Abstract
To determine whether family history of cancer may be a risk factor for the mutator phenotype in colorectal cancer, we recruited 143 consecutive colorectal cancer patients with a family history of accompanying cancers not meeting the Amsterdam criteria. Microsatellite instability (MSI) at 5 markers, hMLH1-promoter methylation, and expression of mismatch repair (MMR) proteins (hMLH1, hMSH2, hMSH6, hMPS1, and hPMS2) were determined. Among the relatives of familial colorectal cancer patients, colorectal cancer was the most common tumor type. Of the proband colorectal cancers, 26 (18.2%) showed high-level MSI (MSI-H); 47 additional tumors with mutator phenotype (32.9%) were identified by hMLH1-promoter methylation and/or loss of MMR protein expression. Mutator phenotype was associated with right-sided colon cancer and the type of accompanying cancer. Family history, which was differentially quantified according to the degree of relatives and the type of accompanying cancers, effectively discriminated MSI-H from microsatellite stable (MSS) and low-level microsatellite instability (MSI-L) and mutator phenotypes. Our findings indicate that familial colorectal cancer may be associated with multiple occurrences of colorectal or accompanying cancers and that family history could be correlated with microsatellite instability.
Keywords: Familial Colorectal Cancer, Family History, Mutator Phenotype, Ncrosatellite Instability
INTRODUCTION
A family history of colorectal cancer is an important determinant of an individual's risk for the disease. The highest degree of familial risk is provided by the dominantly inherited syndromes of colorectal cancer, hereditary non-polyposis colorectal cancer (HNPCC) and familial adenomatous polyposis (FAP), which constitute 1% to 3% of all colorectal cancers, respectively. In these syndromes, the probabilities of an untreated gene carrier developing colorectal cancers are about 80% and 100%, respectively (1, 2). An additional 15% of individuals with colorectal cancer have other affected family members, but their family histories do not fulfill the criteria for either FAP or HNPCC, and they may not appear to follow a recognizable pattern of inheritance (3). These families, categorized as having familial colorectal cancer (4), are at increased risk of developing colorectal cancer, but the familial risk, which is likely to have a genetic component, is difficult to define.
Susceptibility to HNPCC is caused by mutations in one of the genes in the DNA mismatch repair (MMR) gene system. The majority of germline mutations have been detected in hMLH1 and hMSH2, whereas germline mutations in hMSH6, hPMS1, and hPMS2 seem to be rare (5). MMR gene mutations are accompanied by a mutator phenotype, which is also caused by hMLH1-promoter methylation even if not sufficient for complete gene inactivation. Tumors arising in carriers of MMR gene mutations exhibit a characteristic phenotype termed microsatellite instability (MSI), which is characterized by alterations in the length of simple repetitive microsatellite sequences found throughout the genome. MSI frequency seems to be directly proportional to the hereditary basis of a tumor, with frequencies ranging from 15% in sporadic colorectal cancer to 85% in HNPCC, with suspected HNPCC being intermediate (6, 7).
Recent studies have demonstrated that colorectal tumors with high-level sporadic microsatellite instability (MSI-H) share several clinicopathological features with HNPCC tumors (8). In contrast to HNPCC, however, the molecular and clinical characteristics of familial colorectal cancer have not been clearly defined. We found that familial colorectal cancer can be comprehensively explained as multiple occurrences of colorectal and accompanying cancers, inherited by dominant or recessive transmission (9). The risk associated with family history varies greatly according to the age of onset of colorectal cancer in the family members, the number of affected relatives, the closeness of the genetic relationship, and whether cancers have occurred across generations (10). Various criteria have provided screening recommendations for patients with a family history of colorectal tumors (11-14). Most of these criteria, however, include a family history of colorectal cancer but omit other tumor types, and thus do not consider the frequency of their occurrence and their genetic implications.
We have formulated a familial impact value (FIV), in which the number of alleles shared between subjects was found to vary by their degree of relationship and the differential association of colorectal cancer and other accompanying cancers in an ancestry (9). This value could be determined by a simple calculation, suggesting it may be used efficiently as a risk index in familial colorectal cancer. Moreover, we found that FIV differed significantly between MSI-H and MSI-L/MSS tumors. This study was done to confirm our previous results, and to evaluate the clinical usefulness of FIV for selecting relatives of patients with familial colorectal cancer who require genetic testing.
MATERIALS AND METHODS
Patients
Of the 1250 colorectal cancer patients treated at the Asan Medical Center (Seoul, Korea) between November 2003 and June 2005, we recruited 143 consecutive patients (87 men, 56 women; mean age, 56 yr; range, 28-89 yr) with a family history of accompanying cancers. Detailed family histories were obtained through questionnaires and through interviews with patients and their relatives. The questionnaire included cancer history in first- and second-degree relatives and contained questions regarding their age at diagnosis, type of cancer, hospital at which the diagnosis had been made, current age, and current status. Patients with HNPCC meeting the Amsterdam criteria, patients with familial adenomatous polyposis, and patients with a vague family history were excluded. All solid cancers were included as accompanying cancers, except for cancers associated with viral infection, such as primary liver and uterine cervix cancers. Hospital records were used to confirm family history. Patients treated with preoperative radiotherapy were also excluded because of possible alteration of tumor DNA.
Histologically identified normal and tumor samples were freshly obtained from each patient. This study was conducted prospectively, with the approval of the Institutional Review Board for Human Research, and all patients provided written informed consent.
Detection of MSI and hMLH1-promotor methylation
MSI was determined by PCR, using primers amplifying the microsatellite markers BAT25 and BAT 26 for mononucleotide repeats and D5S346, D2S123, and D17S250 for di-nucleotide repeats (7). Tumors were scored as MSI-H (i.e., high MSI) when ≥2 markers showed instability; MSI-L (i.e., low MSI) when 1 marker showed instability; and microsatellite stable (MSS) when none of the assayed markers showed MSI.
Methylation of the hMLH1 promoter region in tumor DNA was determined by methylation-specific PCR, as described previously (15), using 1 µg of genomic DNA that had been denatured with NaOH and treated with sodium bisulfite. Primer sequences were selected to cover the region upstream of the hMLH1 promoter, i.e., nucleotides -716 to -602. Primers for unmethylated DNA were 5'-TTTTGATGTAGATGTTTTATTAGGGTTGT-3'(sense) and 5'-ACCACCTCATCATAACTACCCACA-3'(antisense), whereas those for methylated DNA were 5'-ACGTAGACGTTTTATTAGGGTCGC-3'(sense) and 5'-CCTCATCGTAACTACCCGCG-3'(antisense). Ten µL of each PCR reaction product were loaded directly onto nondenaturing 6% polyacrylamide gels, which were stained with ethidium bromide and visualized under UV illumination. DNA from the colon cancer cell line SW48, which is completely methylated at the hMLH1-promoter region, was used as a positive control, whereas DNA from normal tissue was used as a negative control.
Immunohistochemical staining
Immunohistochemical staining for hMLH1, hMSH2, hMSH6, hPMS1, and hPMS2 proteins was performed in all 143 familial colorectal cancers as described previously (9), using diluted monoclonal antibodies to hMLH1 (G168-15; BD PharMingen, San Diego, CA, U.S.A.), hMSH2 (G219-1129, BD PharMingen), hMSH6 (clone 44, BD Transduction Laboratories, San Jose, CA, U.S.A.), hPMS1 (sc-615, Santa Cruz Biotechnology Inc., Santa Cruz, CA, U.S.A.), and hPMS2 (sc-618, Santa Cruz Biotechnology). Samples were also stained with diluted monoclonal antibody to P53 (clone DO-7, Dako, Glostrup, Denmark). Normal colonic epithelium and lymphocytes, which exhibit strong nuclear staining for hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6, were used as positive controls. The percentage of positively stained cells in a sample was divided into two grades for MMR proteins (i.e., negative expression for ≤10% and positive expression for >10% nuclear staining) and into four grades for p53 (<10%, 10-<30%, 30-<50%, and ≥50%).
Calculation of FIV
The FIV score reflecting the severity of familial risk of colorectal cancer was based on the results of a meta-analysis (16) and suspected HNPCC criteria proposed by the Korean Hereditary Tumor Registry (17). Each first-degree relative with colorectal or HNPCC-associated cancer was scored as 4 points, each second-degree relative with colorectal or HNPCC-associated cancer was scored as 3 points, each first-degree relative with another accompanying cancer was scored as 2 points, and each second-degree relative with another accompanying cancer was scored as 1 point. Points were doubled for an affected relative under 50 yr of age. FIV for each patient was calculated as the sum of colorectal and accompanying cancers in each family multiplied by the relative degree. FIVt was defined as the sum of all accompanying cancers, and FIVc as the sum of HNPCC-associated cancers, according to the Amsterdam criteria.
Statistical methods
The Altman's nomogram, assuming a putative MSI-H incidence of 15-40%, was used to determine the sample size to obtain an 80% power to detect MSI-H in familial colorectal cancer. Cross-table analysis using the Fisher's exact test compared the respective mutator phenotype with MSI, hMLH1-promoter methylation, mismatch repair protein expression, and clinicopathologic variables. FIV was compared with MSI using the unpaired Student's t test. All analyses were performed using SPSS software (ver. 11, SPSS Inc., Chicago, IL, U.S.A.), and the significance level was set at 5%.
RESULTS
Characteristics of family history and clinicopathological features
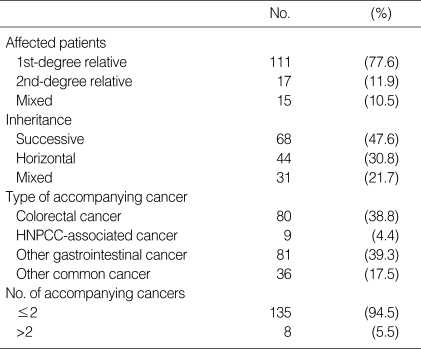
There were 206 colorectal and other cancers present in the first- and/or second-degree relatives of the 143 colorectal cancer patients in this study. The 206 cancers consisted of 80 colorectal cancers, 69 gastric cancers, 9 HNPCC-associated cancers (small bowel, urinary tract, and endometrial cancers), 12 other digestive cancers (biliary, pancreatic, and esophageal cancers), and 36 other common cancers (lung, breast, and ovarian cancers). Among the 143 probands, 111 (77.6%) had at least one affected first-degree relative, 15 (10.5%) had at least one affected second-degree relative, and 15 (10.5%) had affected first- and second-degree relatives (Table 1).
Table 1.
Family history of patients with familial colorectal cancer
Correlation of MSI status, hMLH1-promoter methylation, MMR protein expression, and mutator phenotypes with clinicopathological parameters
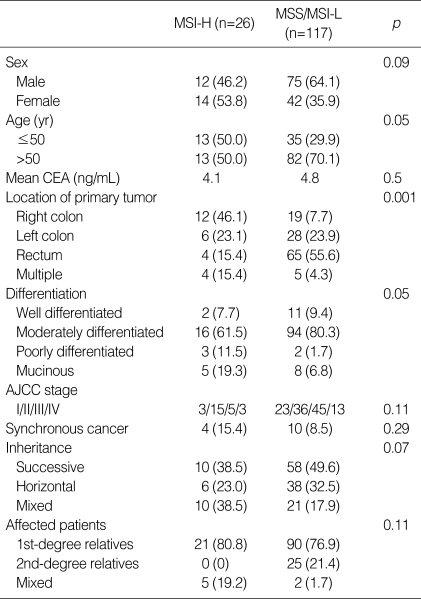
Of the 143 colorectal cancers, 26 (18.2%) were MSI-H and 3 (2.1%) were MSI-L. Younger age at onset, right-sided colon cancer, and mucinous cancer showed significant associations with MSI-H (p=0.05-0.001) (Table 2). The number of patients with MSI-H tumors was significantly higher in families affected with colorectal or HNPCC-associated cancers than in those with other common cancers (p<0.001)
Table 2.
Comparison MSI-H and MSS/MSI-L cancers (%)
MSI-H, microsatellite-high frequency; MSS, microsatellite stable; MSI-L, microsatellite-low frequency; CEA, carcinoembryonic antigen; AJCC, American Joint Committee for Cancer.
hMLH1-promoter methylation was observed in 26 patients (18.2%). Eleven of 26 MSI-H tumors (42.3%) showed hMLH1-promoter methylation compared with 15 of 117 MSS/MSI-L tumors (12.8%; p<0.001).
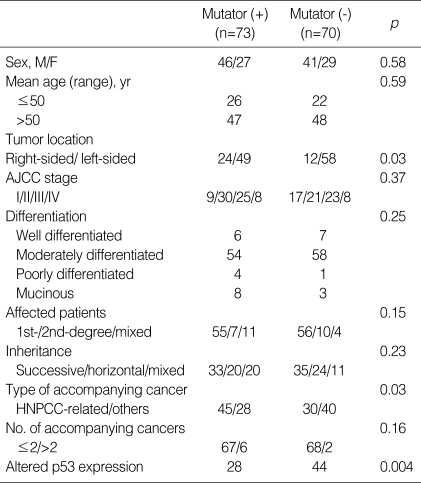
Of the 143 colorectal cancer patients, 73 (51%) exhibited mutator phenotype, defined as a tumor with MSI-H, hMLH1-promoter methylation, or loss of MMR protein expression. This phenotype showed significant associations with right-sided colon cancer, lower expression of altered P53 protein, and a greater number of accompanying HNPCC-associated cancers (Table 3).
Table 3.
Comparison of clinicopathologic characteristics regarding mutator phenotype
FIV as a function of MSI status and mutator phenotype
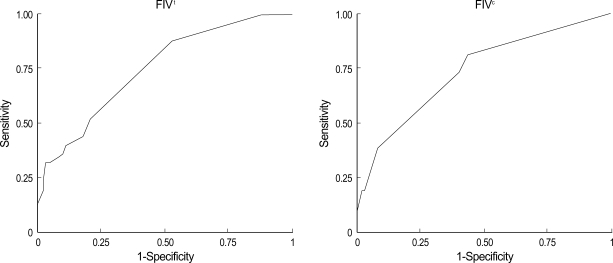
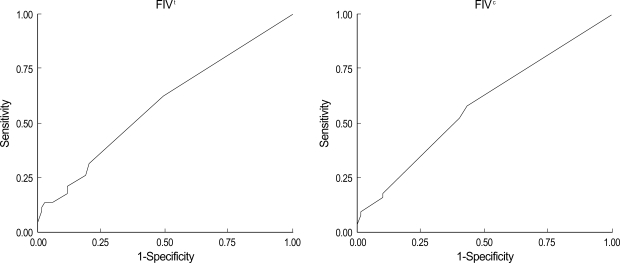
FIVt was significantly higher in patients with MSI-H (6.9±4.7) than in those with MSS/MSI-L (3.6±2.4) tumors when any categories of accompanying cancers were included (p<0.001). At a cut-off value of 3.5, FIVt showed 88.5% sensitivity and 54% specificity for MSI status, whereas FIVc showed 73% sensitivity and 60% specificity (Fig. 1). FIV was linearly correlated with the presence of the mutator phenotype, but this correlation was weaker than that with MSI status. At a cut-off value of 2.5, FIVt showed 64.4% sensitivity and 47.1% specificity for detection of mutator phenotype, whereas, at a cut-off value of 1.5, FIVc showed 57% sensitivity and 57.1% specificity (Fig. 2).
Fig. 1.
Association between the familial impact value (FIV), a quantification of family history according to the type of cancer and the relationship of the affected relatives, and MSI status on receiver operating characteristic (ROC) curves. FIVt, all accompanying cancers; FIVc, colorectal and HNPCC-associated cancers.
Fig. 2.
Association between the familial impact value (FIV) and mutator phenotype on receiver operating characteristic (ROC) curves. FIVt, all accompanying cancers; FIVc, colorectal and HNPCC-associated cancers.
Classification according to the revised Bethesda guidelines
Of our 143 patients, 84 (58.7%) met the revised Bethesda guidelines, whereas 49 (41.3%) did not (18). Patients meeting the revised Bethesda guidelines showed a higher incidence of MSI-H tumors (23.8% vs. 10.2%; p=0.04) and promoter methylation (21.4% vs. 13.6%; p=0.04) than those not meeting the guidelines. However, loss of mismatch protein expression did not differ significantly between these two groups. Mean FIVt (5.3±3.7 vs. 2.7±1.3; p=0.002) and FIVc (4.0±4.2 vs. 1.2±1.8; p<0.001) were significantly higher in patients meeting the revised Bethesda guidelines than those not meeting the guidelines.
DISCUSSION
MSI-H has been shown to have a dominant impact on the global molecular phenotype in colorectal cancer. We found that 18.2% of the tumors in this study were MSI-H, similar to the proportion in our previous study (9) and within the range of 12% to 18% reported in sporadic colorectal cancers (19, 20), but significantly lower than that reported in patients with HNPCC and suspected HNPCC (7, 21, 22). The relatively high incidence of MSI-H tumors might have been due to the higher incidence of distal cancer in Korea than in Western countries and to the high rate of rectal cancer in our patient cohort.
We found that over 70% of the accompanying cancers in first- and second-degree relatives of color cancer patients were gastric and colorectal cancers, whereas 4% were HNPCCassociated cancers. Gastric, ovarian, urinary tract, small bowel, hepatobiliary, and skin cancers have been found to be associated with HNPCC, with brain tumors and prostate cancers occasionally reported in HNPCC-kindreds (23, 24). When all accompanying cancers were included, together with colorectal and HNPCC-associated cancers, FIV showed high correlation with MSI status, suggesting that, in patients with familial colorectal cancer, most solid cancers should be considered as associated cancers.
In contrast to the Amsterdam criteria, the revised Bethesda guidelines included the variable type of HNPCC-related cancers and constituted a useful approach for identifying patients at risk for HNPCC. Introducing the MSI determination as an initial screening test for colorectal cancers enables the molecular detection of HNPCC in large populations. Both the MSI test and immunostaining have been shown to be highly effective for selecting patients who should be tested for hMSH2/hMLH1 germline mutations (25).
Although the revised Bethesda guidelines were formulated to detect HNPCC patients, they did not include many patients at risk for familial colorectal cancer. We found that 58.7% of our patients met the revised Bethesda guidelines; in patients not fulfilling these criteria, however, testing for MSI-H tumors should not be neglected. Moreover, patients not fulfilling the revised Bethesda criteria showed similar findings of immunohistochemical staining for MMR protein expression as patients fulfilling the criteria. Therefore, some patients not meeting the revised Bethesda guidelines may have familial colorectal cancer.
Of our familial colorectal cancer patients, 51% had a mutator phenotype. In HNPCC, molecular mechanisms other than the MMR pathway suggest the possibility of shared environmental carcinogens and the presence of HNPCC phenocopies exclusive of unidentified MMR alterations (26, 27). In agreement with our previous study (9), we found that the number of patients with mutator phenotype did not differ with respect to the inheritance pattern of accompanying cancers, i.e., successive or horizontal generation. These results suggest that, in familial colorectal cancer, associations among horizontal generations would be similar to those among successive generations. Although the mutator could be correlated with family history, expressed as FIV, this correlation was weaker than that between MSI and FIV, suggesting that family history should first be considered for determining the MSI status.
During the past decade the genetic etiology of all of the high-penetrance inherited colon cancer syndromes has been determined. Genetic testing to confirm the diagnosis and to test asymptomatic relatives has become a part of standard care for persons and families with these syndromes. For clinicians, the most difficult aspect of genetic testing may be to know when and whom to test. Familial clustering of colorectal cancer is generally recognized, even when cases are not part of defined genetic syndromes such as FAP and HNPCC. The strength of the relationship between colorectal cancer and family history varies according to the age at diagnosis in the index case, the type of relative, and the number of relatives affected.
Although the increased risk of colorectal cancer is associated with a family history of the disease (16), it is difficult to quantify the level of risk in a clinically meaningful way. Our use of FIV assumed that the number of alleles shared between subjects varied by the degree of relationship and the differential association with colorectal and other accompanying cancers. This value, which differed significantly between MSI-H and MSI-L/MSS tumors, could be determined by a simple calculation, suggesting it may be used as a risk index in familial colorectal cancer. The current standard for assessing for DNA mismatch repair competency is molecular MSI testing (28). Screening by family history is simpler, suggesting it may be the primary approach for identifying families at risk. Since FIV showed high sensitivity for the MSI status, it might be suitable for screening candidates for MSI testing.
One potential weakness of this study is the lack of verification of family history. To rule out any possibility of sporadic colorectal cancers, we attempted to complete a pedigree for each patient by interviewing patients and their family members. A family history of solid cancers, as recorded in medical records, is often inaccurate and may reduce the value of the scoring system. Another aspect of family history that limits its value as a risk predictor is the size of the family. Small families are less likely to show the presence of an inherited trait because there are fewer people at risk, a limitation that applies to the use of family history in any context. Here we did not record the family size, but perhaps this should be a routine part of future family history documentation. Another potential limitation to our study may arise from inaccurate results of immunohistochemical staining or methylation-specific PCR. In the present study, we found that 40.1% of non-MSH-H tumors had mutator phenotypes (47/117), and 13% of MSS/MSI-L cancers showed hypermethylation. This suggested the possibility of environmental effects might have altered the position of the amplified fragment within the hMLH1 promoter and its correlation to gene silencing. In addition, we did not perform direct hMLH1/hMSH2 germline mutation analysis. The aim of this study, however, was to evaluate the usefulness of FIV for selecting patients requiring genetic testing in familial colorectal cancer.
We found that familial colorectal cancer may be associated with multiple occurrences of colorectal and accompanying cancers. About half of the patients did not exhibit a mutator phenotype, indicating that a molecular genetic mechanism other than MMR alterations remains to be identified in familial colorectal cancer. FIV, which can be determined by simple calculations, well reflected the MSI status and may be used to identify familial colorectal cancer patients with a mutator phenotype. Even in the absence of mutator phenotype status, this information may identify familial colorectal cancer patients requiring genetic testing for MMR genes.
Footnotes
This study was supported by a Korea Research Foundation Grant (KRF-2004-014-E00063).
References
- 1.Bulow S. Results of national registration of familial adenomatous polyposis. Gut. 2003;52:742–746. doi: 10.1136/gut.52.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin KM, Shashidharan M, Thorson AG, Ternent CA, Blatchford GJ, Christensen MA, Watson P, Lemon SJ, Franklin B, Karr B, Lynch J, Lynch HT. Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg. 1998;2:67–71. doi: 10.1016/s1091-255x(98)80105-4. [DOI] [PubMed] [Google Scholar]
- 3.McGrath DR, Spigelman AD. Hereditary colorectal cancer: keeping it in the family--the bowel cancer story. Intern Med J. 2002;32:325–330. doi: 10.1046/j.1445-5994.2002.00224.x. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Rahman WM, Peltomaki P. Molecular basis and diagnostics of hereditary colorectal cancers. Ann Med. 2004;36:379–388. doi: 10.1080/07853890410018222. [DOI] [PubMed] [Google Scholar]
- 5.Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 6.Pedroni M, Tamassia MG, Percesepe A, Roncucci L, Benatti P, Lanza G, Jr, Gafa R, Di Gregorio C, Fante R, Losi L, Gallinari L, Scorcioni F, Vaccina F, Rossi G, Cesinaro AM, Ponz de Leon M. Microsatellite instability in multiple colorectal tumors. Int J Cancer. 1999;81:1–5. doi: 10.1002/(sici)1097-0215(19990331)81:1<1::aid-ijc1>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 7.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 8.Shia J, Ellis NA, Paty PB, Nash GM, Qin J, Offit K, Zhang XM, Markowitz AJ, Nafa K, Guillem JG, Wong WD, Gerald WL, Klimstra DS. Value of histopathology in predicting microsatellite instability in hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer. Am J Surg Pathol. 2003;27:1407–1417. doi: 10.1097/00000478-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Kim JC, Lee KH, Ka IH, Koo KH, Roh SA, Kim HC, Yu CS, Kim TW, Chang HM, Gong GY, Kim JS. Characterization of mutator phenotype in familial colorectal cancer patients not fulfilling Amsterdam Criteria. Clin Cancer Res. 2004;10:6159–6168. doi: 10.1158/1078-0432.CCR-04-0651. [DOI] [PubMed] [Google Scholar]
- 10.Slattery ML, Levin TR, Ma K, Goldgar D, Holubkov R, Edwards S. Family history and colorectal cancer: predictors of risk. Cancer Causes Control. 2003;14:879–887. doi: 10.1023/b:caco.0000003840.94591.76. [DOI] [PubMed] [Google Scholar]
- 11.Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J, Ganiats T, Levin T, Woolf S, Johnson D, Kirk L, Litin S, Simmang C. Gastrointestinal Consortium Panel. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology. 2003;124:544–560. doi: 10.1053/gast.2003.50044. [DOI] [PubMed] [Google Scholar]
- 12.Helm JF, Sandler RS. Colorectal cancer screening. Med Clin North Am. 1999;83:1403–1422. doi: 10.1016/s0025-7125(05)70172-3. [DOI] [PubMed] [Google Scholar]
- 13.Bradshaw N, Holloway S, Penman I, Dunlop MG, Porteous ME. Colonoscopy surveillance of individuals at risk of familial colorectal cancer. Gut. 2003;52:1748–1751. doi: 10.1136/gut.52.12.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Church JM. A scoring system for the strength of a family history of colorectal cancer. Dis Colon Rectum. 2005;48:889–896. doi: 10.1007/s10350-004-0880-9. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 16.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001;96:2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 17.Park JG, Vasen HF, Park KJ, Peltomaki P, Ponz de Leon M, Rodriguez-Bigas MA, Lubinski J, Beck NE, Bisgaard ML, Miyaki M, Wijnen JT, Baba S, Lynch HT. Suspected hereditary nonpolyposis colorectal cancer: International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) criteria and results of genetic diagnosis. Dis Colon Rectum. 1999;42:710–716. doi: 10.1007/BF02236922. [DOI] [PubMed] [Google Scholar]
- 18.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 20.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 21.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 22.Samowitz WS, Curtin K, Lin HH, Robertson MA, Schaffer D, Nichols M, Gruenthal K, Leppert MF, Slattery ML. The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology. 2001;121:830–838. doi: 10.1053/gast.2001.27996. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF, Puisieux A. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet. 1999;105:79–85. doi: 10.1007/s004399900064. [DOI] [PubMed] [Google Scholar]
- 24.Soravia C, van der Klift H, Brundler MA, Blouin JL, Wijnen J, Hutter P, Fodde R, Delozier-Blanchet C. Prostate cancer is part of the hereditary non-polyposis colorectal cancer (HNPCC) tumor spectrum. Am J Med Genet A. 2003;121:159–162. doi: 10.1002/ajmg.a.20106. [DOI] [PubMed] [Google Scholar]
- 25.Piñol V, Castells A, Andreu M, Castellví-Bel S, Alenda C, Llor X, Xicola RM, Rodríguez-Moranta F, Payá A, Jover R, Bessa X Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 26.Moslein G, Tester DJ, Lindor NM, Honchel R, Cunningham JM, French AJ, Halling KC, Schwab M, Goretzki P, Thibodeau SN. Microsatellite instability and mutation analysis of hMSH2 and hMLH1 in patients with sporadic, familial and hereditary colorectal cancer. Hum Mol Genet. 1996;5:1245–1252. doi: 10.1093/hmg/5.9.1245. [DOI] [PubMed] [Google Scholar]
- 27.Charbonnier F, Olschwang S, Wang Q, Boisson C, Martin C, Buisine MP, Puisieux A, Frebourg T. MSH2 in contrast to MLH1 and MSH6 is frequently inactivated by exonic and promoter rearrangement in hereditary nonpolyposis colorectal cancer. Cancer Res. 2002;62:848–853. [PubMed] [Google Scholar]
- 28.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]