Abstract
We previously found that plasma RBP4 levels were strongly associated with metabolic syndrome components. This study aimed to determine whether RBP4 variants are associated with the metabolic syndrome components and plasma RBP4 levels, and to investigate whether the associations between plasma RBP4 and the metabolic syndrome components are causal. Five tagSNPs were tested for their associations with plasma RBP4 levels and metabolic syndrome components in a population-based sample of 3,210 Chinese Hans. A possible causal relationship between plasma RBP4 levels and hypertriglyceridemia was explored by Mendelian randomization. Plasma RBP4 levels were significantly associated with rs10882273 (βz −0.10SD[−0.17, −0.03], P = 0.0050), rs3758538 (βz −0.13SD[−0.24, −0.02], P = 0.0249) in all participants, and with rs17108993 in Shanghai participants (βz −0.19SD[−0.32, −0.05], P = 0.0061). The single nucleotide polymorphism (SNP) rs3758538 was significantly associated with hypertriglyceridemia (OR 0.62[0.45–0.85], P = 0.0026) and triglycerides (βz −0.19SD[−0.30, −0.07], P = 0.001) in all participants. In Mendelian randomization analysis, the observed effect size of association between rs3758538 and hypertriglyceridemia was different from the expected effect size (P = 0.0213). This is the first study to show that the RBP4 variants are significantly associated with plasma RBP4 levels and hypertriglyceridemia risk in Chinese Hans. However, results of Mendelian randomization do not support the hypothesis that RBP4 levels are causally related to hypertriglyceridemia risk.
Keywords: Mendelian randomization, SNP
Retinol-binding protein 4 (RBP4), initially known as the specific carrier of retinol, has recently been identified as a new adipokine that is robustly associated with visceral fat and insulin resistance (1). The potential link between RBP4 and insulin resistance was first suggested by the observations that serum RBP4 levels were elevated in adipose-Glut4−/− mice and completely normalized by the anti-diabetic agent rosiglitazone (2), whereas genetic deletion of Rbp4 (Rbp4−/− mice) improved insulin sensitivity (2). These findings in mice were strongly supported by clinical data in humans from the same group (3), showing that serum RBP4 levels were elevated in insulin-resistant individuals with obesity, impaired glucose tolerance, type 2 diabetes, and even in lean nondiabetic individuals with a strong family history of type 2 diabetes. Several recent genetic association studies also found an association between common RBP4 variants and insulin resistance or type 2 diabetes (4–7). However, results of subsequent clinical and cross-sectional studies were inconsistent, as many failed to confirm the inverse correlation between RBP4 and insulin resistance, or some found opposite results (8–13). Apparently, the role of RBP4 as a mediator of insulin resistance in humans remains to be clarified.
Growing evidence suggests that RBP4 may play a more important role in lipid metabolism than insulin resistance. For example, most of the previous human studies that confirmed the association of RBP4 levels with insulin resistance also observed significant associations with lipid levels, in particular with triglyceride, HDL-cholesterol, and LDL-cholesterol (3, 9, 14, 15). Others observed associations of RBP4 with increased triglycerides levels and with pro-atherogenic lipoproteins or key enzymes of lipoprotein metabolism, but not with insulin resistance marker (9, 16–18). Since hypertriglyceridemia plays an important role in the pathogenesis of cardiovascular disease, circulating RBP4 levels might emerge as a suitable target for therapeutic intervention in cardiovascular disease if the association between circulating RBP4 and hypertriglyceridemia is causal. However, the observational nature of epidemiological association studies does not allow inference of the causal direction between two related traits. It is, therefore, of interest to determine whether elevated circulating RBP4 levels causally contribute to an unfavorable lipid profile and, therefore, to the pathogenesis of cardiovascular disease. This determination can be achieved by Mendelian randomization (19, 20), an epidemiological approach for assessing the direction of causality, in an unbiased way, between putative risk factors and a disease. According to Mendelian randomization, genetic variants in the RBP4 gene are randomly transmitted to the offspring and largely free from reverse causation and confounding. If the association between high circulating RBP4 levels and hypertriglyceridemia or other components of metabolic syndrome is causal, then the genetic variants associated with circulating RBP4 levels also should be associated with risk of hypertriglyceridemia or other components of metabolic syndrome for individuals carrying these variants to the extent predicted by the magnitudes of the associations between genetic variant and circulating RBP4 levels and between the circulating RPB4 and elevated triglyceride levels. Otherwise, the causality is refuted. Therefore, a reliable association between genetic variant in RBP4 gene and plasma RBP4 levels is one of the key assumptions for performing Mendelian randomization.
One of our previous studies based on the same population indicated that the increased levels of circulating RBP4 were strongly and positively associated with body mass index (BMI), waist circumference, blood pressure, plasma triglyceride, total- and LDL-cholesterol levels, and negatively with HDL-cholesterol levels (21). The aims of the present study are (1) to determine whether common variants in the RBP4 gene are associated with circulating RBP4 levels and with the various components of metabolic syndrome, and (2) to investigate the triangular relationship among plasma RBP4 levels, RBP4 gene variants, and the risk of the metabolic syndrome–related traits to assess the possible causal direction that we observed in a Chinese Han population.
RESEARCH DESIGN AND METHODS
Study population
The study population consisted of 3,210 unrelated Chinese Hans from 50 to 70 years of age (1,423 men and 1,787 women) from the Study on Nutrition and Health of Aging Population in China. All participants underwent a complete physical examination including standard anthropometric measurements, overnight fasting blood sample collection, and completion of questionnaires about medical history, nutrition, and physical activity. Height and weight were measured with participants dressed in light-weight clothing without shoes, and BMI was calculated as [weight (kg) / height2 (m2)]. Waist circumference (cm) was measured midway between the lowest rib and the iliac crest to the nearest 0.1 cm, after inhalation and exhalation. Blood pressure was measured by using an electronic blood pressure monitor (Omron HEM-705CP, OMRON Healthcare Inc., Vernon Hills, Illinois) on the right arm of the participant, who was in a comfortable seated position after at least a 5-min rest. Participants were asked to avoid vigorous exercise, eating, drinking, smoking, or long exposure to cold or hot temperatures for 1 h before the measurement. Three measurements were taken, and the mean of the last two measurements was used for analysis. The metabolic syndromes were defined by the updated National Cholesterol Education Program Adult Treatment Panel III criteria (NCEP-ATP III) for Asian-Americans as presenting three or more of the following components: (1) waist circumferences 90 cm or greater in men or 80 cm or greater in women; (2) triglycerides 1.7 mmol/L or greater; (3) HDL-C less than 1.03 mmol/L in men or less than 1.30 mmol/L in women; (4) fasting plasma 5.6 mmol/L or greater or previously diagnosed type 2 diabetes or on oral anti-diabetic medication; (5) blood pressure 130/85 mmHg or greater or current use of anti-hypertensive drugs. The study design and recruitment protocol of this population-based cohort has been described in detail elsewhere (22) and was approved by the Institutional Review Board of the Institute for Nutritional Sciences. Written informed consent was obtained from all participants. The phenotypic characteristics of the study population are shown (see supplementary Table I).
Biochemical assays
Plasma RBP4 protein levels were measured in duplicate by an in-house–developed sandwich ELISA method, utilizing affinity-chromatography purified polyclonal and monoclonal antibodies generated against recombinant human RBP4 protein. The assay system was subsequently cross-validated by western blotting. The intra-assay CV was 1.8–7.6% and inter-assay was 3.7–8.8% (21). Fasting glucose, triglycerides, and HDL-cholesterol were measured enzymatically on an automatic analyzer (Hitachi 7080, Japan) with reagents purchased from Wako Pure Chemical Industries (Osaka, Japan) (22).
Selection of tagSNPs and genotyping
The tagSNPs for RBP4 were selected by using Tagger program implemented in Haploview V3.2 (http://www.broad.mit.edu/mpg/haploview) from the HapMap genotype data/phase II Mar08 for the Asians combined (JPT + CHB populations). According to the HapMap data, there are a total of 25 SNPs in the genomic region from ∼5kb upstream to ∼5kb downstream of RBP4 gene of which 10 are monomorphic. For the remaining 15 polymorphic single nucleotide polymorphisms (SNPs) [minor allele frequency (MAF) ⩾ 5%], tagSNPs were selected with Tagger using a pairwise approach with an r2 threshold ⩾ 0.8. This yielded 4 tagSNPs (rs17108973, rs3758538, rs17108993, and rs11187549) that capture all common variants in this gene region. We genotyped these 4 tagSNPs and an additional 3 SNPs (rs10882273, nonHapMap SNP rs3758539, and rs34571439) that were previously reported to be associated with type 2 diabetes or its related traits in Caucasians (4, 6, 7). SNP genotyping was performed with the GenomeLab™ SNPstream® Genotyping System (Beckman Coulter) according to the manufacturer's protocol. The success rate of genotyping was greater than 94%, and the concordance rate was greater than 99% based on 12% duplicate samples for each SNP. All the variants were in Hardy-Weinberg equilibrium (P > 0.21). The estimates of pairwise linkage disequilibrium (LD) between the 7 SNPs are shown (see supplementary Table II). Since the SNP rs17108973, rs10882273, and rs34571439 are in strong LD with each other (r2 > 0.84), only rs10882273 was used for further analyses.
Statistical analyses
Hardy-Weinberg equilibrium was tested using a likelihood ratio test. TagSNP selection and LD estimation were performed with Haploview V3.2. Logistic regression models were applied to assess the association between RBP4 variants and metabolic syndrome components in case-control analyses. Generalized linear models were used to assess the associations between plasma RBP4 and triglyceride levels, as well as the associations of RBP4 variants with circulating RBP4 levels and the metabolic syndrome–related quantitative traits, including BMI, waist circumference, fasting glucose, fasting triglycerides, HDL-cholesterol, systolic blood pressure (SBP) and diastolic blood pressure (DBP). The dominant model was used for analyses of the SNPs (rs3758538 and rs17108993) with the small number of homozygous carriers of minor alleles (n < 12); otherwise, additive genetic model was applied. We excluded individuals with known diabetes or who were receiving glucose-lowering treatment (n = 267) from the quantitative trait analyses for fasting glucose. Individuals with self-reported dyslipidemia diagnoses status or lipid-lowering medication (n = 479) were excluded from the lipid-related quantitative trait analyses. We adjusted the blood pressure by adding 10 mmHg to SBP and 5 mmHg to DBP, respectively, for individuals who were receiving blood pressure–lowering treatment (23). The association studies were performed either in the whole population for the SNPs with similar genotype distribution between Beijing and Shanghai, or in Beijing and Shanghai subpopulations separately for the remaining three SNPs (rs10882273, rs3758539, and rs17108993) with different genotype distribution between Beijing and Shanghai (see supplementary Table III). Meta-analyses were then conducted to evaluate the combined effects size across the two sub-populations under an additive or dominant genetic model. Triglycerides and BMI was log-transformed to reach normal distribution. All continuous traits were standardized to sex-specific Z-scores before analyses and effect sizes are presented as standardized βs (Z-score).
We used the triangulation approach (Mendelian randomization) to explore the potential causal relationship between plasma RBP4 and triglyceride levels. We applied a stepwise approach including the following analyses: (1): plasma RBP4 levels–hypertriglyceridemia risk; (2) RBP4 gene–plasma RBP4 levels; (3) observed RBP4 gene–hypertriglyceridemia risk; and (4) expected RBP4 gene–hypertriglyceridemia risk. The expected effect size of the RBP4 gene–hypertriglyceridemia association was calculated by multiplying the magnitudes of RBP4 gene–plasma RBP4 association and of plasma RBP4–hypertriglyceridemia association. The associations were estimated using logistic regression or generalized linear regression as described above. Data management and statistical analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC) unless otherwise indicated. All reported P values were nominal and two-sided.
RESULTS
We first examined whether the genetic variants in the RBP4 gene were associated with plasma RBP4 levels. As shown in Table 1, the minor C-alleles of SNP rs10882273 (βz = −0.10SD [−0.17, −0.03], P = 0.0050) and rs3758538 (βz = −0.13SD [−0.24, −0.02], P = 0.0249) were all significantly associated with reduced plasma RBP4 levels in the total population, whereas the minor G-allele of the SNP rs17108993 was significantly associated with decreased plasma RBP4 levels in the Shanghai subpopulation only (βz = −0.19SD [−0.32, −0.05], P = 0.0061).
TABLE 1.
Associations between RBP4 variants and plasma RBP4 levels
| Beijing (n = 1,574) |
Shanghai (n = 1,636) |
Beijing + Shanghai (n = 3,210) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Genotype | Mean (SE) | β (95% CI) | P | Mean (SE) | β (95% CI) | P | Mean (SE) | β (95% CI) | P | P(heter) |
| rs10882273 (+11880 T>C) | TT | 43.0 (0.4) | 38.5 (0.3) | 40.7 (0.2) | |||||||
| TC | 41.5 (0.6) | −0.10 (−0.20, −0.01) | 0.0364 | 37.4 (0.6) | −0.09 (−0.19, 0.00) | 0.06 | 39.4 (0.4) | −0.10 (−0.17, −0.03) | 0.0050a | 0.85 | |
| CC | 41.7 (2.0) | 36.4 (2.1) | 39.1 (1.4) | ||||||||
| rs3758539(−803 G>A) | GG | 42.7 (0.4) | 38.4 (0.3) | 40.5 (0.2) | |||||||
| GA | 42.1 (0.6) | −0.05 (−0.16, 0.05) | 0.34 | 37.4 (0.6) | −0.08 (−0.19, 0.02) | 0.13 | 39.8 (0.4) | −0.07 (−0.14, 0.01) | 0.08a | 0.69 | |
| AA | 41.1 (2.3) | 36.7 (2.3) | 38.9 (1.6) | ||||||||
| rs3758538 (−1265 A>C)b | AA | 42.5 (0.3) | −0.15 (−0.31, 0.02) | 0.08 | 38.3 (0.3) | −0.10 (−0.26, 0.06) | 0.21 | 40.4 (0.2) | −0.13 (−0.24, −0.02) | 0.0249c | 0.67 |
| AC+CC | 40.9 (0.9) | 37.1 (0.9) | 38.9 (0.6) | ||||||||
| rs17108993 (−3248 C>G)b | CC | 42.4 (0.3) | −0.01 (−0.18, 0.16) | 0.93 | 38.4 (0.3) | −0.19 (−0.32, −0.05) | 0.0061 | 40.4 (0.2) | −0.10 (−0.28, 0.07) | 0.24d | 0.10 |
| CG+GG | 42.3 (1.0) | 36.2 (0.7) | 39.1 (0.6) | ||||||||
| rs11187549(−3839 G>A) | GG | 41.7 (0.6) | 37.7 (0.5) | 39.7 (0.4) | |||||||
| GA | 42.3 (0.4) | 0.06 (−0.01, 0.13) | 0.10 | 38.2 (0.4) | −0.01 (−0.07, 0.06) | 0.81 | 40.2 (0.3) | 0.03 (−0.02, 0.07) | 0.29c | 0.16 | |
| AA | 43.0 (0.6) | 37.4 (0.6) | 40.2 (0.4) | ||||||||
BMI, body mass index; SNP, single nucleotide polymorphism; RBP4, retinol-binding protein 4. Effect sizes β (95% CI) are relative to the generalized linear regression model when using the normalized trait and represent the change in plasma RBP4 levels in SD units, on average, for each additional copy of minor allele. P values corresponded to standardized effects and were adjusted for age and BMI.
P values were from fix-effect model in meta-analyses.
Dominant model was applied. Otherwise, additive model was used.
When Beijing and Shanghai participants were combined, analyses were performed by pooling all 3,210 subjects together for SNPs (rs3758538 and rs11187549) with no significant difference in genotype distribution. Otherwise, meta-analyses were conducted to evaluate the combined effects size for SNPs with significant differences in genotype distribution.
P values were from random-effect model in meta-analyses.
As suggested in our previous study (21), plasma RBP4 levels (sex-specific Z-scores) were strongly associated with increased risk of hypertriglyceridemia (odds ratio [OR] 1.80 [1.64–1.96], P = 2.53E-38), obesity (OR 1.38 [1.24–1.54], P = 4.08E-9)/central obesity (OR 1.35 [1.25–1.45], P = 1.15E-14), low HDL-cholesterol (OR 1.11 [1.03–1.21], P = 0.0066) and hypertension (OR 1.18 [1.08–1.28], P= 0.0001) in this population-based sample of Chinese Hans (Table 2). We next performed six case-control studies for each SNP to examine whether they were associated with individual components of the metabolic syndrome, including obesity/central obesity, hypertriglyceridemia, low HDL-cholesterol, hyperglycemia, and hypertension. As shown in Table 2, the minor C-allele of SNP rs3758538 was significantly associated with low risk of hypertriglyceridemia not only in the meta-analyses that combined all individuals (OR 0.62 [0.45–0.85], P = 0.0026), but also in Beijing (OR 0.65 [0.43–0.97], P = 0.0372) and Shanghai (OR 0.60 [0.37–0.96], P = 0.0325) subpopulations separately. The minor G-allele of SNP rs17108993 was significantly associated with low risk of hypertension in Shanghai subpopulation (OR 0.68 [0.50–0.94], P = 0.0185) and marginally in the meta-analyses that combined all individuals (OR 0.78 [0.61–1.00], P = 0.05). We found no significant association between the remaining three SNPs (rs10882273, rs3758539, and rs11187549) and any of the components of metabolic syndrome.
TABLE 2.
Case-control analyses for associations with obesity and metabolic syndrome components in Chinese Hans with the criteria of NCEP ATP III for Asian-Americans
| Beijing |
Shanghai |
Beijing + Shanghai |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total number | Case (MAF) | OR (95% CI) | P | Case (MAF) | OR (95% CI) | P | OR (95% CI) | P | P(heter) | |
| RBP4-metabolic syndrome components | ||||||||||
| Obesity | 1989 | 320 | 1.34 (1.17 − 1.53) | 2.07E-5 | 154 | 1.48 (1.23 − 1.76) | 1.91E-5 | 1.38 (1.24 − 1.54) | 4.08E-9 | |
| Central obesity | 3210 | 867 | 1.31 (1.18 − 1.45) | 3.28E-7 | 664 | 1.41 (1.26 − 1.58) | 1.21E-9 | 1.35 (1.25 − 1.45) | 1.15E-14 | |
| Hypertriglyceridemia | 3210 | 423 | 1.62 (1.44 − 1.82) | 6.50E-16 | 354 | 2.06 (1.79 − 2.36) | 1.25E-24 | 1.80 (1.64 − 1.96) | 2.53E-38 | |
| Low HDL-C | 3210 | 621 | 1.08 (0.97 − 1.20) | 0.19 | 721 | 1.16 (1.03 − 1.30) | 0.0122 | 1.11 (1.03 − 1.21) | 0.0066 | |
| Hyperglycemia | 3210 | 851 | 1.00 (0.91 − 1.10) | 0.99 | 451 | 1.05 (0.94 − 1.19) | 0.39 | 1.02 (0.95 − 1.10)) | 0.56 | |
| Hypertension | 3210 | 1188 | 1.19 (1.06 − 1.35) | 0.0043 | 1033 | 1.17 (1.04 − 1.32) | 0.0111 | 1.18 (1.08 − 1.28) | 0.0001 | |
| RBP4-metabolic syndrome components | ||||||||||
| rs10882273 (+11880 T>C)a | ||||||||||
| Obesity | 315 (0.17) | 1.12 (0.85 − 1.47) | 0.42 | 151 (0.13) | 1.03 (0.72 − 1.47) | 0.87 | 1.08 (0.87 − 1.35) | 0.46 | 0.72 | |
| Central obesity | 853 (0.17) | 1.03 (0.85 − 1.26) | 0.75 | 643 (0.13) | 1.06 (0.86 − 1.32) | 0.58 | 1.05 (0.90 − 1.21) | 0.54 | 0.85 | |
| Hypertriglyceridemia | 416 (0.17) | 1.03 (0.82 − 1.28) | 0.82 | 346 (0.13) | 1.06 (0.82 − 1.37) | 0.64 | 1.04 (0.88 − 1.23) | 0.63 | 0.84 | |
| Low HDL-C | 607 (0.17) | 1.02 (0.82 − 1.26) | 0.87 | 703 (0.13) | 0.92 (0.73 − 1.15) | 0.45 | 0.97 (0.83 − 1.13) | 0.69 | 0.51 | |
| Hyperglycemia | 836 (0.17) | 1.00 (0.82 − 1.21) | 0.96 | 432 (0.12) | 0.85 (0.66 − 1.08) | 0.19 | 0.94 (0.80 − 1.09) | 0.39 | 0.32 | |
| Hypertension | 1166 (0.17) | 1.06 (0.84 − 1.35) | 0.63 | 998 (0.13) | 0.92 (0.73 − 1.15) | 0.45 | 0.98 (0.83 − 1.16) | 0.83 | 0.38 | |
| rs3758539 (-803 G>A)a | ||||||||||
| Obesity | 317 (0.15) | 1.23 (0.93 − 1.63) | 0.16 | 145 (0.11) | 1.13 (0.77 − 1.66) | 0.53 | 1.19 (0.95 − 1.50) | 0.13 | 0.74 | |
| Central obesity | 858 (0.14) | 1.05 (0.85 − 1.31) | 0.63 | 634 (0.11) | 1.12 (0.88 − 1.42) | 0.36 | 1.08 (0.92 − 1.27) | 0.33 | 0.71 | |
| Hypertriglyceridemia | 421 (0.13) | 0.90 (0.70 − 1.14) | 0.37 | 336 (0.11) | 1.05 (0.79 − 1.39) | 0.75 | 0.96 (0.80 − 1.15) | 0.96 | 0.41 | |
| Low HDL-C | 613 (0.14) | 0.92 (0.74 − 1.16) | 0.49 | 686 (0.10) | 0.97 (0.76 − 1.25) | 0.83 | 0.94 (0.80 − 1.12) | 0.51 | 0.76 | |
| Hyperglycemia | 841 (0.14) | 1.02 (0.82 − 1.25) | 0.88 | 437 (0.09) | 0.94 (0.72 − 1.22) | 0.63 | 0.98 (0.94 − 1.16) | 0.85 | 0.65 | |
| Hypertension | 1170 (0.14) | 1.03 (0.80 − 1.33) | 0.84 | 986 (0.10) | 0.93 (0.73 − 1.20) | 0.59 | 0.98 (0.82 − 1.17) | 0.81 | 0.60 | |
| rs3758538 (-1265 A>C)b | ||||||||||
| Obesity | 300 (0.06) | 0.90 (0.58 − 1.40) | 0.64 | 142 (0.05) | 0.97 (0.53 − 1.75) | 0.91 | 0.94 (0.66 − 1.35) | 0.74 | 0.85 | |
| Central obesity | 814 (0.06) | 0.85 (0.61 − 1.19) | 0.35 | 626 (0.05) | 0.83 (0.58 − 1.19) | 0.32 | 0.86 (0.67 − 1.09) | 0.21 | 0.93 | |
| Hypertriglyceridemia | 395 (0.04) | 0.65 (0.43 − 0.97) | 0.0372 | 341 (0.04) | 0.60 (0.37 − 0.96) | 0.0325 | 0.62 (0.45 − 0.85) | 0.0026 | 0.80 | |
| Low HDL-C | 577 (0.06) | 0.76 (0.54 − 1.11) | 0.17 | 683 (0.05) | 0.81 (0.56 − 1.17) | 0.26 | 0.79 (0.61 − 1.02) | 0.07 | 0.86 | |
| Hyperglycemia | 797 (0.06) | 1.07 (0.77 − 1.48) | 0.70 | 434 (0.04) | 0.81 (0.54 − 1.21) | 0.31 | 0.96 (0.74 − 1.23) | 0.72 | 0.30 | |
| Hypertension | 1112 (0.06) | 0.87 (0.59 − 1.28) | 0.49 | 980 (0.05) | 0.98 (0.68 − 1.41) | 0.91 | 0.93 (0.71 − 1.22) | 0.61 | 0.67 | |
| rs17108993 (-3248 C>G)a,b | ||||||||||
| Obesity | 314 (0.05) | 0.93 (0.59 − 1.47) | 0.77 | 152 (0.04) | 0.56 (0.30 − 1.05) | 0.07 | 0.78 (0.54 − 1.13) | 0.19 | 0.20 | |
| Central obesity | 854 (0.05) | 0.95 (0.67 − 1.34) | 0.77 | 657 (0.06) | 0.88 (0.65 − 1.20) | 0.41 | 0.91 (0.72 − 1.15) | 0.42 | 0.74 | |
| Hypertriglyceridemia | 412 (0.05) | 1.07 (0.73 − 1.58) | 0.73 | 352 (0.06) | 0.99 (0.68 − 1.43) | 0.94 | 1.03 (0.78 − 1.34) | 0.86 | 0.76 | |
| Low HDL-C | 606 (0.05) | 0.97 (0.67 − 1.41) | 0.88 | 712 (0.06) | 1.14 (0.83 − 1.56) | 0.42 | 1.07 (0.84 − 1.35) | 0.60 | 0.53 | |
| Hyperglycemia | 834 (0.05) | 0.93 (0.66 − 1.31) | 0.68 | 447 (0.06) | 0.89 (0.63 − 1.25) | 0.49 | 0.91 (0.71 − 1.16) | 0.44 | 0.84 | |
| Hypertension | 1164 (0.05) | 0.97 (0.65 − 1.46) | 0.89 | 1022 (0.06) | 0.68 (0.50 − 0.94) | 0.0185 | 0.78 (0.61 − 1.00) | 0.05 | 0.18 | |
| rs11187549 (-3839 G>A) | ||||||||||
| Obesity | 304 (0.50) | 1.05 (0.86 − 1.28) | 0.64 | 152 (0.45) | 0.84 (0.66 − 1.08) | 0.18 | 0.96 (0.82 − 1.13) | 0.63 | 0.18 | |
| Central obesity | 833 (0.49) | 1.04 (0.89 − 1.20) | 0.64 | 642 (0.47) | 0.93 (0.81 − 1.08) | 0.33 | 0.98 (0.88 − 1.09) | 0.68 | 0.31 | |
| Hypertriglyceridemia | 403 (0.49) | 1.04 (0.88 − 1.23) | 0.65 | 347 (0.46) | 0.96 (0.81 − 1.14) | 0.65 | 1.00 (0.89 − 1.13) | 0.98 | 0.52 | |
| Low HDL-C | 598 (0.48) | 0.95 (0.82 − 1.16) | 0.55 | 699 (0.48) | 0.98 (0.84 − 1.13) | 0.76 | 0.97 (0.87 − 1.08) | 0.52 | 0.83 | |
| Hyperglycemia | 810 (0.48) | 0.98 (0.85 − 1.13) | 0.77 | 440 (0.46) | 0.97 (0.83 − 1.13) | 0.69 | 0.97 (0.87 − 1.08) | 0.56 | 0.92 | |
| Hypertension | 1146 (0.47) | 0.88 (0.74 − 1.05) | 0.17 | 1004 (0.47) | 0.94 (0.80 − 1.09) | 0.41 | 0.91 (0.81 − 1.02) | 0.11 | 0.62 | |
BMI, body mass index; RBP4, retinol-binding protein 4. Obesity and normal weight were defined as BMI ⩾ 28 kg/m2 and BMI < 24 kg/m2 according to Chinese criteria. Overweight participants (n = 1221, 24 kg/m2 ⩽ BMI < 28 kg/m2) were excluded from the analyses for obesity. The metabolic syndromes was defined by the updated National Cholesterol Education Program Adult Treatment Panel III criteria (NCEP-ATP III) for Asian-Americans: (1) waist circumference ⩾ 90 cm (men); ⩾ 80 cm (women); (2) triglycerides ⩾ 1.7 mmol/L; (3) HDL-C < 1.03 mmol/L (men); < 1.30 mmol/L (women); (4) fasting plasma ⩾ 5.6 mmol/L or previously diagnosed type 2 diabetes or oral anti-diabetic medication; (5) blood pressure ⩾ 130 / 85 mm Hg or current use of anti-hypertensive drugs. P values were adjusted for age, sex, and BMI (where appropriate).
When Beijing and Shanghai participants combined, meta-analyses were conducted to evaluate the combined effects size for SNPs with significant differences in genotype distribution. Otherwise, analyses were performed by pooling all 3,210 subjects together.
Dominant model was applied. Otherwise, additive model was used.
We then tested for associations between the SNPs and the metabolic syndrome-related quantitative traits (BMI, waist circumference, fasting triglyceride, HDL-cholesterol, glucose, systolic and diastolic blood pressure). Consistently, the minor C-alleles of SNP rs3758538 were significantly associated with the decreased levels of plasma triglyceride in the total population (βz −0.19SD [−0.30, −0.07], P = 0.001), as well as in the Beijing (βz −0.17SD [−0.34, −0.00], P = 0.045) and Shanghai (βz −0.20SD [−0.35, −0.05], P = 0.009) subpopulations separately (Table 3). None of the other SNPs were found to be associated with any of the six quantitative traits, although the minor A-allele of SNP rs11187549 exhibited significant association with decreased levels of fasting glucose (P = 0.034) in the whole population as well as with BMI (P = 0.0185) and waist circumference (P = 0.0338) in Shanghai subpopulation (Table 3).
TABLE 3.
Quantitative traits analyses for associations between RBP4 variants and components of metabolic syndromes or BMI
| Beijing |
Shanghai |
Beijing + Shanghai |
|||||
|---|---|---|---|---|---|---|---|
| β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | P(heter) | |
| RBP4-metabolic syndrome componentsa | |||||||
| BMI | 0.11 (0.07, 0.16) | 2.12E-6 | 0.16 (0.11, 0.21) | 2.09E-10 | 0.13 (0.10, 0.17) | 1.73E-14 | |
| Waist circumference | 0.14 (0.10, 0.19) | 1.87E-9 | 0.22 (0.17, 0.27) | 1.65E-17 | 0.17 (0.14, 0.21) | 1.59E-23 | |
| Triglyceride | 0.24 (0.19, 0.29) | 1.26E-21 | 0.33 (0.28, 0.37) | 5.01E-40 | 0.28 (0.25, 0.32) | 3.34E-57 | |
| HDL-C | −0.04 (−0.09, 0.02) | 0.18 | −0.04 (−0.09, 0.01) | 0.15 | −0.04 (−0.07, −0.00) | 0.0489 | |
| Glucose | −0.02 (−0.06, 0.02) | 0.34 | −0.00 (−0.04, 0.03) | 0.77 | −0.01 (−0.04, 0.01) | 0.31 | |
| SBP | 0.06 (0.02, 0.11) | 0.0047 | 0.07 (0.02, 0.11) | 0.0047 | 0.07 (0.03, 0.10) | 6.82E-5 | |
| DBP | 0.08 (0.04, 0.13) | 0.0006 | 0.11 (0.06, 0.16) | 1.44E-5 | 0.10 (0.06, 0.13) | 2.61E-8 | |
| rs10882273 (+11880 T>C)b | |||||||
| BMI | 0.01 (−0.08, 0.11) | 0.76 | 0.01 (−0.09, 0.11) | 0.82 | 0.00 (−0.07, 0.07) | 0.95 | 0.81 |
| Waist circumference | −0.02 (−0.11, 0.08) | 0.69 | 0.01 (−0.09, 0.11) | 0.88 | −0.01 (−0.08, 0.06) | 0.85 | 0.70 |
| Triglyceride | 0.05 (−0.05, 0.15) | 0.35 | −0.01 (−0.11, 0.09) | 0.81 | 0.02 (−0.05, 0.09) | 0.63 | 0.40 |
| HDL-C | 0.00 (−0.10, 0.10) | 0.98 | 0.06 (−0.04, 0.16) | 0.26 | 0.03 (−0.04, 0.10) | 0.44 | 0.15 |
| Glucose | 0.03 (−0.04, 0.10) | 0.41 | −0.04 (−0.12, 0.04) | 0.32 | −0.00 (−0.05, 0.05) | 0.95 | 0.20 |
| SBP | 0.05 (−0.04, 0.14) | 0.30 | 0.01 (−0.08, 0.10) | 0.88 | 0.03 (−0.04, 0.09) | 0.40 | 0.54 |
| DBP | 0.05 (−0.04, 0.15) | 0.26 | −0.01 (−0.10, 0.09) | 0.89 | 0.02 (−0.04, 0.09) | 0.48 | 0.38 |
| rs3758539 (-803 G>A)b | |||||||
| BMI | 0.06 (−0.04, 0.17) | 0.21 | −0.00 (−0.11, 0.10) | 0.94 | 0.03 (−0.04, 0.11) | 0.38 | 0.36 |
| Waist circumference | 0.01 (−0.09, 0.11) | 0.88 | 0.00 (−0.11, 0.11) | 0.99 | 0.00 (−0.07, 0.08) | 0.90 | 0.92 |
| Triglyceride | 0.00 (−0.11, 0.11) | 0.99 | 0.02 (−0.09, 0.13) | 0.74 | 0.01 (−0.07, 0.08) | 0.82 | 0.84 |
| HDL-C | 0.05 (−0.06, 0.15) | 0.39 | 0.06 (−0.05, 0.18) | 0.28 | 0.05 (−0.02, 0.13) | 0.17 | 0.81 |
| Glucose | 0.01 (−0.07, 0.09) | 0.77 | −0.05 (−0.13, 0.04) | 0.29 | −0.01 (−0.07, 0.04) | 0.62 | 0.41 |
| SBP | 0.02 (−0.08, 0.12) | 0.68 | 0.03 (−0.08, 0.13) | 0.61 | 0.02 (−0.05, 0.09) | 0.52 | 0.93 |
| DBP | −0.01 (−0.11, 0.09) | 0.81 | −0.02 (−0.12, 0.09) | 0.74 | −0.01 (−0.09, 0.06) | 0.69 | 0.95 |
| rs3758538 (-1265 A>C)c | |||||||
| BMI | −0.07 (−0.22, 0.09) | 0.42 | 0.02 (−0.15, 0.18) | 0.85 | −0.02 (−0.14, 0.09) | 0.71 | 0.48 |
| Waist circumference | −0.02 (−0.18, 0.14) | 0.77 | −0.08 (−0.24, 0.08) | 0.33 | −0.04 (−0.16, 0.07) | 0.45 | 0.63 |
| Triglyceride | −0.17 (−0.34, −0.00) | 0.045 | −0.20 (−0.35, −0.05) | 0.009 | −0.19 (−0.30, −0.07) | 0.001 | 0.81 |
| HDL-C | 0.11 (−0.06, 0.28) | 0.20 | 0.10 (−0.07, 0.26) | 0.25 | 0.10 (−0.01, 0.22) | 0.08 | 0.90 |
| Glucose | 0.03 (−0.11, 0.17) | 0.71 | −0.03 (−0.13, 0.06) | 0.50 | −0.00 (−0.09, 0.08) | 0.98 | 0.48 |
| SBP | −0.03 (−0.18, 0.12) | 0.73 | 0.08 (−0.07, 0.23) | 0.28 | 0.03 (−0.08, 0.13) | 0.61 | 0.31 |
| DBP | −0.05 (−0.20, 0.11) | 0.57 | −0.03 (−0.18, 0.13) | 0.75 | −0.04 (−0.15, 0.07) | 0.52 | 0.86 |
| rs17108993 (-3248 C>G)b,c | |||||||
| BMI | −0.05 (−0.22, 0.11) | 0.54 | −0.03 (−0.17, 0.11) | 0.64 | −0.04 (−0.15, 0.07) | 0.45 | 0.87 |
| Waist circumference | −0.07 (−0.24, 0.09) | 0.39 | −0.10 (−0.25, 0.04) | 0.15 | −0.09 (−0.20, 0.02) | 0.10 | 0.77 |
| Triglyceride | −0.10 (−0.27, 0.08) | 0.28 | 0.00 (−0.13, 0.13) | 0.98 | −0.04 (−0.14, 0.07) | 0.51 | 0.38 |
| HDL-C | −0.02 (−0.19, 0.15) | 0.84 | −0.07 (−0.22, 0.07) | 0.30 | −0.05 (−0.16, 0.06) | 0.35 | 0.62 |
| Glucose | 0.00 (−0.14, 0.15) | 0.94 | −0.01 (−0.10, 0.08) | 0.85 | −0.00 (−0.08, 0.07) | 0.91 | 0.87 |
| SBP | 0.05 (−0.10, 0.21) | 0.49 | 0.01 (−0.12, 0.14) | 0.94 | 0.03 (−0.07, 0.13) | 0.61 | 0.63 |
| DBP | 0.01 (−0.15, 0.18) | 0.87 | 0.04 (−0.09, 0.18) | 0.53 | 0.03 (−0.07, 0.14) | 0.56 | 0.79 |
| rs11187549 (-3839 G>A) | |||||||
| BMI | 0.02 (−0.05, 0.09) | 0.62 | −0.08 (−0.15, −0.01) | 0.0185 | −0.03 (−0.08, 0.01) | 0.17 | 0.05 |
| Waist circumference | −0.01 (−0.08, 0.06) | 0.73 | −0.07 (−0.14, −0.01) | 0.0338 | −0.04 (−0.09, 0.00) | 0.07 | 0.22 |
| Triglyceride | −0.04 (−0.12, 0.04) | 0.31 | 0.00 (−0.06, 0.07) | 0.89 | −0.01 (−0.06, 0.03) | 0.56 | 0.38 |
| HDL-C | −0.01 (−0.09, 0.06) | 0.73 | −0.03 (−0.10, 0.04) | 0.41 | −0.02 (−0.07, 0.03) | 0.41 | 0.77 |
| Glucose | −0.05 (−0.11, 0.01) | 0.12 | −0.03 (−0.07, 0.01) | 0.18 | −0.04 (−0.08, −0.00) | 0.034 | 0.58 |
| SBP | −0.02 (−0.09, 0.04) | 0.46 | −0.02 (−0.08, 0.04) | 0.51 | −0.02 (−0.07, 0.02) | 0.29 | 0.93 |
| DBP | −0.02 (−0.09, 0.05) | 0.60 | −0.00 (−0.07, 0.06) | 0.93 | −0.01 (−0.06, 0.03) | 0.60 | 0.74 |
BMI, body mass index; DBP, diastolic blood pressure; RBP4, retinol-binding protein 4; SBP, systolic blood pressure. Data of BMI and triglyceride were log-transformed before calculation of sex-corrected Z scores. All βs (95% CI) were presented in SD units. P values corresponded to standardized effects and adjusted for age and BMI (only adjusted for age in analyses for waist circumference). Participants with known diabetes or receiving glucose-lowering treatment (n = 267) were excluded from analyses for associations with fasting glucose; individuals with dyslipidemia diagnoses based on self-report or those using lipid-lowering medication (n = 479) were excluded from the analyses for associations with triglyceride and HDL-C; Blood pressures of individuals with current anti-hypertension treatment were adjusted by adding 10 mmHg to SBP and 5 mmHg to DBP, respectively.
Correlations between plasma RBP4 and metabolic syndrome components have been previously reported (21).
When Beijing and Shanghai participants combined, meta-analyses were conducted to evaluate the combined effects size for SNPs with significant differences in genotype distribution. Otherwise, analyses were performed by pooling all 3,210 subjects together.
Dominant model was applied. Otherwise, additive model was used.
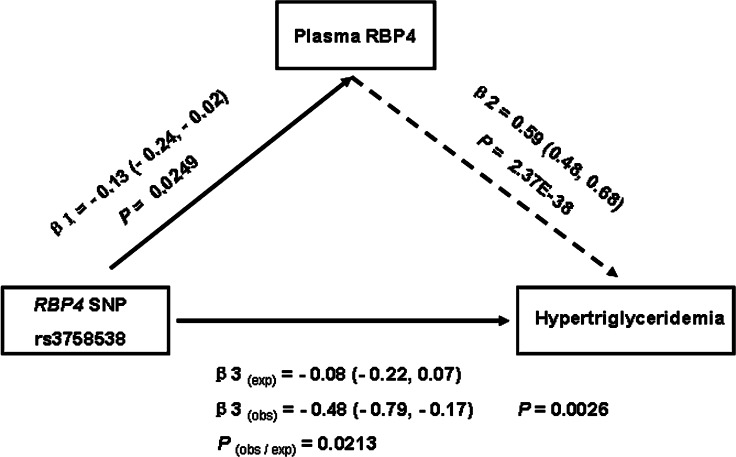
The significant associations of SNP rs3758538 with plasma RBP4 levels and hypertriglyceridemia allow us to further investigate whether the elevated risk of hypertriglyceridemia was attributable to the increased RBP4 levels by using the triangulation approach. As shown in Fig. 1, the observed effect size per C-allele of rs3758538 on the risk of hypertriglyceridemia was −0.48 SD (95% CI: −0.79, −0.17), which is significantly different from the expected effect size (−0.08 SD, 95% CI: −0.22, −0.07) (P for difference = 0.0213), which was calculated based on the observed associations between rs3758538 and RBP4 levels (−0.13 SD, 95% CI: −0.24, −0.02) and between BRP4 levels and triglyceridemia (0.59 SE, 95% CI: 0.48, 0.68) (Fig. 1). Moreover, adjustment for plasma RBP4 values did not materially change the observed effect size per C-allele of rs3758538 on the risk of hypertriglyceridemia (−0.43 SD, 95% CI: −0.75, −0.11) (P for difference = 0.86). These results largely exclude the possibility that the association between RBP4-rs3758538 C-allele and risk of hypertriglyceridemia is mediated by plasma RBP4 levels. Because triglyceride levels have a continuous distribution, classification of hypertriglyceridemia is somewhat arbitrary. Therefore, we reevaluated the triangulation relation of genetic variant, plasma RBP4, and elevated plasma triglycerides levels by taking triglycerides as a continuous variable and obtained similar results (data not shown). These results do not support the hypothesis that increased plasma RBP4 levels are the causal factor for hypertriglyceridemia.
Fig. 1.
Results of Mendelian randomization study. β1: the effect size of association between rs3758538 and plasma RBP4 levels (shown in Table 1); β2: the effect size of association between RBP4 levels and hypertriglyceridemia; β3(obs): the observed effect size of association between SNP rs3758538 and hypertriglyceridemia; β3(exp): the expected effect size of association between SNP rs3758538 and hypertriglyceridemia; P(obs/exp): P value for difference between β3(obs) and β3(exp). The β2 and β3(obs) were derived from natural log transformation of the ORs for the associations between plasma RBP4 and hypertriglyceridemia (β2 = In1.80 = 0.59) and between SNP rs3758538 and hypertriglyceridemia (β3(obs) = In0.62 = −0.48) (Table 2), and β3(exp) was calculated by multiplying the magnitudes of rs3758538-RBP4 and RBP4-hypertriglyceride (β3 = β1 × β2).
DISCUSSION
In this population-based sample of Chinese Hans, we confirmed that plasma RBP4 levels were strongly and independently associated with increased risk of hypertriglyceridemia, central obesity, low HDL-cholesterol, and hypertension. Genetic association studies suggested that both the SNP rs3758538 (−1265A>C) and rs10882273 (+11880T>C) were significantly associated with plasma RBP4 levels in the total sample, but only RBP4-rs3758538 C-allele also exhibited significant association with reduced fasting levels of plasma triglycerides, conferring a decreased risk of hypertriglyceridemia. However, the results of further Mendelian randomization analyses (the triangulation approach) by taking SNP rs3758538 as an instrumental variable suggested that the association between circulating RBP4 levels and hypertriglyceridemia risk was not causal.
Previously, a genetic variant (−803G>A, rs3758539) in RBP4 gene was shown to influence the transcription efficiency in a hepatocarcinoma cell line as well as the binding efficiency of hepatocyte nuclear factor 1 α to the motif, and its A-allele was associated with circulating RBP4 levels in the diabetic patients of Mongolians (4). This variant might, therefore, represent a good instrument for determining causal effects of circulating RBP4 on the metabolic syndrome components if carrying of the A-allele could indeed expose individuals to a long-term elevation in plasma RBP4 levels. However, we failed to replicate the previously reported association between SNP rs3758539 and plasma RBP4 levels in the present study. The reason for the discrepancies between our results and that observed in Mongolians is unclear, but our result is consistent with a recent study in German Caucasians where no significant impact of SNP rs3758539 on either RBP4 mRNA expression in adipose tissue or on serum RBP4 concentration was found (7). Yet, we found that the minor alleles of both the SNP rs3758538 and rs10882273 were significantly associated with lower plasma RBP4 levels, a novel finding that was not reported in the previous studies. Therefore, we investigated whether these two variants could be used to clarify the potential causal effects of plasma RBP4 levels on hypertriglyceridemia or other components of metabolic syndrome.
We found that RBP4 rs3758538 C-allele was significantly associated with reduced risk of hypertriglyceridemia, and individuals carrying this allele had significantly lower fasting levels of plasma triglycerides. The plausible mechanism by which the RBP4 rs3758538 C-allele protects against hypertriglyceridemia and lowers plasma triglyceride concentration remains to be elucidated. What seems plausible is that the rs3758538 C-allele might influence the transcription efficiency of RBP4 gene and, consequently, lead to plasma RBP4 levels and lower risk for hypertriglycerdemia, assuming the observational association between plasma RBP4 levels and risk of hypertriglycerdemia is causal and unbiased. However, we found no association of SNP rs10882273 with hypertriglyceridemia or fasting levels of plasma triglycerides, despite this variant being more strongly associated with plasma RBP4 levels than rs3758538. This implies that either the plasma RBP4–hypertriglyceridemia is not causal or the SNP rs10882273–plasma RBP4 association is subject to confounding by other environmental factors. Consistent with associations with plasma RBP4 levels, no significant association with hypertriglyceridemia was observed for the remaining three SNPs, including the rs3758539 (−803G>A). Therefore, the rs3758538 was used to explore the causal effects between plasma RBP4 levels and hypertrglyceridemia risk.
We believe this is the first study to explore the causality of association between circulating RBP4 levels and hypertriglyceridemia risk by the triangulation approach (Mendelian randomization). The assumptions underlying the triangulation approach is that genetic variants in the RBP4 gene are randomly transmitted to the offspring due to the random assortment of genes at the time of gamete formation and conception, and consequently, their associations with circulating RBP4 levels and disease outcome should not be subject to reverse causation and confounding (24, 25). If the association between plasma RBP4 and hypertriglyceridemia is unconfounded and causal, the genetic variants that expose carriers to a long-term elevation in plasma RBP4 levels should confer an increased risk of hypertriglyceridemia proportional to the difference in plasma RBP4 attributable to the risk allele. Therefore, the observed effect sizes of RBP4 genotypes on hypertriglyceridemia risk would be similar to the expected effect sizes, which are estimated by the magnitude of the association between RBP4 genotypes and plasma RBP4 levels and of the association between plasma RBP4 levels and risk of hypertriglyceridemia, and adjustment for the plasma RBP4 values would markedly attenuate or abolish the association between the RBP4 genotypes and hypertriglyceridemia risk. We found that the observed effect sizes for the association between SNP rs3758538 and hypertriglyceridemia risk was significantly higher than the expected effect sizes, which suggested that the association between plasma RBP4 levels and hypertriglyceridemia risk is potentially due to confounding by other risk factors. Moreover, adjustment for plasma RBP4 concentration only slightly attenuated the effect sizes of the SNP rs3758538 C-allele on hypertriglyceridemia risk, which largely excluded the possibility that SNP rs3758538 increased the risk of hypertriglyceridemia by modulating circulating RBP4 levels. Therefore, results of the Mendelian randomization do not support the hypothesis that the elevated circulating RBP4 levels are causally related to increased risk of hypertriglyceridemia. Because this is the first study to examine the Mendelian randomization for RBP4 SNP and levels in relation to the seven metabolic traits, we chose to report all the results without adjustment for multiple testing to ensure that all potentially useful associations are available for subsequent (meta-analysis) studies.
Theoretically, several issues may lead to false-negative results in assessing causality using Mendelian randomization (20, 24, 26). First, the genetic variant used as the instrumental variable could influence the disease risk through its effects on the physiological function of the protein, rather than through its influence on plasma concentration. But this seems highly unlikely for the present study. The SNP rs3758538 is located in the promoter region of RBP4 gene, and allelic variation in this SNP does not result in the amino acid change. In addition, no variant in the RBP4 coding region is in high LD with SNP rs3758538, according to the HapMap HCB database. Therefore, the impact of SNP rs3758538 on RBP4 protein is mainly in concentration rather than function, although its exact function remains to be determined. Second, the variant chosen as instrument is in LD with other variants of nearby genes that are actually responsible for the genotype–disease-risk association or has pleiotropic effects. We failed to find any variant in the 300-kb flanking regions of RBP4 gene that is in LD with the RBP4 rs3758538 in the HapMap CHB database. Therefore, it is more likely the observed association is mediated through the RBP4 gene rather than through a secondary locus, but we are unable to reliably exclude the possibility that the rs3758538 variant has unrecognized pleiotropic effects, and the biological function remains to be elucidated. Third, there is population stratification in the study sample. In this study, all participants are ancestrally homogeneous of Chinese Hans, and all the analyses were performed for Beijing and Shanghai, separately, and the meta-analyses were conducted to evaluate the combined effects size of the two sub-populations. Thus, population stratification is unlikely to be a major confounding of the genotype-hypertriglyceridemia association and the genotype–plasma-RBP4 associations in this study.
A limitation of this study is that the association between SNP rs3758538 and plasma RBP4 level is only nominally significant. No previous study is available to demonstrate that this variant is indeed a valid measure of lifelong difference in circulating RBP4 levels. Another limitation is that the sample size in this study may not be big enough to clarify whether small differences in plasma RBP4 levels attributable to the RBP4 variants cause hypertriglyceridemia and provide definite evidence for the causal effect (19). Therefore, further studies on potential functional effect of SNP rs3758538 and genetic association studies with much larger sample sizes are required to provide definite evidence for the causal relationship between circulating RBP4 levels and hypertriglyceridemia.
In summary, our results demonstrate that genetic variation in the RBP4 gene is associated with plasma RBP4 levels and hypertriglyceridemia risk in this population of Chinese Hans. However, results of Mendelian randomization do not support a causal association of plasma RBP4 levels with risk of hypertriglyceridemia or elevated levels of plasma RBP4. Much larger genetic studies are required to obtain precise estimates and completely rule out the possibility that circulating RBP4 levels are causally related to hypertriglyceridemia and elevated levels of plasma triglycerides.
Acknowledgments
The authors are grateful to all participants of the Study on Nutrition and Health of Aging Population in China. We also thank our colleagues at the laboratory and the local CDC staffs of Beijing and Shanghai for their assistance with data collection.
Abbreviations
BMI, body mass index
DBP, diastolic blood pressure
LD, linkage disequilibrium
MAF, minor allele frequency
OR, odds ratio
RBP4, retinol-binding protein 4
SBP, systolic blood pressure
SNP, single nucleotide polymorphism
This study was funded by the National Natural Science Foundation of China (Grant 30571562); the Knowledge Innovation Program Pilot Project of the Chinese Academy of Sciences (KSCX2-YW-R-73); the National Basic Research Program of China (973 Program 2006CB503902); the Major Projects of Knowledge Innovation Program (KSCX2-YW-R-116 and KSCX1-YW-02); and the Shanghai-Unilever Research Development Fund (CH-2006-0941).
Published, JLR Papers in Press, March 14, 2009.
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
References
- 1.von Eynatten M., and P. M. Humpert. 2008. Retinol-binding protein-4 in experimental and clinical metabolic disease. Expert Rev. Mol. Diagn. 8 289–299. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q., T. E. Graham, N. Mody, F. Preitner, O. D. Peroni, J. M. Zabolotny, K. Kotani, L. Quadro, and B. B. Kahn. 2005. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 436 356–362. [DOI] [PubMed] [Google Scholar]
- 3.Graham T. E., Q. Yang, M. Bluher, A. Hammarstedt, T. P. Ciaraldi, R. R. Henry, C. J. Wason, A. Oberbach, P. A. Jansson, U. Smith, et al. 2006. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N. Engl. J. Med. 354 2552–2563. [DOI] [PubMed] [Google Scholar]
- 4.Munkhtulga L., K. Nakayama, N. Utsumi, Y. Yanagisawa, T. Gotoh, T. Omi, M. Kumada, B. Erdenebulgan, K. Zolzaya, T. Lkhagvasuren, et al. 2007. Identification of a regulatory SNP in the retinol binding protein 4 gene associated with type 2 diabetes in Mongolia. Hum. Genet. 120 879–888. [DOI] [PubMed] [Google Scholar]
- 5.van Hoek M., A. Dehghan, M. C. Zillikens, A. Hofman, J. C. Witteman, and E. J. Sijbrands. 2008. An RBP4 promoter polymorphism increases risk of type 2 diabetes. Diabetologia. 51 1423–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig R. L., W. S. Chu, and S. C. Elbein. 2007. Retinol binding protein 4 as a candidate gene for type 2 diabetes and prediabetic intermediate traits. Mol. Genet. Metab. 90 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovacs P., M. Geyer, J. Berndt, N. Kloting, T. E. Graham, Y. Bottcher, B. Enigk, A. Tonjes, D. Schleinitz, M. R. Schon, et al. 2007. Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes. 56 3095–3100. [DOI] [PubMed] [Google Scholar]
- 8.Janke J., S. Engeli, M. Boschmann, F. Adams, J. Bohnke, F. C. Luft, A. M. Sharma, and J. Jordan. 2006. Retinol-binding protein 4 in human obesity. Diabetes. 55 2805–2810. [DOI] [PubMed] [Google Scholar]
- 9.von Eynatten M., P. M. Lepper, D. Liu, K. Lang, M. Baumann, P. P. Nawroth, A. Bierhaus, K. A. Dugi, U. Heemann, B. Allolio, et al. 2007. Retinol-binding protein 4 is associated with components of the metabolic syndrome, but not with insulin resistance, in men with type 2 diabetes or coronary artery disease. Diabetologia. 50 1930–1937. [DOI] [PubMed] [Google Scholar]
- 10.Takashima N., H. Tomoike, and N. Iwai. 2006. Retinol-binding protein 4 and insulin resistance. N. Engl. J. Med. 355 1392 (author reply 1394–1395). [DOI] [PubMed] [Google Scholar]
- 11.Erikstrup C., O. H. Mortensen, and B. K. Pedersen. 2006. Retinol-binding protein 4 and insulin resistance. N. Engl. J. Med. 355 1393–1394 (author reply 1394–1395). [PubMed] [Google Scholar]
- 12.Broch M., J. Vendrell, W. Ricart, C. Richart, and J. M. Fernandez-Real. 2007. Circulating retinol-binding protein-4, insulin sensitivity, insulin secretion, and insulin disposition index in obese and nonobese subjects. Diabetes Care. 30 1802–1806. [DOI] [PubMed] [Google Scholar]
- 13.Promintzer M., M. Krebs, J. Todoric, A. Luger, M. G. Bischof, P. Nowotny, O. Wagner, H. Esterbauer, and C. Anderwald. 2007. Insulin resistance is unrelated to circulating retinol binding protein and protein C inhibitor. J. Clin. Endocrinol. Metab. 92 4306–4312. [DOI] [PubMed] [Google Scholar]
- 14.Takebayashi K., M. Suetsugu, S. Wakabayashi, Y. Aso, and T. Inukai. 2007. Retinol binding protein-4 levels and clinical features of type 2 diabetes patients. J. Clin. Endocrinol. Metab. 92 2712–2719. [DOI] [PubMed] [Google Scholar]
- 15.Lee D. C., J. W. Lee, and J. A. Im. 2007. Association of serum retinol binding protein 4 and insulin resistance in apparently healthy adolescents. Metabolism. 56 327–331. [DOI] [PubMed] [Google Scholar]
- 16.Hutchison S. K., C. Harrison, N. Stepto, C. Meyer, and H. J. Teede. 2008. Retinol-binding protein 4 and insulin resistance in polycystic ovary syndrome. Diabetes Care. 31 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zugaro A., C. Pandolfi, A. Barbonetti, M. R. Vassallo, A. D'Angeli, S. Necozione, M. S. Colangeli, S. Francavilla, and F. Francavilla. 2007. Retinol binding protein 4, low birth weight-related insulin resistance and hormonal contraception. Endocrine. 32 166–169. [DOI] [PubMed] [Google Scholar]
- 18.Aeberli I., R. Biebinger, R. Lehmann, D. L'Allemand, G. A. Spinas, and M. B. Zimmermann. 2007. Serum retinol-binding protein 4 concentration and its ratio to serum retinol are associated with obesity and metabolic syndrome components in children. J. Clin. Endocrinol. Metab. 92 4359–4365. [DOI] [PubMed] [Google Scholar]
- 19.Sandhu M. S., S. L. Debenham, I. Barroso, and R. J. Loos. 2008. Mendelian randomisation studies of type 2 diabetes: future prospects. Diabetologia. 51 211–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheehan N. A., V. Didelez, P. R. Burton, and M. D. Tobin. 2008. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 5 e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Q., Z. Yu, X. Ye, F. Zhao, P. Huang, F. B. Hu, O. H. Franco, J. Wang, H. Li, Y. Liu, et al. 2007. Elevated retinol-binding protein 4 levels are associated with metabolic syndrome in Chinese people. J. Clin. Endocrinol. Metab. 92 4827–4834. [DOI] [PubMed] [Google Scholar]
- 22.Ye X., Z. Yu, H. Li, O. H. Franco, Y. Liu, and X. Lin. 2007. Distributions of C-reactive protein and its association with metabolic syndrome in middle-aged and older Chinese people. J. Am. Coll. Cardiol. 49 1798–1805. [DOI] [PubMed] [Google Scholar]
- 23.Tobin M. D., N. A. Sheehan, K. J. Scurrah, and P. R. Burton. 2005. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat. Med. 24 2911–2935. [DOI] [PubMed] [Google Scholar]
- 24.Davey Smith G., and S. Ebrahim. 2003. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32 1–22. [DOI] [PubMed] [Google Scholar]
- 25.Smith G. D., D. A. Lawlor, R. Harbord, N. Timpson, I. Day, and S. Ebrahim. 2007. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 4 e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nitsch D., M. Molokhia, L. Smeeth, B. L. DeStavola, J. C. Whittaker, and D. A. Leon. 2006. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am. J. Epidemiol. 163 397–403. [DOI] [PubMed] [Google Scholar]