CAD. These patterns were not seen among Caucasians. Further, we observed an ethnically distinct pattern of linkage disequilibrium between the apoC-I and apoE genes. Among Caucasians, apo ɛ3 homozygosity was strongly associated with apoC-I H1 homozygosity, while the association was less pronounced in African Americans.
In our previous studies, we have demonstrated a strong degree of linkage disequilibrium between and the apoE the apoC-I loci in Caucasians. The lesser degree of linkage disequilibrium present in African Americans prompted us to evaluate the effect of apoC-I genotypes in African-American and Caucasians patients with two different phenotypes, presence or absence of CAD. Our study design allowed us to assess two different types of comparisons in each of the ethnic groups: (a) a comparison across apoC-I genotypes provided information for a given phenotype (presence or absence of CAD); and (b) a comparison within a given genotype (H1 homozygotes or H2-carriers) allowed a direct comparison between two different phenotypes (presence or absence of CAD). Regarding the first comparison across apoC-I genotype, the H2 allele was associated with a protective anti-atherogenic pattern in subjects without CAD in African Americans but not in Caucasians. For the second comparison, we observed differences in the relationship between apoC-I genotypes and CAD phenotype both within and between the ethnic groups. While African-American H1 homozygotes with CAD had higher levels of parameters associated with the metabolic syndrome, H2 carriers with CAD had a more pronounced pro-atherogenic lipid pattern, each genotype compared with subjects carrying corresponding genotypes without CAD. Interestingly, a different pattern for H1 homozygotes was seen among Caucasians, with higher levels of parameters associated with metabolic syndrome in the CAD group.
To the best of our knowledge, there are no studies simultaneously addressing the effect of apoC-I and apoE gene polymorphisms on CAD across African American and Caucasian ethnicity. Previous studies have shown that apoC-I decreases apoE-mediated binding of lipoproteins to VLDL and LDL receptors (4, 29, 30). These mechanisms may contribute to the well-known inhibitory effect of apoC-I on clearance of TG-rich lipoproteins (1–4). While the high degree of linkage disequilibrium between the apoE and apoC-I genes loci in Caucasians suggests that effects of apoC-I polymorphisms independent of apoE gene variation might be difficult to establish, studies among African Americans with a lesser degree of linkage disequilibrium are more informative. Indeed, our findings among African Americans suggest that the apoC-I H2 allele might convey cardioprotective effects, as levels of total and LDL cholesterol and apoB were lower among H2 carriers in the absence of CAD. Whether this is due to any direct effect of the apoC-I H2 allele on lipoprotein interactions with the LDL receptor remains to be shown. Interestingly, we found that taking LDL cholesterol levels into account uncovered a cardioprotective effect of the apoC-I H2 allele. This was only seen among African Americans. However, this potential advantage among H2-carriers might be lost in the presence of CAD. While many factors are likely to contribute to these results, one can speculate that mechanisms beyond apoC-I genetic variability resulting in an increase in apoB-containing lipoproteins contribute to the CAD phenotype. Gautier et al., recently reported that apoC-I inhibited cholesteryl ester transfer protein (CETP) activity and that immunodepletion of apoC-I produced a substantial increase in cholesteryl ester transfer rates (31). Furthermore, earlier studies have demonstrated that apoC-I enhanced lecithin cholesterol acyltransferase (LCAT) activity (32, 33). However, any effect of genetic variation of apoC-I on these effects has not been studied. It might also be speculated that the location of hpa polymorphism in the promoter of apoC-I gene might result in the altered expression levels and, therefore, effect on lipoprotein parameters. We have previously demonstrated in vitro gene-expression studies that H2 sequence may decrease the binding of a negatively acting transcription factor, leading to overexpression of apoC-I (18). This may produce a functional effect on lipoprotein levels, but confirmation is needed in other populations. Taken together with our findings, an effect of apoC-I on lipoprotein remnant clearance may potentially contribute to the observed increase in apoB-containing lipoproteins among African-American H2-carriers with CAD.
In our study, the impact of the apoC-I H2 allele on atherogenic lipoproteins differed in subjects with and without CAD. As the apoC-I genotype showed a less pronounced pattern of linkage disequilibrium with the apoE gene in African Americans, we were able to further analyze the independent effect of apoC-I polymorphism separately for apoE genotypes in this ethnic group. We observed significant differences of the apoC-I H2-allele on plasma lipoprotein levels in apo ɛ4 carriers across CAD phenotype, and the results were similar to findings observed for the entire group. Interestingly, the H2-allele showed a protective anti-atherogenic effect (e.g., lower total and LDL cholesterol and apoB levels) compared with H1 homozygotes in subjects without CAD. No difference was seen among subjects with CAD. When comparing differences between genotypes among African Americans, ɛ4/H2 carriers with CAD had a pronounced pro-atherogenic lipid pattern, while a pattern more typical of insulin resistance was seen for H1 homozygotes with CAD, compared with subjects carrying the same genotype without CAD. In the ɛ4 group, the apoC-I H2 allele had a protective effect on atherogenic lipoproteins in the absence of CAD. This protective effect decreased and was no longer significant in the presence of CAD. This difference was limited to African-American apo ɛ4 carriers and was not seen among African-American apo ɛ3 homozygotes. Notably, the apo ɛ4-allele frequency is considerably higher among African Americans than Caucasians (16). Taken together, our results suggest the presence of genotype-phenotype relationships for the apoC-I and apoE genes with respect to CAD, suggestive of more adverse constellations of risk factors for some genotypes in African Americans. Moreover, the results suggest that the impact of genetic variability of these genes might be modulated by disease phenotype. Ours is one of the first studies to report relationships between genetic variability of the apo C-I gene and phenotypes associated with increased cardiovascular disease or risk in a specific ethnic group. For African Americans, we report on effects of apo C-I genetic variability when taking apoE allelic variation into account, suggesting an effect of the apo C-I gene on cardiovascular risk beyond the apoE gene. Further studies are needed to extend these findings to other populations.
The present study has several limitations. Subjects in our study were recruited from patients scheduled for coronary angiography, and for some genotypes, the number was relatively small. As the apo ɛ4 genotype has been associated with CAD, a potential source of error might be a differing distribution of apoE genotypes among our subjects compared with the population at large. Arguing against this possibility, the apoE allele frequency pattern was similar to that previously described for African-American and Caucasian populations (10, 11). Given the relatively small sample size, we did not do statistical analyses separately for men and women.
In summary, the apoC-I polymorphism had an ethnically distinct pattern of allele frequency, linkage disequilibrium, and effect on lipoprotein phenotype. In African Americans, but not in Caucasians, apoC-I genotypes were associated with variation in lipoprotein phenotypes, and in particular, the apoC-I H2-allele was associated with an anti-atherogenic pattern.
Acknowledgments
E. Anuurad was the recipient of an American Heart Association postdoctoral fellowship (0725125Y), and M. Yamasaki was supported by funds from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations
Apo, apolipoprotein
CAD, coronary artery disease
CETP, cholesteryl ester transfer protein
CVD, cardiovascular disease
HOMA-IR, homeostasis model assessment-insulin resistance
LCAT, lecithin cholesterol acyltransferase
LRP, LDL receptor-related protein
TG, triglyceride
This study was supported by Grants 49735 (T.A.P.) and 62705 (L.B.) from the National Heart, Lung and Blood Institute of the National Institutes of Health. This work was supported in part by Grant RR-024146 from the Clinical and Translational Science Center, University of California at Davis.
Published, JLR Papers in Press, February 27, 2009.
References
- 1.Conde-Knape K., A. Bensadoun, J. H. Sobel, J. S. Cohn, and N. S. Shachter. 2002. Overexpression of apoC-I in apoE-null mice: severe hypertriglyceridemia due to inhibition of hepatic lipase. J. Lipid Res. 43 2136–2145. [DOI] [PubMed] [Google Scholar]
- 2.Windler E., Y. Chao, and R. J. Havel. 1980. Regulation of the hepatic uptake of triglyceride-rich lipoproteins in the rat. Opposing effects of homologous apolipoprotein E and individual C apoproteins. J. Biol. Chem. 255 8303–8307. [PubMed] [Google Scholar]
- 3.Quarfordt S. H., G. Michalopoulos, and B. Schirmer. 1982. The effect of human C apolipoproteins on the in vitro hepatic metabolism of triglyceride emulsions in the rat. J. Biol. Chem. 257 14642–14647. [PubMed] [Google Scholar]
- 4.Jong M. C., K. W. van Dijk, V. E. Dahlmans, H. Van der Boom, K. Kobayashi, K. Oka, G. Siest, L. Chan, M. H. Hofker, and L. M. Havekes. 1999. Reversal of hyperlipidaemia in apolipoprotein C1 transgenic mice by adenovirus-mediated gene delivery of the low-density-lipoprotein receptor, but not by the very-low-density-lipoprotein receptor. Biochem. J. 338 281–287. [PMC free article] [PubMed] [Google Scholar]
- 5.Shachter N. S., T. Ebara, R. Ramakrishnan, G. Steiner, J. L. Breslow, H. N. Ginsberg, and J. D. Smith. 1996. Combined hyperlipidemia in transgenic mice overexpressing human apolipoprotein Cl. J. Clin. Invest. 98 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jong M. C., V. E. Dahlmans, P. J. van Gorp, K. W. van Dijk, M. L. Breuer, M. H. Hofker, and L. M. Havekes. 1996. In the absence of the low density lipoprotein receptor, human apolipoprotein C1 overexpression in transgenic mice inhibits the hepatic uptake of very low density lipoproteins via a receptor-associated protein-sensitive pathway. J. Clin. Invest. 98 2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jong M. C., M. J. Gijbels, V. E. Dahlmans, P. J. Gorp, S. J. Koopman, M. Ponec, M. H. Hofker, and L. M. Havekes. 1998. Hyperlipidemia and cutaneous abnormalities in transgenic mice overexpressing human apolipoprotein C1. J. Clin. Invest. 101 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn J. S., M. Tremblay, R. Batal, H. Jacques, L. Veilleux, C. Rodriguez, L. Bernier, O. Mamer, and J. Davignon. 2002. Plasma kinetics of VLDL and HDL apoC-I in normolipidemic and hypertriglyceridemic subjects. J. Lipid Res. 43 1680–1687. [DOI] [PubMed] [Google Scholar]
- 9.Mahley R. W. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240 622–630. [DOI] [PubMed] [Google Scholar]
- 10.Moore R. J., R. M. Chamberlain, and F. R. Khuri. 2004. Apolipoprotein E and the risk of breast cancer in African-American and non-Hispanic white women: a review. Oncology. 66 79–93. [DOI] [PubMed] [Google Scholar]
- 11.de Knijff P., and C. M. van Duijn. 1998. Role of APOE in dementia: a critical reappraisal. Haemostasis. 28 195–201. [DOI] [PubMed] [Google Scholar]
- 12.Utermann G., M. Hees, and A. Steinmetz. 1977. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 269 604–607. [DOI] [PubMed] [Google Scholar]
- 13.van Bockxmeer F. M., and C. D. Mamotte. 1992. Apolipoprotein epsilon 4 homozygosity in young men with coronary heart disease. Lancet. 340 879–880. [DOI] [PubMed] [Google Scholar]
- 14.Stengard J. H., J. Pekkanen, C. Ehnholm, A. Nissinen, and C. F. Sing. 1996. Genotypes with the apolipoprotein epsilon4 allele are predictors of coronary heart disease mortality in a longitudinal study of elderly Finnish men. Hum. Genet. 97 677–684. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes L. U., C. Gerdes, K. Kervinen, M. Savolainen, I. C. Klausen, P. S. Hansen, Y. A. Kesaniemi, and O. Faergeman. 2000. The apolipoprotein epsilon4 allele determines prognosis and the effect on prognosis of simvastatin in survivors of myocardial infarction: a substudy of the Scandinavian simvastatin survival study. Circulation. 101 1366–1371. [DOI] [PubMed] [Google Scholar]
- 16.Anuurad E., J. Rubin, G. Lu, T. A. Pearson, S. Holleran, R. Ramakrishnan, and L. Berglund. 2006. Protective effect of apolipoprotein E2 on coronary artery disease in African Americans is mediated through lipoprotein cholesterol. J. Lipid Res. 47 2475–2481. [DOI] [PubMed] [Google Scholar]
- 17.Smit M., E. van der Kooij-Meijs, L. P. Woudt, L. M. Havekes, and R. R. Frants. 1988. Exact localization of the familial dysbetalipoproteinemia associated HpaI restriction site in the promoter region of the APOC1 gene. Biochem. Biophys. Res. Commun. 152 1282–1288. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., L. Berglund, R. Ramakrishnan, R. Mayeux, C. Ngai, S. Holleran, B. Tycko, T. Leff, and N. S. Shachter. 1999. A common Hpa I RFLP of apolipoprotein C–I increases gene transcription and exhibits an ethnically distinct pattern of linkage disequilibrium with the alleles of apolipoprotein E. J. Lipid Res. 40 50–58. [PubMed] [Google Scholar]
- 19.Seixas S., M. J. Trovoada, and J. Rocha. 1999. Haplotype analysis of the apolipoprotein E and apolipoprotein C1 loci in Portugal and Sao Tome e Principe (Gulf of Guinea): linkage disequilibrium evidence that APOE*4 is the ancestral APOE allele. Hum. Biol. 71 1001–1008. [PubMed] [Google Scholar]
- 20.Cohn J. S., M. Tremblay, L. Boulet, H. Jacques, J. Davignon, M. Roy, and L. Bernier. 2003. Plasma concentration and lipoprotein distribution of ApoC-I is dependent on ApoE genotype rather than the Hpa I ApoC-I promoter polymorphism. Atherosclerosis. 169 63–70. [DOI] [PubMed] [Google Scholar]
- 21.Anuurad E., G. Lu, J. Rubin, T. A. Pearson, and L. Berglund. 2007. ApoE genotype affects allele-specific apo[a] levels for large apo[a] sizes in African Americans: the Harlem-Basset Study. J. Lipid Res. 48 693–698. [DOI] [PubMed] [Google Scholar]
- 22.McGowan M. W., J. D. Artiss, D. R. Strandbergh, and B. Zak. 1983. A peroxidase-coupled method for the colorimetric determination of serum triglycerides. Clin. Chem. 29 538–542. [PubMed] [Google Scholar]
- 23.Allain C. C., L. S. Poon, C. S. Chan, W. Richmond, and P. C. Fu. 1974. Enzymatic determination of total serum cholesterol. Clin. Chem. 20 470–475. [PubMed] [Google Scholar]
- 24.Warnick G. R., J. Benderson, and J. J. Albers. 1982. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin. Chem. 28 1379–1388. [PubMed] [Google Scholar]
- 25.Friedewald W. T., R. I. Levy, and D. S. Fredrickson. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18 499–502. [PubMed] [Google Scholar]
- 26.Levy J. C., D. R. Matthews, and M. P. Hermans. 1998. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 21 2191–2192. [DOI] [PubMed] [Google Scholar]
- 27.Adler A. I., J. C. Levy, D. R. Matthews, I. M. Stratton, G. Hines, and R. R. Holman. 2005. Insulin sensitivity at diagnosis of Type 2 diabetes is not associated with subsequent cardiovascular disease (UKPDS 67). Diabet. Med. 22 306–311. [DOI] [PubMed] [Google Scholar]
- 28.Hixson J. E., and D. T. Vernier. 1990. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 31 545–548. [PubMed] [Google Scholar]
- 29.Windler E. E., P. T. Kovanen, Y. S. Chao, M. S. Brown, R. J. Havel, and J. L. Goldstein. 1980. The estradiol-stimulated lipoprotein receptor of rat liver. A binding site that membrane mediates the uptake of rat lipoproteins containing apoproteins B and E. J. Biol. Chem. 255 10464–10471. [PubMed] [Google Scholar]
- 30.Sehayek E., and S. Eisenberg. 1991. Mechanisms of inhibition by apolipoprotein C of apolipoprotein E-dependent cellular metabolism of human triglyceride-rich lipoproteins through the low density lipoprotein receptor pathway. J. Biol. Chem. 266 18259–18267. [PubMed] [Google Scholar]
- 31.Gautier T., D. Masson, J. P. de Barros, A. Athias, P. Gambert, D. Aunis, M. H. Metz-Boutigue, and L. Lagrost. 2000. Human apolipoprotein C–I accounts for the ability of plasma high density lipoproteins to inhibit the cholesteryl ester transfer protein activity. J. Biol. Chem. 275 37504–37509. [DOI] [PubMed] [Google Scholar]
- 32.Soutar A. K., C. W. Garner, H. N. Baker, J. T. Sparrow, R. L. Jackson, A. M. Gotto, and L. C. Smith. 1975. Effect of the human plasma apolipoproteins and phosphatidylcholine acyl donor on the activity of lecithin: cholesterol acyltransferase. Biochemistry. 14 3057–3064. [DOI] [PubMed] [Google Scholar]
- 33.Asztalos B. F., E. J. Schaefer, K. V. Horvath, S. Yamashita, M. Miller, G. Franceschini, and L. Calabresi. 2007. Role of LCAT in HDL remodeling: investigation of LCAT deficiency states. J. Lipid Res. 48 592–599. [DOI] [PubMed] [Google Scholar]