Abstract
Plastid genome content and arrangement are highly conserved across most land plants and their closest relatives, streptophyte algae, with nearly all plastid introns having invaded the genome in their common ancestor at least 450 million years ago. One such intron, within the transfer RNA trnK-UUU, contains a large open reading frame that encodes a presumed intron maturase, matK. This gene is missing from the plastid genomes of two species in the parasitic plant genus Cuscuta but is found in all other published land plant and streptophyte algal plastid genomes, including that of the nonphotosynthetic angiosperm Epifagus virginiana and two other species of Cuscuta. By examining matK and plastid intron distribution in Cuscuta, we add support to the hypothesis that its normal role is in splicing seven of the eight group IIA introns in the genome. We also analyze matK nucleotide sequences from Cuscuta species and relatives that retain matK to test whether changes in selective pressure in the maturase are associated with intron deletion. Stepwise loss of most group IIA introns from the plastid genome results in substantial change in selective pressure within the hypothetical RNA-binding domain of matK in both Cuscuta and Epifagus, either through evolution from a generalist to a specialist intron splicer or due to loss of a particular intron responsible for most of the constraint on the binding region. The possibility of intron-specific specialization in the X-domain is implicated by evidence of positive selection on the lineage leading to C. nitida in association with the loss of six of seven introns putatively spliced by matK. Moreover, transfer RNA gene deletion facilitated by parasitism combined with an unusually high rate of intron loss from remaining functional plastid genes created a unique circumstance on the lineage leading to Cuscuta subgenus Grammica that allowed elimination of matK in the most species-rich lineage of Cuscuta.
Introduction
Introns within organellar genes have unique features. Unlike those in eukaryotic nuclear genes, they do not rely on spliceosomes for excision from RNA transcripts, and unlike the similarly structured self-splicing introns of prokaryotes, they typically require other trans-acting factors for efficient splicing in vivo [1], [2]. Land plant plastid genomes usually contain between 17 and 20 of these introns, all of which are classified as group II based on putative folding structure except for a single group I intron within the transfer RNA gene trnL-UAA[3]. Plastid group II introns are further subdivided structurally into two classes, group IIA and group IIB. All of these group II introns, with the exception of the second of two group II introns within clpP, seemingly trace their origin to a shared common ancestor of charophycean algae and all land plants[4].
Only one transcribed open reading frame has been identified within any plastid intron, the presumed intron maturase matK, consistently found within trnK-UUU. Although matK has been shown to be an essential factor for the splicing of the trnK intron within which it is contained[5], its involvement in the splicing of other plastid introns is poorly understood[6]. The plastid genome of the nonphotosynthetic, parasitic angiosperm Epifagus encodes only four proteins not involved in transcription or translation and lacks a functional trnK gene[7]. However, the trnK pseudogene retains a complete open reading frame for matK which is evolving under selective constraint, indicating matK is essential for other functions beyond splicing the trnK intron in that species[8]. A parallel pattern of trnK loss with retention of matK is seen in the photosynthetic streptophyte alga Zygnema circumcarinatum [4]. Various studies have shown that without translation of plastid-encoded proteins, seven group IIA introns in the plastid genome remain in an unspliced transcript form, whereas group IIB introns are largely unaffected and have been shown in maize to primarily rely upon a nuclear-encoded factor, crs2, for splicing[3], [9], [10]. An eighth group IIA intron, clpP intron 2, is present in the chloroplast genomes of most land plants but was not examined in those studies because it is not present in grasses. Excision of the only group I intron, trnL-UAA, is unaffected by any of these factors, as is splicing of the second of two group IIB introns found within ycf3 [10]. Reliance of seven group IIA introns upon a plastid-encoded factor for splicing indicates a role for matK in splicing introns other than the trnK intron within which it resides.
Like Epifagus (Orobanchaceae), members of the genus Cuscuta (Convolvulaceae) are parasitic plants that have undergone substantial gene loss from their plastid genomes [11]. However, at least some members of the genus retain a largely intact plastid genome and contain chlorophyllous tissues [12], albeit in a localized form less crucial to the parasites' survival relative to fully autotrophic plants[13]. Losses of three group IIA introns from the plastid genomes of various Cuscuta species were reported more than a decade ago [14], [15], and presence of the intron found within the 3′ locus of the trans-spliced rps12 gene was shown to be polymorphic in the genus[16]. More recently, the sequencing of four complete plastid genomes from the genus Cuscuta, two from subgenus Monogyna and two from subgenus Grammica, shows that intron content between the two subgenera differs greatly; specifically, both matK and all group IIA introns except the second intron of clpP are lost from the plastid genomes of the closely related members of subgenus Grammica [11], [17]. Intron 2 of clpP, acquired in the common ancestor of land plants millions of years after matK and the other seven plastid group IIA introns [4], was shown to be properly transcribed and translated in Cuscuta gronovii in the absence of plastid matK [17].
In this study we sampled across the taxonomic range of Cuscuta in order to ascertain the distribution of matK and plastid introns in the genus. For those taxa that still contain matK, we investigated whether or not significant changes in selective constraint occurred on branches where intron loss has occurred. Finally, we conducted similar branchXsite tests on an equal sample size of the variously parasitic family Orobanchaceae, where loss of most plastid introns is known to have occurred at least in Epifagus.
Results
Using PCR assays that gave clear positive or negative results based on band size, we surveyed for the presence of matK at the trnK-UUU locus along with all known group IIA introns and three group IIB introns (one in trnG-UCC and two within ycf3) from a variety of Cuscuta species representing all three currently recognized subgenera (Table 1). In cases of tRNA introns, we used sequence reads to confirm presence or absence of the gene and intron, as tRNA exons are generally shorter than 40 nucleotides in length.
Table 1. Intron distribution in relevant taxa.
| Taxon | Subgenus | Group IIA | Group IIB | ||||||||
| trnK-UUU | atpF | * trnV-UAC | rpl2 | 3′rps12 | trnI-GAU | trnA-UGC | clpP intron 2 | trnG-UCC | ycf3 (both) | ||
| Nicotiana tabacum | + | + | + | + | + | + | + | + | + | + | |
| Ipomoea purpurea | + | + | + | − | + | + | + | + | + | + | |
| Cuscuta exaltata | Monogyna | × | + | +/× | − | + | + | + | + | + | + |
| C. reflexa | Monogyna | × | + | +/× | − | + | + | + | + | + | + |
| C. japonica | Monogyna | × | + | +/× | − | + | + | + | + | + | + |
| C. lupuliformis | Monogyna | × | + | +/× | − | + | + | + | + | + | + |
| C. europaea | Cuscuta | × | + | × | − | + | + | + | + | × | + |
| C. epilinum | Cuscuta | × | + | × | − | + | + | + | × | × | + |
| C. nitida | Cuscuta | × | − | × | − | + | × | × | + | × | − |
| C. indecora | Grammica | × | − | × | − | − | × | × | + | × | − |
| C. umbellata | Grammica | × | − | × | − | − | × | × | + | × | − |
| C. tasmanica | Grammica | × | − | × | − | − | × | × | + | × | − |
| C. rostrata | Grammica | × | − | × | − | − | × | × | + | × | − |
| C. gronovii | Grammica | × | − | × | − | − | × | × | + | × | − |
| C. obtusiflora | Grammica | × | − | × | − | − | × | × | + | × | − |
| Epifagus | × | × | × | + | + | × | × | + | × | × | |
trnV introns in Cuscuta subg. Monogyna have deletions that may render them pseudogenes.
Intron presence or absence is shown for Nicotiana tabacum (Solanaceae), Ipomoea purpurea and Cuscuta spp. (Convolvulaceae), and Epifagus virginiana (Orobanchaceae). Nicotiana, Ipomoea, and Cuscuta are classified in the order Solanales, while Epifagus virginiana is in the closely related order Lamiales. Subgeneric taxonomic classifications are listed for Cuscuta spp. Nicotiana intron distribution is typical of most angiosperms. “+” indicates intron present, “−” indicates precise intron loss from an intact gene, and “X” indicates loss of gene (and intron) from the plastid genome. Intron data for Nicotiana, Ipomoea, Cuscuta exaltata, Cuscuta reflexa, Cuscuta gronovii, Cuscuta obtusiflora, and Epifagus were gleaned from complete genome sequences available on genbank; all other data are based on PCR and PCR sequencing assays.
Although the trnK gene itself is absent across all Cuscuta species, all sampled members of subgenus Monogyna and subgenus Cuscuta retain an open reading frame for matK, paralleling the condition in Epifagus and Zygnema. However, all sampled members of subgenus Grammica, which contains the majority of Cuscuta species, have lost matK from the plastid genome. As predicted under the hypothesis that matK is necessary for splicing of all seven group IIA introns shown to be unspliced in grass plastid translational mutants [3], [9], [10], loss of matK in Cuscuta correlates perfectly with the loss of all of those group IIA introns from the plastid genome. Representatives of subgenus Grammica still possess the group IIA intron within clpP (intron 2), five group IIB introns, and the trnL-UAA group I intron within otherwise normal genes, corroborating prior results that resident plastid matK is not necessary for the splicing of these introns [3], [5], [9], [10], [17].
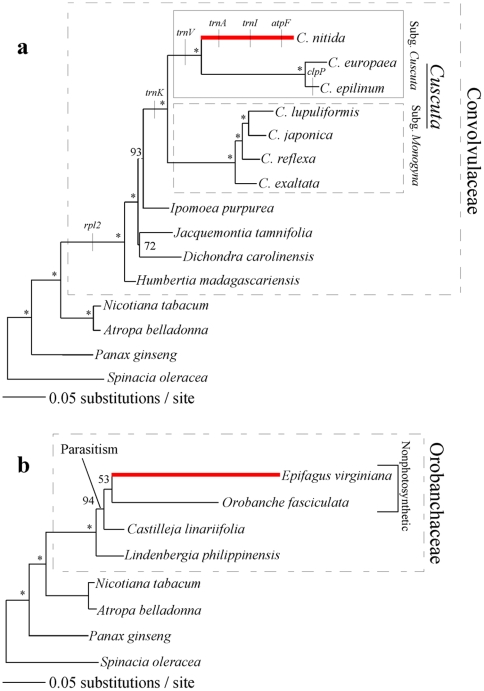
All sampled species of Cuscuta that still possess matK also possess at least four group IIA introns with the exception of Cuscuta nitida, which retains only the 3′ rps12 intron (Table 1, Fig. 1A) and intron 2 of clpP. The open reading frame of matK was partially or fully sequenced for five species in Cuscuta subgenus Monogyna, three species from subgenus Cuscuta, and four species from the otherwise autotrophic family they are derived from within, Convolvulaceae (Morning Glory Family). Using outgroup sequences from available plastid genomes, a well-supported phylogeny was constructed that agrees fully with published relationships within Convolvulaceae and Cuscuta [18], [19] (Fig. 1A). Ipomoea (tribe Convolvuleae) was strongly supported as sister to Cuscuta, although alternative hypotheses at this node could not be rejected in a previous study study[20]. Because our taxon sampling outside of Cuscuta is sparse, we conservatively chose to collapse this node as a polytomy for analyses of selective constraint. We were especially interested in changes in selective constraint within domain X, the portion of matK that has been identified as the putative RNA binding domain [21]. When per-site ratios of nonsynonymous to synonymous nucleotide substitutions (dN/dS = ω) were constrained across the phylogeny, domain X was found to be evolving under stronger purifying selection (ω = 0.21) than the remainder of the gene (ω = 0.392; Table 2, model M0). All sampled species also contained an amino acid consensus motif within domain X (SX3–6TLAXKXK) conserved across land plants and charophytes[22], further suggesting that matK remains functional among all Cuscuta that still possess it.
Figure 1. Phylogenies of Convolvulaceae (a) and Orobanchaceae (b) inferred from full and partial nucleotide sequences of matK.
Branches shown in red denote significant increases in selection within domain X of matK. Trees were produced using Maximum Likelihood with bootstrap values (100 replications) shown at the nodes (bootstrap values of 100 are denoted by asterisks). Taxonomic delimitations of Cuscuta subgenera and Convolvulaceae are boxed and labeled. Group IIA intron losses are mapped on branches where they are inferred to have occurred. The branch joining Ipomoea with Cuscuta species, denoted with a dotted line, was collapsed for analysis due to low support in other studies.
Table 2. Shifting patterns of selection on matK.
| Model (parameters) | Omega ω0, ω1, ω2 | -ln (Likelihood) | Model Comparison* | LRT statistic (df, p) |
| A. Cuscuta nitida lineage - X domain | ||||
| 1. M0 (29) | 0.21 | 1440.306 | --- | --- |
| 2. M3 (31) | 0.089, 0.681 | 1423.843 | M0 vs M3 | 32.926 (2, <<0.001) |
| 3. Discrete BranchXSites (33) | 0.067, 0.623, 4.582 | 1418.591 | M3 vs discrete bXs | 10.505 (2, 0.005) |
| 4. M1a, Nearly Neutral (30) | 0.115, 1.0 | 1425.3 | --- | --- |
| 5. M2a, Pos. Sel. (32) | 0.115, 1.0, 1.0 | 1425.3 | M1a vs M2a | 0 (2,1) |
| 6. Pos. Sel BranchXSites null§ (31) | 0.094, 1.0, 1.0 | 1421.596 | --- | --- |
| 7. Pos. Sel BranchXSites (32) | 0.094, 1.0, 4.371 | 1420.719 | M1a vs Pos. Sel. bXs | 9.161 (2, 0.01) |
| 8. | Null§ vs Pos. Sel. bXs | 1.753 (0∶1, >0.5) | ||
| B. Cuscuta nitida lineage nonX-domain regions | ||||
| 9. M0 (29) | 0.392 | 4875.979 | --- | --- |
| 10. M3 (31) | 0.202, 0.854 | 4839.185 | M0 vs M3 | 73.588 (2,<<0.001) |
| 11. Discrete BranchXSites (33) | 0.207, 0.875, 0 | 4838.633 | M3 vs Discrete bXs | 1.103 (2, 0.576) |
| 12. M1a, Nearly Neutral (30) | 0.233, 1.0 | 4839.67 | --- | --- |
| 13. M2a, Pos. Sel. (32) | 0.238, 1.0, 2.66 | 4839.206 | M1a vs M2a | 0.927 (2, 0.629) |
| 14. Pos. Sel BranchXSites null§ (32) | 0.233, 1.0, 1.0 | 4839.67 | --- | --- |
| 15. Pos. Sel BranchXSites (32) | 0.233, 1.0, 1.0 | 4839.67 | M1a vs Pos. Sel. bXs | 0 (2, 1) |
| 16. | Null§ vs Pos. Sel. bXs | 0 (0∶1, 1) | ||
| C. Epifagus virginiana lineage X-domain | ||||
| 17. M0 (16) | 0.2823 | 990.106 | --- | --- |
| 18. M3 (18) | 0.055, 0.535 | 985.246 | M0 vs M3 | 9.719 (2, 0.008) |
| 19. Discrete BranchXSites (20) | 0, 0.454, 4.077 | 980.044 | M3 vs Discrete bXs | 10.405 (2, 0.006) |
| 20. M1a, Nearly Neutral (17) | 0.204, 1.0 | 985.919 | --- | --- |
| 21. M2a, Pos. Sel. (19) | 0.17218, 1.0, 1.0 | 985.919 | M1a vs M2a | 0 |
| 22. Pos. Sel BranchXSites null§ (18) | 0.1, 1.0, 1.0 | 983.387 | --- | --- |
| 23. Pos. Sel BranchXSites (19) | 0.115, 1.0, 4.54 | 982.839 | M1a vs Pos. Sel. bXs | 6.144 (2, 0.046) |
| 24. | Null§ vs Pos. Sel. bXs | 1.097 (0∶1, >0.05) | ||
| D. Epifagus nonX-domain | ||||
| 25. M0 (16) | 0.47 | 4228.341 | --- | --- |
| 26. M3 (18) | 0.315, 1.43 | 4203.435 | M0 vs M3 | 49.812 (2, <<0.001) |
| 27. Discrete BranchXSites (20) | 0.311, 1.385, 14.885 | 4194.264 | M3 vs Discrete bXs | 18.342 (2, <0.001) |
| 28. M1a, Nearly Neutral (17) | 0.255, 1.0 | 4204.865 | --- | --- |
| 29. M2a, Pos. Sel. (19) | 0.27, 1.0, 4.773 | 4202.478 | M1a vs M2a | 4.774 (2, 0.092) |
| 30. Pos. Sel BranchXSites null§ (18) | 0.237, 1.0, 1.0 | 4203.087 | --- | --- |
| 31. Pos. Sel BranchXSites (19) | 0.261, 1.0, 14.427 | 4195.222 | M1a vs Pos. Sel. bXs | 19.286 (2, <<0.001) |
| 32. | Null § vs Pos. Sel. bXs | 15.73 (0∶1, <<0.001) | ||
Likelihood Ratio Tests indicate shifts in pattern of selection on Cuscuta nitida (Convolvulaceae) and Epifagus virginiana (Orobanchaceae) matK genes following loss of all but one (3′rps12 in Cuscuta nitida) or two (3′rps12 and rpl2 in Epifagus virginiana) group IIA introns from the plastid genome. Models and parameters are described in text. Models with significantly improved likelihoods relative to null hypothesis are shown in boldface. Clade and branch descriptions refer to relationships depicted in Figure 1.
Models M0, M1a, M2a, and M3 are described in text with branchXsites (bXs) models for M3 and M1a.
Null model 2 for positive selection is BranchXSites model constraining foreground ω2 to neutrality (ω2 = 1.0).
Significant variation in selective constraint across sites within domain X was observed when comparing nested models with a single ratio of dN/dS (M0) versus models with two or three rate ratio classes (M3; Table 2, line 2). We used fitmodel [23] to test whether changes in the pattern of among-site variation in selective constraint varied across the tree, perhaps in association with the loss of specific group II introns (Fig. 1). A Likelihood Ratio Test (LRT) did not yield significantly better support for a model that allowed switching among rate ratio classes across the tree relative to the M3 model (among-site variation in dN/dS ) without switching across the tree (p = 0.32; Fig. 1). We wanted to explore this further by focusing on the branch leading to C. nitida, which has lost all introns that are thought to be spliced by matK (see above) with the exception of the one contained in the 3′ portion of rps12. We used branchXsites models implemented in codeml [24], [25], [26] to test the a priori hypothesis that the pattern of variation in constraint among sites was different on the branch leading to C. nitida (specified as the foreground branch) than the pattern of among-site variation across the rest of the tree (background branches). This approach is analogous to the switching test implemented in fitmodel, but in this case we have an a priori hypothesis that switches among rate ratio classes are concentrated on a single branch. The alternative hypothesis for branchXsites tests of Yang, Nielson and colleagues (implemented in codeml) have fewer parameters than the unconstrained switching model implemented in fitmodel, and thus this approach may have more statistical power when one has well defined hypothesis for the branch on which switching is expected to have occurred. In fact, the branchXsites model fits the domain X data significantly better than the rates across sites model (M3) when the C. nitida branch was specified as the foreground (Tables 2, line 3, and 3, test 1). By contrast, the likelihood for discrete branchXsite model was not significantly different than the rates across sites model (M3) when residues outside of domain X were analyzed (Table 2, line 11).
Returning to analyses of domain X, two branchXsites tests were designed specifically to detect evidence of adaptive evolution[25], [26]. The first test compares the “nearly neutral” model (M1a) with codons evolving under conserved (0<ω0<1) and neutral (ω1 = 1) evolution, with a positive selection branchXsites model that includes a third, positive rate ratio class (ω2>1) for a fraction of sites evolving on the foreground branch. The second, more stringent test compares a branchXsites null model with ω2 = 1 on the foreground branch to the positive selection branchXsites model (i.e. ω2>1 on the foreground branch). In addition, codeml [27] provides a posteriori Bayes empirical Bayes (BEB) estimation of the probability that each site on the foreground branch is evolving under positive selection (ω2>1). The likelihood for the branchXsites positive selection model was significantly better than for the nearly neutral model, and adaptive evolution on the branch leading to C. nitida was strongly supported for one site, position 16 in the domain X alignment (Tables 2, line 7, and 3, test 2). However, we were unable to reject a more stringent null model (ω2 = 1; Table 2, line 8). In summary, these results indicate that loss of three of the final four group IIA introns for which matK has been implicated in splicing has resulted in relaxed or even positive selection for some codons within domain X in C. nitida.
In Epifagus one of only two remaining, putatively matK-spliced plastid group IIA introns is the same 3′ rps12 intron retained in C. nitida; the second is an intron in rpl2 that is not found in any Cuscuta species nor autotrophic relatives in Convolvulaceae[28]. Because Epifagus retains only one additional intron relative to Cuscuta nitida, we used codeml to perform LRT analyses testing whether matK may also be evolving under positive selection in Orobanchaceae, the predominantly parasitic family containing Epifagus. Although knowledge of intron distribution among members of Orobanchaceae is lacking, we gathered matK data from a range of species available on Genbank that likely differ in plastid gene and intron content from Epifagus. Orobanche fasciculata, like Epifagus, is nonphotosynthetic but is known to retain a possibly functional copy of rbcL, the large subunit of the Rubisco protein crucial to the Calvin Cycle[29]. A parasite that retains the ability to photosynthesize (Castilleja linariifolia) and a fully autotrophic sister-group to the parasites (Lindenbergia philippinensis) were also included in the analysis, and the same outgroups were used as for the Convolvulaceae tests. The phylogeny obtained for these species (Fig. 1B) was congruent with published relationships; although the branch joining the two nonphotosynthetic Orobanchaceae sensu strictu taxa, Orobanche fasciculata and Epifagus, has relatively low support in our tree, this relationship is incontrovertibly supported in all other systematic work done on Orobanchaceae to date [30], [31], [32]. As was the case with the Cuscuta/Convolvulaceae result, global dN/dS (ω in M0) was lower for domain X than for the rest of the gene (0.28 vs. 0.47), and branchXsites models with the Epifagus lineage set as the foreground were significantly better than the rates across sites models (M1a and M3; Table 2, line 18). As was also seen in the Cuscuta/Convolvulaceae analysis, positive selection was implicated when the nearly neutral model was set as the null, but not when the more stringent null model (ω2 = 1; Tables 2, lines 23 and 24, and 3, test 4) was imposed. Unlike the Cuscuta/Convolvulaceae analysis, however, both fitmodel and codeml analyses identified shifting levels of constraint across branches for some sites outside of domain X and strong evidence for positive selection on the branch leading to Epifagus (Tables 2, lines 31 and 32).
Discussion
In the evolutionary history of Cuscuta, the previously conserved RNA-binding domain of matK underwent dramatic change in selective pressure after the loss of three of the remaining four group IIA introns for which matK is involved in splicing. In Cuscuta nitida, the RNA-binding domain is evolving under less constraint than in other Cuscuta species and outgroups where multiple group IIA introns spliced by matK are still present. It is possible that constraint on domain X to remain a generalist for group IIA intron binding has been released on the branch leading to Cuscuta nitida, and matK may have subsequently specialized to specifically bind to and splice the 3′ rps12 intron. Alternatively, one of the three introns lost on the branch to Cuscuta nitida may be particularly integral to maintaining constraint on domain X. Results of the branchXsites analyses are suggestive of adaptive evolution in domain X on the Cuscuta nitida lineage, but not conclusive. While the Maximum Likelihood estimations of ω2 were >4.0 for some codons on the Cuscuta nitida lineage, we are not able to reject the hypothesis that these sites are evolving under neutrality (ω2 = 1.0; Table 2, lines 7 and 8). This may be due to insufficient statistical power.
Epifagus, which retains two group IIA introns linked to matK splicing in its plastid genome, also shows a dramatic change in selective constraint of domain X relative to related taxa. If one of the three introns lost on the branch leading to Cuscuta nitida (trnF-GAU, trnA-UGC, and atpF) is primarily responsible for constraint of domain X across streptophytes, that intron may be lost on the branch leading to Epifagus as well. As we saw with the Cuscuta analysis, the Maximum Likelihood estimations of ω were >4.0 for some codons in domain X; however, we are not able to reject the hypothesis that these sites are evolving under neutrality (ω2 = 1.0; Table 2, lines 23 and 24). Interestingly, we were able to reject neutral evolution for some sites in the amino terminal region, outside of domain X on the branch leading to Epifagus (Table 2, line 32). The sites showing significant signal for positive selection (Table 3, test 6) are moderately conserved in the pfam alignment for the matK amino terminal region (positions 224 and 277 in the complete alignment of pfam01824), but no function has been hypothesized for this portion of the matK protein.
Table 3. Results of Branch-sites analyses.
| Site Class* | 0 | 1 | 2a | 2b | Pos. Selec. Sites** (posterior prob.) |
| 1. Cuscuta nitida M3 bXs (X domain) | |||||
| Proportion of sites | 0.56562 | 0.22014 | 0.15421 | 0.06002 | |
| Background ω | 0.06745 | 0.62299 | 0.06745 | 0.62299 | |
| C. nitida lineage ω | 0.06745 | 0.62299 | 4.58222 | 4.58222 | |
| 2. Cuscuta nitida Positive Selection bXs (X domain) | |||||
| Proportion of sites | 0.62212 | 0.17038 | 0.16288 | 0.04461 | 16 E (0.981) |
| Background ω | 0.0938 | 1 | 0.0938 | 1 | |
| C. nitida lineage ω | 0.0938 | 1 | 4.37149 | 4.37149 | |
| 3. Epifagus virginiana M3 bXs (X domain) | |||||
| Proportion of site | 0.33139 | 0.37839 | 0.1355 | 0.15472 | |
| Background ω | 0 | 0.45392 | 0 | 0.45392 | |
| E. virginiana lineage ω | 0 | 0.45392 | 4.07717 | 4.07717 | |
| 4. Epifagus virginiana Positive Selection bXs (X domain) | |||||
| Proportion of site | 0.58225 | 0.16591 | 0.19599 | 0.05585 | None with pp>0.95 |
| Background ω | 0.11534 | 1 | 0.11534 | 1 | |
| E. virginiana lineage ω | 0.11534 | 1 | 4.5397 | 4.5397 | |
| 5. Epifagus virginiana M3 bXs (nonX-domain regions) | |||||
| Proportion of site | 0.75647 | 0.18138 | 0.05013 | 0.01202 | |
| Background ω | 0.31107 | 1.38574 | 0.31107 | 1.38574 | |
| E. virginiana lineage ω | 0.31107 | 1.38574 | 14.88588 | 14.88588 | |
| 6. Epifagus virginiana Positive Selection bXs (nonX-domain regions) | |||||
| Proportion | 0.65688 | 0.27813 | 0.04566 | 0.01933 | 130 P (0.968), 177 F (0.951) |
| Background ω | 0.26096 | 1 | 0.26096 | 1 | |
| E. virginiana lineage ω | 0.26096 | 1 | 14.42669 | 14.42669 | |
Foreground (Cuscuta nitida or Epifagus virginiana lineages) and background omega (ω = dN/dS) parameters for Branch-sites models with significant LRT results (Table 2).
Background and foreground values of ω0, ω1 and ω2 listed in the table.
Loss of tRNA genes is a common phenomenon in the plastid genomes of parasitic plants[33], [34], [35], and Epifagus has also lost the group IIA-containing atpF gene along with all other photosynthetic and chlororespiratory genes[7]. However, there are no cases of intron loss from functional genes in Epifagus. Although sampled members of subgenus Grammica parallel Epifagus in losing all group IIA intron-containing tRNAs, atpF and rps12 remain under purifying selection in Cuscuta despite precise intron losses from these genes. Intron 2 of clpP, a group IIA intron not linked to matK splicing, was uniquely lost by Cuscuta epilinum (Table 1); that species still retains clpP intron 1, a group IIB intron. However, the group IIB introns in ycf3 are also precisely lost from subgenus Grammica and Cuscuta nitida (Table 1), indicating a mechanism for intron loss that is not limited to group IIA introns. Intron losses from intact plastid genes are not unprecedented in land plants[14], [36], [37], but they are sporadic and rare. Such losses are much more frequent in conjugating charophycean algae, perhaps due to higher rates of homologous recombination or levels of reverse transcriptase activity[4]. Independent loss of six introns from five different functional genes in Cuscuta suggests this lineage is much more prone to purge introns from its plastid genomes than other land plants, although the mechanism for this increased rate of intron loss is unclear. Because the rpl2 intron was lost before the evolution of parasitism in Cuscuta [28], the high rate of intron loss from otherwise intact genes in Cuscuta may or may not be related to its parasitic habit.
Loss of matK from the plastid genome of Cuscuta is only possible due to a unique combination of tRNA loss related to heterotrophy and a predisposition for plastid intron loss that is otherwise unknown in land plants. This special situation provides an opportunity to test the prediction that matK is indeed required for splicing of most group IIA introns, but isn't required for the evolutionarily distinct group IIA intron 2 of clpP, group IIB introns, nor the group I intron in trnL-UAA. Since the invasion of the chloroplast genome by all group II introns other than intron 2 of clpP at least 450 million years ago, matK has performed the role of both a cis- and trans- group IIA intron-splicing element in the plastid genome. All plastid genomes retaining any of these group IIA introns in genes necessary for survival must also retain a functional copy of matK; thus, loss of matK from functional plastid genomes is expected to be rare or perhaps even nonexistent in land plants other than Cuscuta. Parallel changes in matK associated with intron loss in two independent lineages of parasitic plants indicate that reduction of generalist splicing requirements may cause the protein to undergo adaptive changes to specialize on remaining intron splicing functions. Alternatively, one of three introns lost on the branch to Cuscuta nitida and possibly also on the branch to Epifagus may be primarily responsible for the high constraint of the RNA-binding domain of matK. Investigation of these and other parasitic lineages, which have evolved as natural plastid gene and intron knockout mutants, will help further understanding of organellar intron and maturase coevolution.
Materials and Methods
Complete plastid genome sequences of Cuscuta obtusiflora, Cuscuta exaltata, and Ipomoea purpurea were used to design primers for this study, assess presence of non-group IIA introns within Cuscuta, to eliminate the possibility of gene transpositions in cases of PCR-detected intron and matK loss, and to verify the presence of only the expected loci for genes examined in this study. Genbank accession numbers and voucher numbers for sequences used for this study are shown in Table 4.
Table 4. Voucher information and matK GenBank accession numbers.
| Species | Voucher # | Genbank accession |
| Cuscuta exaltata | * | NC009963 |
| C. reflexa | # | EU330285 |
| C. japonica | # | EU330283 |
| C. lupuliformis | (PAC) JRM03.0808 | EU330284 |
| C. europaea | (PAC) JRM03.1101 | EU330282 |
| C. epilimum | (PAC) JRM03.1210a | EU330281 |
| C. nitida | * | EU330280 |
| C. indecora | (PAC) JRM03.1103 | matK absent |
| C. umbellata | * | matK absent |
| C. tasmanica | * | matK absent |
| C. rostrata | (PAC) JRM03.1001 | matK absent |
| C. obtusiflora | (PAC) JRM03.0207 | matK absent |
| Ipomoea purpurea | (PAC) JRM03.1203 | NC009808 |
| Jacquemontia tamnifolia | (MO) 00883399 | EU330286 |
| Dichondra carolinensis | # | EU330287 |
| Humbertia madagascariensis | (MO) 3854462 | EU330288 |
| Nicotiana tabacum | N/A | NC001879 |
| Atropa belladona | N/A | NC004561 |
| Epifagus virginiana | N/A | NC001568 |
| Orobanche fasciculata | N/A | AF051990 |
| Castilleja linariifolia | N/A | AF051981 |
| Lindenbergia philippinensis | N/A | AF051994 |
| Panax ginseng | N/A | NC006290 |
| Spinacia oleracea | N/A | NC002202 |
Sequences generated outside of our group are shown in bold. Specimens that lacked enough material for herbarium voucher are denoted by an asterisk (*); photographs of the dissected flowers used for identification are available upon request. Plant material or DNA from other labs where no voucher information was provided were verified by sequence identity to existing vouchered sequences on GenBank and are marked with a #.
Primer combinations to assay intron or matK presence were chosen for ease of band size interpretation on 1% agarose gels stained with ethidium bromide. PCRs for matK and plastid introns were conducted using a combination of published [30], [38], [39], [40] and newly designed primer sequences (Table 5). Most sequencing was performed on a Beckman-Coulter CEQ8000 system according to manufacturers protocol, and the remaining sequences were generated by the Pennsylvania State University Nucleic Acids Facility on an ABI 3730XL.
Table 5. Primer sequences designed for this study.
| atpF-F | 5′-ATGAAMRACGTAACCKATT-3′ |
| atpF-R | 5′-CTCTTTGTAAGGYTTGTTG-3′ |
| ycf3-F | 5′-TCAGGAGAAAAAGAGGCATT-3′ |
| ycf3-R | 5′-GCAATTTCAGAATCTCCCTGTTG-3′ |
| rrn16-endF | 5′-GTGAAGTCGTAACAAGGTAGCCG-3′ |
| rrn23-R1 | 5′-CGTCTCTGGGTGCCTAGGTATCC-3′ |
| clpP-1F | 5′-ATGCCYATTGGTGTTCCAARAG-3′ |
| clpP-C562R | 5′-CCCCTACAACATCRACAAKTCC-3′ |
| trnKConv-endF | 5′-CACTATGTATCATTTGATAACCC-3′ |
| matKConv-54F | 5′-CCTATATCCACTTMTCTTTCAGGAG-3′ |
| matKConv-783F | 5′-GTYTTTGYTAAGGATTTTMAGG-3′ |
| matKConv-801F | 5′-GGCCAACCTAGGCTTGCTCAAGG-3′ |
| matKConv-882R | 5′-TTGAAGCCAGAAKKGATTTTCC-3′ |
| matKConv-1339R | 5′-AGTTCKAGCRCAAGAAAG-3′ |
| matKConv-1423R | 5′-GTTCTTCCGACGTWAAGAATTCTTC-3′ |
| matKConv-1450F | 5′-TTTRTATCRAATAAAGTATATAC-3′ |
| trnKsubgM-F1 | 5′-GGGCGAGTATAAAGAGAGAGGG-3′ |
| matKsubgM-2R | 5′CGTTCAATAATATCAGAATCT-3′ |
| matKsubgM-3F | 5′-CGCGCTTTTTTACAAAGCTTGGG-3′ |
| matKsubgM-ex3R | 5′-CCCAAGCTTTGTAAAAAAGCGCG-3′ |
| matKsubgM-ex4F | 5′-ATCTCAGAATTTACGATCAATTC-3′ |
| matKsubgM-ex5R | 5′-TGTAGAAAGAATTGTAATAAATG-3′ |
| matKsubgM-ex6R | 5′-CGAAGCGTCTTGTACCCAGACCG-3′ |
| matKsubgC-R1 | 5′-GAATCTGAKAARTCGGYCCAACC-3′ |
| matKsubgC-R2 | 5′-CAMGATTTCCARATGAGGGGGG-3′ |
matK primers designed using sequence from subgenus Monogyna are designated by the suffix subgM, ones designed using subgenus Cuscuta sequences by subgC, and ones designed with Convolvulaceae sequences by Conv.
Separate matK phylogenies were estimated for the Cuscuta/Convolvulaceae and Epifagus/Orobanchaceae analyses. Maximum Likelihood (ML) trees were estimated in PAUP*4.0b10 [41] using GTR + gamma models with parameters estimated from the data. The ML trees were used in molecular evolutionary analyses to test for change in constraint on lineages leading to Cuscuta nitida and Epifagus. Likelihood ratio tests were applied to compare a series of nested models including equal constraint (M0 dN/dS = ω), variation in ω across sites (M3, M2a and 1a ) and distinct patterns of variation across sites on foreground and background branches (branchXsites models). Model parameter and likelihood values (Table 2 and 3) were estimated using codeml within the PAML package v.3.15 [27]; http://abacus.gene.ucl.ac.uk/software/paml.html). Foreground branches were specified as those leading to Cuscuta nitida or Epifagus in separate analyses. Sites with Bayes empirical Bayes posterior probabilities >0.95 for ω2>1.0 were estimated in codeml [25].
We also checked for switching among ω rate ratio classes across the Convolvulaceae and Orobanchaceae trees using fitmodel v0.5.2 program [42] www.cebl.auckland.ac.nz/~sguindon/fitmodel.html]. Unlike codeml, fitmodel does not specify foreground and background branches.
Acknowledgments
We would like to thank Dave Geiser, Steve Schaeffer, and Andy Stephenson for critical review of the manuscript. George Yatskievych, Daniel Austin, Julian Hibberd, Andreas Fleischmann, Peter Endress, Kim Steiner, Greg Jordan, and Todd Barkman assisted in providing plant material used in this study. We would also like to thank Tony Omeis and the Pennsylvania State University Greenhouse for assistance in growing plant material.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a National Science Foundation Doctoral Dissertation Improvement Grant to J. McNeal (DEB-0206659) and a Henry W. Popp Fellowship granted to J. McNeal through the Pennsylvania State University Department of Biology. Some sequences were generated from full plastid genome projects funded under National Science Foundation grant DEB-0120709 and under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, by the University of California, Lawrence Berkeley National Laboratory, under contract No. DE-AC02-05CH11231, and analyses were completed with funding from the National Science Foundation Assembling the Tree of Life program (DEB-0830009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lambowitz AM, Perlman PS. Involvement of aminoacyl-transfer RNA-synthetases and other proteins in group-I and group-Ii intron splicing. Trends in Biochemical Sciences. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 2.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns- a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J, Borner T, Hess WR. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Research. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biology. 2005;3 doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J, Hubschmann T, Borner T, Hess WR. Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: Support for matK as an essential splice factor. Journal of Molecular Biology. 1997;270:179–187. doi: 10.1006/jmbi.1997.1115. [DOI] [PubMed] [Google Scholar]
- 6.Liere K, Link G. RNA-binding activity of the matK protein encoded by the chloroplast trnK intron from Mustard (Sinapis alba L.). Nucleic Acids Research. 1995;23:917–921. doi: 10.1093/nar/23.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young ND, dePamphilis CW. Purifying selection detected in the plastid gene matK and flanking ribozyme regions within a group II intron of nonphotosynthetic plants. Molecular Biology and Evolution. 2000;17:1933–1941. doi: 10.1093/oxfordjournals.molbev.a026295. [DOI] [PubMed] [Google Scholar]
- 9.Hubschmann T, Hess WR, Borner T. Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Molecular Biology. 1996;30:109–123. doi: 10.1007/BF00017806. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNeal JR, Kuehl JV, Boore JL, dePamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biology. 2007;7 doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberhausen G, Valentin K, Zetsche K. Organization and sequence of photosynthetic genes from the plastid genome of the holoparasitic flowering plant Cuscuta reflexa. Molecular & General Genetics. 1992;232:154–161. doi: 10.1007/BF00299148. [DOI] [PubMed] [Google Scholar]
- 13.Hibberd JM, Bungard RA, Press MC, Jeschke WD, Scholes JD, et al. Localization of photosynthetic metabolism in the parasitic angiosperm Cuscuta reflexa. Planta. 1998;205:506–513. [Google Scholar]
- 14.Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, et al. Six independent losses of the chloroplast DNA rpl2 intron in Dicotyledons - Molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.1111/j.1558-5646.1991.tb04390.x. [DOI] [PubMed] [Google Scholar]
- 15.Bommer D, Haberhausen G, Zetsche K. A large deletion in the plastid DNA of the holoparasitic flowering plant Cuscuta reflexa concerning two ribosomal-proteins (rpl2, rpl23), one transfer-RNA (trnI) and an orf2280 homolog. Current Genetics. 1993;24:171–176. doi: 10.1007/BF00324682. [DOI] [PubMed] [Google Scholar]
- 16.Freyer R, Neckermann K, Maier RM, Kossel H. Structural and functional-analysis of plastid genomes from parasitic plants - Loss of an intron within the genus Cuscuta. Current Genetics. 1995;27:580–586. doi: 10.1007/BF00314451. [DOI] [PubMed] [Google Scholar]
- 17.Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biology. 2007;7 doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neyland R. A phylogeny inferred from large ribosomal subunit (26S) rDNA sequences suggests that Cuscuta is a derived member of Convolvulaceae. Brittonia. 2001;53:108–115. [Google Scholar]
- 19.McNeal JR, Arumuganathan K, Kuehl JV, Boore JL, dePamphilis CW. Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae). BMC Biology. 2007;5 doi: 10.1186/1741-7007-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefanovic S, Olmstead RG. Testing the phylogenetic position of a parasitic plant (Cuscuta, Convolvulaceae, Asteridae): Bayesian inference and the parametric bootstrap on data drawn from three genomes. Systematic Biology. 2004;53:384–399. doi: 10.1080/10635150490445896. [DOI] [PubMed] [Google Scholar]
- 21.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among groupII intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Research. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders ER, Karol KG, McCourt RM. Occurrence of matK in a trnK group II intron in charophyte green algae and phylogeny of the Characeae. American Journal of Botany. 2003;90:628–633. doi: 10.3732/ajb.90.4.628. [DOI] [PubMed] [Google Scholar]
- 23.Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. Modeling the site-specific variation of selection patterns along lineages. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 28.Stefanovic S, Krueger L, Olmstead RG. Monophyly of the Convolvulaceae and circumscription of their major lineages based on DNA sequences of multiple chloroplast loci. American Journal of Botany. 2002;89:1510–1522. doi: 10.3732/ajb.89.9.1510. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe AD, dePamphilis CW. The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Molecular Biology and Evolution. 1998;15:1243–1258. doi: 10.1093/oxfordjournals.molbev.a025853. [DOI] [PubMed] [Google Scholar]
- 30.Young ND, Steiner KE, dePamphilis CW. The evolution of parasitism in Scrophulariaceae/Orobanchaceae: Plastid gene sequences refute an evolutionary transition series. Annals of the Missouri Botanical Garden. 1999;86:876–893. [Google Scholar]
- 31.Olmstead RG, dePamphilis CW, Wolfe AD, Young ND, Elisons WJ, et al. Disintegration of the Scrophulariaceae. American Journal of Botany. 2001;88:348–361. [PubMed] [Google Scholar]
- 32.Bennett JR, Mathews S. Phylogeny of the parasitic plant family Orobanchaceae inferred from Phytochrome A. American Journal of Botany. 2006;93:1039–1051. doi: 10.3732/ajb.93.7.1039. [DOI] [PubMed] [Google Scholar]
- 33.Taylor GW, Wolfe KH, Morden CW, Depamphilis CW, Palmer JD. Lack of a functional plastid transfer RNA(Cys) gene is associated with loss of photosynthesis in a lineage of parasitic plants. Current Genetics. 1991;20:515–518. doi: 10.1007/BF00334780. [DOI] [PubMed] [Google Scholar]
- 34.Wimpee CF, Morgan R, Wrobel RL. Loss of transfer-RNA genes from the plastid 16S-23S ribosomal-RNA gene spacer in a parasitic plant. Current Genetics. 1992;21:417–422. doi: 10.1007/BF00351704. [DOI] [PubMed] [Google Scholar]
- 35.Lohan AJ, Wolfe KH. A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics. 1998;150:425–433. doi: 10.1093/genetics/150.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downie SR, Llanas E, KatzDownie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA's. Systematic Botany. 1996;21:135–151. [Google Scholar]
- 37.McPherson MA, Fay ME, Chase MW, Graham SW. Parallel loss of a slowly evolving intron from two closely related families in asparagales. Systematic Botany. 2004;29:296–307. [Google Scholar]
- 38.Dumolin-Lapegue S, Pemonge MH, Petit RJ. An enlarged set of consensus primers for the study of organelle DNA in plants. Molecular Ecology. 1997;6:393–397. doi: 10.1046/j.1365-294x.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 39.Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic noncoding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 40.Nickrent DL, Yan OY, Duff RJ, dePamphilis CW. Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Molecular Biology. 1997;34:717–729. doi: 10.1023/a:1005860632601. [DOI] [PubMed] [Google Scholar]
- 41.Swofford DL. PAUP*.Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2002. Version 4.0b10 ed. [Google Scholar]
- 42.Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. Modeling the site-specific variation of selection patterns along lineages. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]