Abstract
Horse ferricytochrome c (cyt c) undergoes exchange of one of its axial heme ligands (Met-80) for one or more non-native ligands under denaturing conditions. We have used 1H NMR spectroscopy to detect two conformations of paramagnetic cyt c with non-native heme ligation through a range of urea concentrations. One non-native form is an equilibrium unfolding intermediate observed under partially denaturing conditions and is attributed to replacement of Met-80 with one or more Lys side chains. The second non-native form, in which the native Met ligand is replaced by a His, is observed under strongly denaturing conditions. Thermodynamic analysis of these data indicates a relatively small ΔG (17 kJ/mol) for the transition from native to the Lys-ligated intermediate and a significantly larger ΔG (47 kJ/mol) for the transition from native to the His-ligated species. Although CD and fluorescence data indicate that the equilibrium unfolding of cyt c is a two-state process, these NMR results implicate an intermediate with His-Lys ligation.
The folding and unfolding of ferricytochrome c (cyt c) is strongly influenced by the binding of protein-donated ligands to its covalently attached heme. Denaturation of cyt c by concentrated guanidine hydrochloride or urea at neutral pH leads to replacement of the native Met-80 heme axial ligand with one or more protein-donated strong-field ligands, whereas the native His-18 heme ligand remains coordinated to the heme (1). Non-native heme ligation in unfolded cyt c has been attributed primarily to His side chains (1, 2), and such bis-His species have been shown to act as kinetic traps of cyt c folding (3–7). Recently, evidence for ligation of the N-terminal amino group to the heme in unfolded yeast cyt c has been reported (8). Despite the significant amount of data on the relationship between heme ligation and cyt c folding, the description of heme ligation in denatured cyt c is not yet complete. Results from kinetics (6, 7) and pH titration studies (8) on cyt c variants have led to the proposal that Lys may act as a heme ligand in denatured cyt c, although such species have not been observed directly. Here, we report the 1H NMR detection of a paramagnetic horse cyt c species with His-Lys heme ligation under partially denaturing conditions at neutral pH. In addition, we have directly detected a form of cyt c with bis-His heme ligation present under strongly denaturing conditions. Despite the coincidence of CD and fluorescence equilibrium denaturation curves for cyt c (9–11), NMR spectroscopy implicates an intermediate with His-Lys heme ligation in equilibrium unfolding.
The complex, heterogeneous nature of unfolded and partially unfolded proteins makes their characterization technically challenging. Cyt c is an attractive subject for unfolding studies because it has a covalently attached heme group (Fig. 1) that allows the use of a number of spectroscopic techniques to monitor its folding and unfolding both at equilibrium and in real time (1–15). Optimally, to fully characterize the folding reaction of cyt c, NMR spectroscopy would be used to perform a more detailed analysis of denatured cyt c. The poor chemical shift dispersion shown in NMR spectra of denatured proteins, however, makes characterization of such states difficult (16). Isotope exchange (17, 18) and nuclear relaxation (19) studies have provided valuable information on structural fluctuations under partially denaturing conditions, but do not allow direct observation of structural contacts in unfolded states. The use of heteronuclear NMR techniques to obtain information on non-native protein structures has been a significant advance. Unfortunately, the poor chemical shift dispersion shown by 1H and 13C nuclei in denatured and partially denatured proteins impedes the detection of structural contacts (16). In our investigation of non-native conformations of cyt c, we have chosen to target paramagnetic forms of denatured cyt c for NMR analysis. 1H NMR spectra of denatured and partially denatured paramagnetic cyt c display well-resolved resonances for heme substituents. The chemical shifts of these substituents provide information about the heme ligation state, even when such conformations make up a small fraction of the sample. These results also point to the possibility of using NMR to further characterize the heme environment of denatured cyt c through detection of nuclear Overhauser effects (NOEs) involving well-resolved hyperfine-shifted resonances.
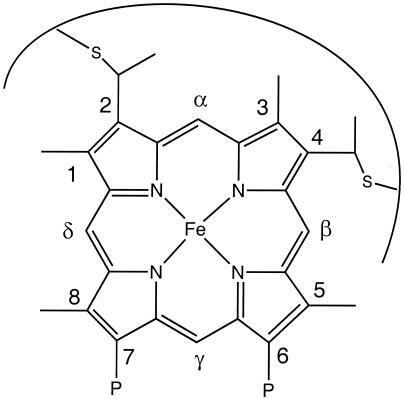
Figure 1.
Structure of c-type heme group and Fischer nomenclature used in the text. The covalent thioether linkages are shown at positions 2 and 4. P denotes propionate group.
Materials and Methods
Preparation of Cyt c and Urea Solutions.
Horse heart cyt c was purchased from Sigma, oxidized with K[Co(EDTA)] prepared by a literature method (20), and purified by FPLC using a Mono S column (Amersham Pharmacia). Cyt c concentration was determined by using an extinction coefficient of 106,100 liters/mol-cm at 410 nm (21). Solutions of urea and urea-d4 (highest quality available, Aldrich) were prepared in 50 mM sodium phosphate in H2O (with 10% D2O added for lock) or D2O, stored at 4°C, and used within 12 h. The uncorrected pH (pH*) of urea solutions was adjusted to 7.0, and the concentration of denaturant was determined by refractive index measurement (22). Urea and protein solutions were combined to give the desired final concentrations, followed by adjustment of pH* to 7.0. Data collection commenced within 6 h of sample preparation. Under the conditions used in these experiments, the amount of degradation of urea to give ammonium cyanate under the conditions used is expected to be negligible (23). The cyanide derivative of horse heart myoglobin (CN-myoglobin) was prepared by addition of a slight excess of sodium cyanide to 3 mM myoglobin (Sigma) in 50 mM phosphate buffer in D2O, pH* 7.0. The pH* was readjusted to 7.0 after mixing, and the Soret band shift to 422 nm confirmed complete cyanide binding. The concentration was measured by using an extinction coefficient of 116,000 liters/mol-cm at 422 nm (24).
Chemical Modification of Cyt c.
To convert all Lys residues in cyt c to homoarginine, a 0.8 mM cyt c solution was incubated with 0.5 M o-methylisourea hydrogen sulfate (Aldrich) for 120 h at pH 11.0, 4°C, according to a literature preparation (25). Modified protein was purified by gel filtration on Sephadex G-25 (Amersham Pharmacia). Amino acid analysis (University of Rochester Microchemical Protein Peptide Core Facility) confirmed that all 19 Lys residues were modified. To modify His residues, chloro(2,2′:6′,2"-terpyridine)platinum(II) chloride ([Pt(trpy)Cl]Cl) was prepared according to a standard literature preparation (26). A sample of 2 mM cyt c was incubated with equimolar [Pt(trpy)Cl]Cl for 72 h in 0.1 M sodium acetate, pH 5.0, 22°C, followed by chromatography on CM Sepharose (Amersham Pharmacia) as described in detail (27). UV-vis spectra of fractions collected from the CM Sepharose column indicated that unmodified, singly modified, and doubly modified products eluted as expected (27). The presence of a 342-nm band indicated modification by the reagent, and its ratio to the protein Soret band indicated whether the fraction collected contained singly or doubly modified protein (27). The fraction containing the doubly modified protein, bis-(2,2′:6′,2"-terpyridine)platinum(II)-ferricytochrome c ([Pt(trpy)]2cyt c), was collected and used in NMR experiments.
Collection and Analysis of 1H NMR Spectra.
1H NMR spectra were collected on a Varian 500-MHz spectrometer at 30°C, 45°C, or 55°C with presaturation of the residual solvent residue and repetition rates of 4 s-1 or 0.5 s-1. One-dimensional (1D) NOE difference spectra were collected by using a 3 s-1 repetition rate at 30°C and 45°C. Difference spectra were obtained by alternately collecting eight scans with the decoupler applied to the peak of interest and a reference point in the spectrum, followed by subtraction of the resulting free induction decays (FIDs). Magnetization transfer experiments were performed with the same pulse sequence as the 1D NOE but at 55°C. Data were processed by using felix 97 (Molecular Simulations). Exponential multiplication (20–50 Hz) was applied to FIDs before Fourier transform to enhance detection of broad resonances. Chemical shifts were referenced to 3-(trimethylsilyl)-1-propanesulfonic acid (DSS) via the residual 1H2HO or H2O signal. All samples used for integration had a 1.5 mM protein concentration. Integration was performed by using 3 mM CN-myoglobin as an external standard in a coaxial insert. Integrals of native and non-native heme methyl peaks in varying concentrations of urea-d4 were related to the integrals of the native heme methyl peaks in the absence of urea via the resolved CN-myoglobin heme 5-CH3 peak (28). Measurement at each urea concentration was repeated at least three times, and the results were averaged.
Results and Discussion
Detection of Non-Native Heme Resonances in the Presence of Urea.
The chemical shifts of the substituents of low-spin ferric hemes (S = 1/2) are extremely sensitive to heme axial ligation. In native cyt c, the interaction of the axial ligands (His-18 and Met-80) with the iron is the primary factor that determines the asymmetry of unpaired electron spin density on the heme, resulting in the characteristic chemical shift pattern observed for the four heme methyl groups (29–32). Replacing Met-80 with other ligands such as a lysine (as in the alkaline form of cyt c, ref. 33), cyanide (34), imidazole (35), or pyridine (36) causes the heme resonance shift pattern to change entirely (Table 1). Here, the relationship between heme substituent shifts and heme ligation is used to probe cyt c heme ligation in the presence of denaturant.
Table 1.
Chemical shifts for selected heme substituents of cyt c derivatives
| Cyt c derivative | 8-CH3 | 5-CH3 | 3-CH3 | 1-CH3 | δ-meso | Temp, °C | Ref. |
|---|---|---|---|---|---|---|---|
| Native cyt c* | 34.5 | 10.2 | 31.7 | 7.2 | 2.7 | 30 | (32) |
| Species L | 23.9 | 22.2 | 11.9 | 13.2 | −5.6 | 30 | tw |
| Alkaline horse cyt c† | 23.4 | 22.1 | 12.9 | 13.2 | −5.4‡ | 30 | (33) |
| CN-Met80Ala cyt c | 22.5 | 19.5 | 11.3 | 15.4 | −4.3 | 30 | (34) |
| Species H | 25.7 | 18.6§ | 13.5 | nd | nd | 30 | tw |
| Im-cyt c | 24.2 | 14.1 | 16.7 | 10.7 | nr | 46 | (35) |
| py-cyt c | 28.6 | 20.5 | 17.2 | 14.7 | nr | 32 | (36) |
nd, not detected; nr, not reported; tw, this work.
Shifts measured at 30°C. The shifts of 1-CH3 and δ-meso were measured from cross peaks between δ-meso/8-CH3, and δ-meso/1-CH3 in a two-dimensional NOE spectroscopy spectrum (R.M. and K.L.B., unpublished work).
Assignments for one of the two conformers. Chemical shifts measured for horse cyt c in 50 mM sodium phosphate, pH 10.5.
δ-meso for the alkaline form was assigned by 1D NOE (R.M. and K.L.B., unpublished work).
Tentative assignment.
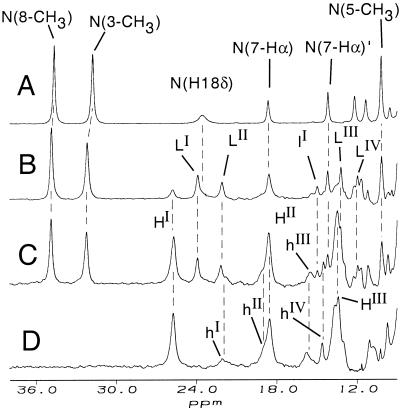
Upon subjecting horse cyt c to partially denaturing conditions [3–8 M urea-d4, 50 mM sodium phosphate, D2O, pH* of 7.0, 30°C (the midpoint for unfolding using CD spectroscopy at this temperature is 7.2 M urea, R.M., unpublished results)] well-resolved non-native resonances appear at the expense of those caused by the native protein (Fig. 2 B–D). Upon fully denaturing the protein by exposure to > 9 M urea-d4, only selected non-native peaks (i.e., at 25.7 and 18.6 ppm) remain whereas other non-native (i.e., at 23.9 and 22.2 ppm) and native resonances are no longer observed (Fig. 2D). The species that gives rise to the non-native resonances present in moderate but not in high concentrations of urea-d4 will be referred to as species L. At a lower urea concentration and higher temperature (5.4 M urea at 55°C; Fig. 3A) species L is highly populated, with well-resolved resonances. Four of these resonances (23.9, 22.2, 13.2, and 11.9 ppm at 30°C) have intensities of three relative to the others (−5.6 and 14.9 ppm), and thus are attributed to heme methyl groups and are labeled LI, LII, LIII, and LIV. Species L resonances with relative intensity one are labeled with a lowercase l (lI, lII) in the figures. The species that persists in > 9 M urea-d4 we denote species H, and its resonances of intensity 3 and 1 are labeled in an analogous fashion (HI, HII, hI, hII, etc.) The spectra observed in all concentrations of urea-d4 did not change over 1 week, indicating that the sample was at equilibrium. Reversibility of unfolding was verified by the observation that the spectrum of cyt c refolded by removal of urea is indistinguishable from that of native.
Figure 2.
500-MHz 1H NMR spectra (50 mM sodium phosphate, 90% D2O/10% H2O, pH* 7.0, 30°C) of 2 mM horse cyt c and (A) no denaturant added (native state), (B) 6.3 M urea-d4, (C) 7.0 M urea-d4, and (D) 9.2 M urea-d4. Resonances attributed to native (His-Met ligated) cyt c are labeled N(assignment) in A. Well-resolved resonances with relative intensity of three attributed to the non-native species L (or H) are labeled LI, LII, etc. (or HI, HII, etc.). Resolved non-native resonances with relative intensity of one are labeled with lowercase letters (hI or lI, etc.).
Figure 3.
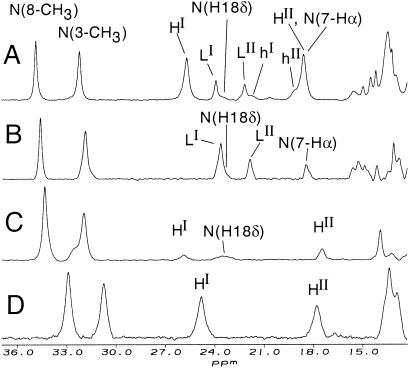
Assignment of selected 1H NMR (500 MHz) resonances for species L. Spectra are of 4 mM cyt c, 50 mM sodium phosphate, pH* 7.0. (A) Reference 1H NMR spectrum, 90% H2O/10% D2O, 55°C, 5.4 M urea. (B and C) Difference spectra showing saturation transfer observed (90% H2O/10% D2O, 55°C, 5.4 M urea) upon irradiating the native protein resonances at (B) 32.1 ppm (native 8-CH3) and (C) 29.8 ppm (native 3-CH3), allowing assignment of LI and LIV in species L to heme 8-CH3 and 3-CH3, respectively. (D) Reference spectrum, 90% D2O/10% H2O, 40°C, 5.5 M urea-d4. (E) 1D NOE difference spectrum (90% D2O/10% H2O, 40°C, 5.5 M urea-d4) showing the NOEs observed upon saturating the δ-meso resonance (lII).
Assignment of Non-Native Heme Resonances.
The resonances attributed to non-native species L are detected under partially denaturing conditions but not under strongly denaturing conditions. This observation indicates that species L is an equilibrium unfolding intermediate. The significant change in hyperfine shifts from native suggests that species L has non-native heme ligation. To aid in determining its heme ligation state, assignments of the heme methyl resonances for species L were made through 1D NOE and magnetization transfer experiments (Fig. 3). Magnetization transfer from the well-resolved heme 8-CH3 and 3-CH3 resonances of native cyt c at 55°C allows assignment of resonances LI and LIV to 8-CH3 and 3-CH3 of the non-native form L, respectively (Fig. 3 B and C). Irradiation of an upfield one-proton non-native resonance (lII, at −5.6 ppm at 30°C) results in strong NOEs of similar intensity for two of the heme methyls, LI and LIII (Fig. 3E). As LI already has been assigned to heme 8-CH3, this allows assignment of LIII to heme 1-CH3 and of lII to the heme δ-meso. The remaining heme methyl, LII, then can be assigned to heme 5-CH3.
Resonances for species H were assigned by saturation transfer experiments performed on partially denatured modified protein in which all 19 Lys residues were converted to homoarginine (Har) to give guanidinated cyt c [(Lys19→Har19)-cyt c]. This derivative exhibits a simplified NMR spectrum under partially denaturing conditions, which improved quality of saturation transfer data [preparation and further studies of (Lys19→Har19)-cyt c are discussed below]. Irradiation of resonance HI in this derivative results in saturation transfer to native heme 8-CH3, allowing assignment of HI. Assignment of HIII was made through observation of weak saturation transfer upon irradiating native methyl 3-CH3. The saturation transfer experiment aided in the detection of HIII at 13.5 ppm (3-CH3 for species H) which is not well resolved in 1D spectra. Irradiating resonance HII did not yield reliable saturation transfer results because of overlap with the native heme propionate resonance (7-Hα). The remaining heme methyl group (HIV) is not resolved for species H. Although resonance HII could not be assigned by saturation transfer, the observation that heme 5-CH3 shifts are always larger than heme 1-CH3 shifts for S = 1/2 ferric heme proteins (30) allows its tentative assignment to 5-CH3.
Relationship Between Heme Resonance Assignments and Heme Ligation.
Low-spin ferric heme proteins with similar type and orientation of axial ligands have similar heme methyl shift patterns (Table 1). This is attributed to the dominant role played by the heme axial ligands in determining the unpaired electron spin density pattern on the heme and, thus, heme substituent contact shifts (29–31). In cyt c, the proximity of the native His-18 ligand to the covalent Cys thioether linkages to the heme (Cys-14 and Cys-17) causes it to remain strongly bound to the heme iron upon denaturation at neutral pH. In the cyt c heme octapeptide (MP-8), which consists of the heme and residues 14–21, it has been determined by NMR that the native His-18 remains bound at an angle nearly the same as in native cyt c despite the absence of most of the polypeptide (37). Changes in heme substituent shifts for cyt c, therefore, are attributed to replacement of Met-80 with non-native ligands, whereas the His-18 maintains its native orientation. The pattern of heme methyl shifts for species L bears a striking resemblance to that observed for the alkaline form of cyt c, known to have two conformers in which the axial Met ligand is replaced with Lys side chains (Table 1) (33, 38). It also bears a strong similarity to the cyanide adducts of Met-80–Ala cyt c (34) and MP-8 (37). Species L, alkaline cyt c, and these cyt c derivatives all have heme methyl shifts ranging from 25 to 10 ppm with an order of 8-CH3 > 5-CH3 > 1-CH3 > 3-CH3, as well as an upfield-shifted heme δ-meso resonance (Table 1) (32–37). Its similarities with the NMR properties of these characterized paramagnetic proteins indicate that the cyt c unfolding intermediate in urea is ligated by the native His-18 and a second ligand with cylindrical symmetry. The only such available donor in horse cyt c that would give a low-spin heme is an amino group. Because the N terminus of horse cyt c is acetylated (39), the remaining candidate is a deprotonated Lys side chain. The heme in species L thus is proposed to be ligated by the native His-18 and a deprotonated Lys.
As bis-His heme ligation has previously been established in denatured cyt c (2), His is expected to be the non-native heme ligand in species H, which persists under strongly denaturing conditions. The assignments (HI and HIII to 8-CH3 and 3-CH3) and proposed assignment (HII to 5-CH3) for the heme methyl resonances are indeed similar to those observed for the pyridine (36) and imidazole (35) derivatives of cyt c (Table 1). Note that the ordering of 5-CH3 and 3-CH3 shifts in imidazole-cyt c is reversed from that observed for the pyridine derivative and proposed for species H. This is consistent with small differences in the angle at which these planar ligands bind with respect to the heme in-plane axes (30).
Investigations of Chemically Modified Derivatives.
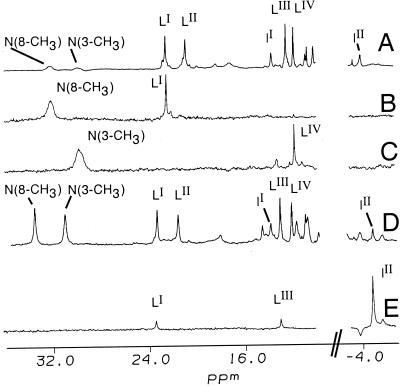
To test the proposal that species H and L observed by NMR have bis-His and His-Lys heme ligation, respectively, derivatives of cyt c were prepared in which His or Lys residues were modified to render them incapable of binding the heme. To make a variant in which non-native His are prevented from acting as heme ligands, His-26 and His-33 both were modified with chloro(2,2′:6′,2"-terpyridine)platinum(II) chloride ([Pt(trpy)Cl]Cl) to give [Pt(trpy)]2cyt c (bis-(2,2′:6′,2"-terpyridine)platinum(II)-cyt c) (27). The 1H NMR chemical shifts of the heme substituents for [Pt(trpy)Cl]2cyt c are similar to those observed for native, indicating that the protein's structure is not significantly perturbed by modification. The platinum-modified protein, when partially denatured by urea, shows hyperfine-shifted peaks LI and LII (Fig. 4B), supporting the proposal that species L does not have bis-His ligation. In addition, resonances attributed to species H are absent for partially denatured [Pt(trpy)Cl]2cyt c, supporting the assignment of species H to a bis-His species.
Figure 4.
500-MHz 1H NMR spectra (50 mM sodium phosphate, 90% D2O/10% H2O, pH* 7.0) of (A) cyt c in 7.0 M urea-d4, 30°C. (B) Bis-(2,2′:6′,2"-terpyridine)platinum(II)-ferricytochrome c ([Pt(trpy)]2cyt c) in 7.2 M urea-d4, 30°C. (C) (Lys19→Har19)-cyt c in 7.0 M urea-d4, 30°C. (D) (Lys19→Har19)-cyt c in 7.0 M urea-d4, 45°C. Raising the temperature to 45°C in D increases population of species H relative to native.
To prepare a cyt c variant lacking Lys side chains, all 19 Lys residues in cyt c were converted to Har by chemical guanidination to give (Lys19→Har19)-cyt c (25). Native cyt c and (Lys19→Har19)-cyt c have nearly the same heme resonance shifts, verifying that guanidination does not significantly perturb the protein fold. Upon partial urea denaturation, this variant gives rise to resonances attributed to species H but not to species L, supporting the assignment of His-Lys heme ligation to species L (Fig. 4 C and D). The NMR spectra of both [Pt(trpy)Cl]2cyt c and (Lys19→Har19)-cyt c were examined over a range of urea concentrations (5.0–7.4 M) and temperatures (20–55°C) to verify that the inability to observe species H (for [Pt(trpy)Cl]2cyt c) or species L [for (Lys19→Har19)-cyt c] is not a result of a perturbation of the energetics of the conformational change for the variant as compared with native.
Thermodynamic Analysis.
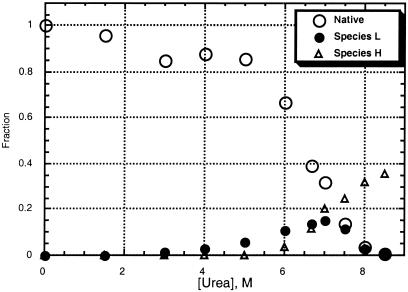
The fractional populations of each of the observable conformations (native, species L, and species H) were measured over a range of urea-d4 concentrations by integration of resolved heme methyl resonances (Fig. 5; for details, see Materials and Methods). Note that the sum of the measured fractional populations is clearly less than one at high urea concentrations. This indicates that there is at least one unobservable species (species O, for “other”) present that is substantially populated under denaturing conditions. As denatured cyt c previously has been shown to be low-spin with bis-His heme ligation (1–3) and no resonances indicative of high-spin species were observed in these experiments, we propose that species O is a bis-His-ligated species in which there is substantial disorder in the angle at which the non-native His binds the heme, leading to lack of observable hyperfine-shifted resonances.
Figure 5.
Plot of fractional populations of native protein, species L, and species H, as a function of urea-d4 concentration.
The possibility that species O and the other species present here consist of protein dimers or oligomers was considered. The shifts of native protein, species L, and species H were found to be independent of protein concentration (0.2–4.0 mM protein in 7 or 8 M urea-d4). In addition, the fractional populations of species H and L do not increase with increasing protein concentration. These observations indicate that species H and L are monomeric. However, the fractional population of species O does increase with increasing protein concentration (for concentrations above 1.5 mM), indicating that it consists at least in part of protein dimers or oligomers. For the protein concentrations used in NMR studies (1.5 mM), small angle x-ray scattering results suggest that the amount of aggregated protein present should be minimal (40). In addition, our data show no significant increase in the fraction of species O (fO) between 0.2 and 1.5 mM protein in 7 or 8 M urea, suggesting minimal formation of aggregates in 1.5 mM samples. The fractional population of species O was found to be 0.28, 0.29, and 0.59 for 0.2 mM, 1.5 mM, and 4.0 mM cyt c in 8 M urea (B.S.R., unpublished observations).
The fractional populations determined through integration of NMR resonances (native, species L, and species H) were fit to a four-state thermodynamic model. The population of species O was defined as fO = 1 − fN − fL − fH (fN, fL, and fH are the measured populations of native, species L, and species H, respectively). The fractional populations can be expressed in terms of the free energies of the transitions from the native state to each of the others (L, H, and O), where ΔGXD is the free energy change for a transition from the native state to state X at a given denaturant concentration. The dependence of the free energy on urea concentration for each state is assumed to be linear (ΔGXD = ΔGXH2O − mX[urea]) (41), and the data were fit by linear least squares to extract mX values and ΔGXH2O (ΔGX in the absence of denaturant). The resulting values of ΔGL and mL are 17 kJ/mol and 2.2 kJ/(mol-M), and the values obtained for ΔGH and mH are 47 kJ/mol and 6.5 kJ/(mol-M).
Examination of Fig. 5 clearly reveals that species L is an equilibrium unfolding intermediate. The detection of an intermediate is consistent with previous studies using small angle x-ray scattering and UV-vis spectroscopy. At least one intermediate state for equilibrium cyt c unfolding by guanidine hydrochloride was reported by researchers using small angle x-ray scattering (40). Others have noted that loss of the 695-nm band, attributed to a Met-80 sulfur → Fe(III) charge transfer, slightly precedes loss of CD signal upon guanidine hydrochloride denaturation and that this may indicate deviation from two-state unfolding because of ligand replacement (42). Species L also may serve as an intermediate on the kinetic folding pathway; recent results implicate a Lys-ligated intermediate in the slow folding of cobalt-substituted cyt c (7). The small values of ΔGL and mL suggest that species L has a structure quite similar to that of the native protein (41); this is consistent with the lack of evidence for an intermediate in CD studies. Replacement of Met-80 by one or more nearby Lys (Lys-72, -73, or -79) could be achieved without substantial disruption to the overall protein fold. Interestingly, the value for ΔGL is similar in magnitude to that measured for a low-energy local unfolding of cyt c in low concentrations of guanidine hydrochloride by isotope exchange studies (25.1 kJ/mol) (17). This local unfolding was attributed to unfolding of the small loop consisting of residues 70–85 (i.e., including Met-80 and three Lys). Experiments designed to identify sequence specifically the Lys ligand(s) in species L are in progress.
The high intrinsic pKa of Lys (10.8) makes it an unlikely heme ligand at neutral pH. The Lys residues proposed as candidates for ligation to the heme iron in species L are the same as those proposed to bind heme in the alkaline form of the protein, which forms with a pK of 9 (43). In the alkaline form of Saccharomyces cerevisiae iso-1-cytochrome c, Lys-73 and -79 have been identified as the non-native heme ligands (38). By analogy, Lys-73 and -79 (as well as Lys-72, which is trimethylated in S. cerevisiae cyt c but not in horse cyt c) are candidates for heme ligands in the alkaline form of horse cyt c. Given that the pK for the formation of the alkaline form is substantially lower than that for the side chain of a Lys, it has been proposed that deprotonation of another group (yet to be identified) acts to trigger a conformational change that leads to loss of Met ligation and introduction of the Lys into the heme vicinity. The formation of the strong Lys-Fe bond may then drive Lys deprotonation (43). The lowering of the pK of the alkaline transition to values as low as 7.2 by mutating the heme pocket residue Phe-82 indicates that perturbation of heme pocket structure may drive displacement of Met by Lys, even near neutral pH (44). We propose that a similar conformational change occurs for cyt c under mildly denaturing conditions.
NMR analysis of denatured cyt c has allowed the direct detection of a form of cyt c with bis-His heme ligation. The ΔGH measured here (47 kJ/mol) is similar to the ΔG determined for the global unfolding reaction by isotope exchange (53.6 kJ/mol) (17). The presence of observable hyperfine-shifted peaks for species H indicates that the heme ligation geometry is well defined in this species. In other words, species H represents a denatured form of cyt c with a well-defined non-native structural (heme-ligand) contact. NMR analysis of the urea denaturation of cyt c thus has shown that it involves at least four states, three of which (native, species L, and species H) have well-defined heme ligation geometry.
Acknowledgments
We thank Andrew Vetter for his assistance with CD and NMR experiments and James McGarrah for assistance in preparing [Pt(trpy)Cl]Cl. This work was supported by National Science Foundation-Research Experience for Undergraduates Grant CHE-9619935 and the University of Rochester.
Abbreviations
- cyt c
ferricytochrome c
- Har
homoarginine
- (Lys19→Har19)-cyt c
guanidinated cyt c
- NOE
nuclear Overhauser effect
- pH*
uncorrected pH
- 1D
one-dimensional
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150239397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150239397
References
- 1.Babul J, Stellwagen E. Biopolymers. 1971;10:2359–2361. doi: 10.1002/bip.360101125. [DOI] [PubMed] [Google Scholar]
- 2.Muthukrishnan K, Nall B T. Biochemistry. 1991;30:4706–4710. doi: 10.1021/bi00233a010. [DOI] [PubMed] [Google Scholar]
- 3.Elöve G A, Bhuyan A K, Roder H. Biochemistry. 1994;33:6925–6935. doi: 10.1021/bi00188a023. [DOI] [PubMed] [Google Scholar]
- 4.Sosnick T R, Mayne L, Hiller R, Englander S W. Nat Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi S, Yeh S-R, Das T K, Chan C-K, Gottfried D S, Rousseau D L. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 6.Colón W, Wakem L P, Sherman F, Roder H. Biochemistry. 1997;36:12535–12541. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 7.Tezcan F A, Winkler J R, Gray H B. J Am Chem Soc. 1999;121:11918–11919. [Google Scholar]
- 8.Hammack B, Godbole S, Bowler B E. J Mol Biol. 1998;275:719–724. doi: 10.1006/jmbi.1997.1493. [DOI] [PubMed] [Google Scholar]
- 9.Chan C-K, Hu Y, Takahashi S, Rousseau D L, Eaton W A, Hofrichter J. Proc Natl Acad Sci USA. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamada D, Kuroda Y, Kataoka M, Aimoto S, Yoshimura T, Goto Y. J Mol Biol. 1996;256:172–186. doi: 10.1006/jmbi.1996.0075. [DOI] [PubMed] [Google Scholar]
- 11.Myer Y P, MacDonald L H, Verma B C, Pande A. Biochemistry. 1980;19:199–207. doi: 10.1021/bi00542a030. [DOI] [PubMed] [Google Scholar]
- 12.Shastry M C R, Sauder J R, Roder H. Acc Chem Res. 1998;31:717–725. [Google Scholar]
- 13.Yeh S-R, Han S, Rousseau D L. Acc Chem Res. 1998;31:727–736. [Google Scholar]
- 14.Englander S W, Sosnick T R, Mayne L C, Shtilerman M, Qi P X, Bai Y. Acc Chem Res. 1998;31:737–744. [Google Scholar]
- 15.Telford J R, Wittung-Stafshede P, Gray H B, Winkler J R. Acc Chem Res. 1998;31:755–763. [Google Scholar]
- 16.Yao J, Dyson H J, Wright P E. FEBS Lett. 1997;419:285–289. doi: 10.1016/s0014-5793(97)01474-9. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Sosnick T R, Mayne L, Englander S W. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda Y, Endo S, Nagayama K, Wada A. J Mol Biol. 1995;247:682–688. doi: 10.1006/jmbi.1995.0272. [DOI] [PubMed] [Google Scholar]
- 19.Brutscher B, Brüschweiler R, Ernst R R. Biochemistry. 1997;36:13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 20.Dwyer F P, Gyarfas E C, Mellor D P. J Phys Chem. 1955;59:296–297. [Google Scholar]
- 21.Margoliash E, Frohwirt N. Biochem J. 1959;71:570–572. doi: 10.1042/bj0710570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace C N, Shirley B A, Thompson J A. In: Protein Structure: A Practical Approach. Creighton T E, editor. Oxford: IRL; 1989. pp. 311–322. [Google Scholar]
- 23.Stark G R, Stein W H, Moore S. J Biol Chem. 1960;235:3177–3181. [Google Scholar]
- 24.Hanania G I H, Yeghiayan A, Cameron B F. Biochem J. 1966;98:189–192. doi: 10.1042/bj0980189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hettinger T P, Harbury H A. Proc Natl Acad Sci USA. 1964;52:1469–1476. doi: 10.1073/pnas.52.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howe-Grant M, Lippard S J. Inorg Synth. 1980;20:102–103. [Google Scholar]
- 27.Ratilla E M A, Brothers II H M, Kostic̀ N M. J Am Chem Soc. 1987;109:4592–4599. [Google Scholar]
- 28.Lecomte J T J, La Mar G N. Biochemistry. 1985;24:7388–7395. doi: 10.1021/bi00346a054. [DOI] [PubMed] [Google Scholar]
- 29.La Mar G N, Walker F A. In: The Porphyrins. Dolphin D, editor. New York: Academic; 1979. pp. 61–157. [Google Scholar]
- 30.Shokhirev N V, Walker F A. J Biol Inorg Chem. 1998;3:581–594. [Google Scholar]
- 31.Senn H, Böhme H, Wüthrich K. Biochim Biophys Acta. 1984;789:311–323. [Google Scholar]
- 32.Keller R M, Wüthrich K. Biochim Biophys Acta. 1978;533:195–208. doi: 10.1016/0005-2795(78)90564-0. [DOI] [PubMed] [Google Scholar]
- 33.Hong X, Dixon D W. FEBS Lett. 1989;246:105–108. doi: 10.1016/0014-5793(89)80262-5. [DOI] [PubMed] [Google Scholar]
- 34.Bren K L, Gray H B, Banci L, Bertini I, Turano P. J Am Chem Soc. 1995;117:8067–8073. [Google Scholar]
- 35.Shao W, Sun H, Yao Y, Tang W. Inorg Chem. 1995;34:680–687. [Google Scholar]
- 36.Shao W, Sun H, Yao Y, Tang W. Inorg Chem. 1993;32:6112–6114. [Google Scholar]
- 37.Low D W, Gray H B, Duus J Ø. J Am Chem Soc. 1997;119:1–5. [Google Scholar]
- 38.Rosell F I, Ferrer J C, Mauk A G. J Am Chem Soc. 1998;120:11234–11245. [Google Scholar]
- 39.Moore G R, Pettigrew G W. Cytochrome c: Evolutionary, Structural, and Physicochemical Aspects. New York: Springer; 1990. pp. 115–127. [Google Scholar]
- 40.Segel D J, Fink A L, Hodgson K O, Doniach S. Biochemistry. 1998;37:12443–12451. doi: 10.1021/bi980535t. [DOI] [PubMed] [Google Scholar]
- 41.Myers J D, Pace C N, Sholtz J M. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godbole S, Dong A, Garbin K, Bowler B E. Biochemistry. 1997;36:119–126. doi: 10.1021/bi961915m. [DOI] [PubMed] [Google Scholar]
- 43.Wilson M T, Greenwood C. In: Cytochrome c: A Multidisciplinary Approach. Scott R A, Mauk A G, editors. Sausalito, CA: University Science Books; 1996. pp. 611–634. [Google Scholar]
- 44.Pearce L L, Gärtner A L, Smith M, Mauk A G. Biochemistry. 1989;28:3152–3156. doi: 10.1021/bi00434a006. [DOI] [PubMed] [Google Scholar]