Abstract
Neuroblastoma serves as a paradigm for utilising tumour genomic data for determining patient prognosis and treatment allocation. However, before the establishment of the International Neuroblastoma Risk Group (INRG) Task Force in 2004, international consensus on markers, methodology, and data interpretation did not exist, compromising the reliability of decisive genetic markers and inhibiting translational research efforts. The objectives of the INRG Biology Committee were to identify highly prognostic genetic aberrations to be included in the new INRG risk classification schema and to develop precise definitions, decisive biomarkers, and technique standardisation. The review of the INRG database (n=8800 patients) by the INRG Task Force finally enabled the identification of the most significant neuroblastoma biomarkers. In addition, the Biology Committee compared the standard operating procedures of different cooperative groups to arrive at international consensus for methodology, nomenclature, and future directions. Consensus was reached to include MYCN status, 11q23 allelic status, and ploidy in the INRG classification system on the basis of an evidence-based review of the INRG database. Standardised operating procedures for analysing these genetic factors were adopted, and criteria for proper nomenclature were developed. Neuroblastoma treatment planning is highly dependant on tumour cell genomic features, and it is likely that a comprehensive panel of DNA-based biomarkers will be used in future risk assignment algorithms applying genome-wide techniques. Consensus on methodology and interpretation is essential for uniform INRG classification and will greatly facilitate international and cooperative clinical and translational research studies.
Keywords: neuroblastoma, treatment planning, genomic, translational, international consensus, INRG, genetic risk factors
Modern treatment strategies adjust the therapy of cancer patients according to the predicted biological behaviour of the individual tumour. This is especially important for neuroblastoma patients, as a subset of neuroblastic tumours will undergo spontaneous regression (in infants and in young children) or maturation (in children), whereas others will rapidly progress despite intensive multimodality therapy. This clinical heterogeneity has been known for decades (D’Angio et al, 1971; Evans et al, 1976). However, patient age and tumour stage alone cannot reliably predict tumour behaviour. Over the past two decades, tumour histology, (Shimada et al, 1999) status of the MYCN oncogene, (Brodeur et al, 1984; Seeger et al, 1985), and tumour cell DNA content (ploidy; Look et al, 1991; Ladenstein et al, 2001) have each been shown to be independently predictive of patient outcome in large retrospective and prospective studies. Besides these two genetic markers, recently 11q aberrations were also included in the INRG pretreatment risk classification (Cohn et al, 2009). However, the optimal combination of these and additional recently described prognostic biomarkers to build a treatment stratification algorithm has not been determined yet. Here, we present the consensus of the INRG Biology Committee on which markers should be used currently, on the standardised operating procedures (SOPs; partly on the basis of SIOPEN activities (Ambros et al, 2003)) for analysing neuroblastoma tumour molecular diagnostics and genetic nomenclature in this rapidly evolving field.
Genetic features of neuroblastic tumours associated with favourable clinical behaviour
Favourable clinical behaviour of neuroblastic tumours is classically characterised by a propensity to undergo spontaneous regression or maturation without cytotoxic therapy (Figure 1). These tumours almost always show whole chromosome gains with few, if any, segmental chromosome aberrations and without gene amplifications. The DNA content is usually in the near-triploid (penta/hexaploid) range (Ambros et al, 1995, 1996; Brodeur, 2003; George et al, 2007; Mosse et al, 2007; Schleiermacher et al, 2007). This is also the case for the neuroblastic/ganglionic cell population in Schwann cell stroma-rich tumours, whereas the Schwann cell population itself has a diploid DNA content (Ambros et al, 1996; Bourdeaut et al, 2008).
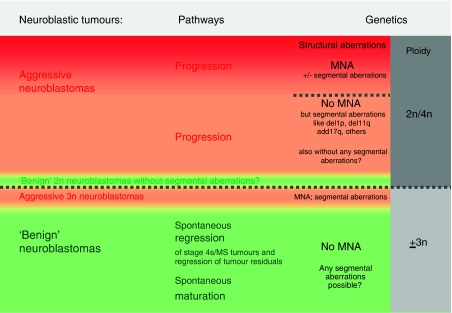
Figure 1.
Biological pathways and genetic features in neuroblastic tumours. Tumour cell ploidy (grey columns) can be used to subdivide neuroblastoma tumours into two broad groups (separated by the long punctuated line). Although the ploidy subgroups roughly correspond to the biologic subgroups (aggressive neuroblastomas marked by a red background – either with MYCN amplification in dark red and separated by a short punctuated line from neuroblastomas without MYCN amplification vs less aggressive behaving neuroblastomas indicated by a green background), they do not totally match. Although aggressive near-triploid neuroblastomas (in red below the long punctuated line) have been observed, it is less clear if ‘benign’ diploid neuroblastomas without any structural aberrations (in green above the long punctuated line) occur. ‘Benign’ clinical behaviour refers either to spontaneous regression/maturation without any therapy or with surgery only (no cytotoxic therapy).
Although tumour cell hyperdiploidy (usually near-triploid DNA content) is frequently associated with spontaneous maturation (Ambros et al, 1996) and possibly also with spontaneous regression, this ploidy level alone cannot guarantee benign tumour behaviour as near-triploid tumours may also have segmental chromosomal aberrations (i.e., gains or losses of only parts of chromosomes) and MYCN amplification, which can lead to clinically aggressive behaviour.
Genetic features of neuroblastic tumours associated with unfavourable clinical behaviour
Neuroblastomas with an unfavourable clinical behaviour (Figure 1) have a high propensity for locally invasive growth and widespread metastatic dissemination through the lymphatic and haematogenous systems. These tumours frequently show segmental aberrations and high-level amplification of the MYCN locus is detected in a substantial subset (Schwab et al, 1983; Brodeur et al, 1984; Seeger et al, 1985). MYCN amplification has been shown to be strongly associated with rapid tumour progression and poor prognosis in patients of all ages, with any stage of disease (Brodeur et al, 1984; Seeger et al, 1985; Ambros et al, 1995; Perez et al, 2000). As expected, MYCN amplification was also highly predictive of worse outcome in the INRG cohort of patients (Cohn et al, 2009). Frequently, adjacent or more distantly located genes, such as DDX1, NAG, and ALK, are coamplified with MYCN, but amplification events in the absence of MYCN amplification are rare. Whether these coamplifications have a prognostic impact awaits clarification (Manohar et al, 1995; Squire et al, 1995; George et al, 1997; De Preter et al, 2002; Weber et al, 2004).
Although MYCN status is central to the risk stratification systems in all cooperative clinical trial groups, it is important to emphasise that the majority of metastatic neuroblastomas do not show amplification of this oncogene (see also Figure 1). Other chromosomal aberrations (and a diploid DNA content) have been assumed to predict unfavourable tumour behaviour, including deletion at the chromosomal region 1p36.3 or 11q23, (Maris et al, 1995; Caron et al, 1996b; Attiyeh et al, 2005) as well as unbalanced gain of the long arm of chromosome 17 (17q21 to 17qter; Caron, 1995; Bown et al, 1999; Spitz et al, 2003). In addition, some studies have shown that deletions on chromosome 3p, 4p, 9p, and 12p also have prognostic significance (Caron et al, 1996a; Thompson et al, 2001; Schleiermacher et al, 2007). As 11q deletions are inversely associated with MYCN amplification, this aberration has emerged as a powerful biomarker of outcome in cases without MYCN amplification (Attiyeh et al, 2005). Statistical analysis of the INRG database has confirmed this finding, and on the basis of these studies, 11q status has been included as a prognostic criterion in the INRG classification system. (Cohn et al, 2009). Recent publications clearly show the potential of comprehensive genome-wide approaches to further refine the prognostic accuracy of somatically acquired chromosomal alterations, (Vandesompele et al, 2005; Michels et al, 2007; Mosse et al, 2007; Schleiermacher et al, 2007; Tomioka et al, 2008). These studies have shown that in tumours without MYCN amplification, segmental chromosome aberrations are associated with clinically aggressive disease. These findings have been extended in a series of 493 MYCN amplified and non-amplified neuroblastomas; in tumours with only whole-chromosome copy number variations, there were no disease-related deaths. In contrast, the presence of segmental alterations (see Table 3) with or without MYCN amplification was the strongest predictor of relapse (Janoueix-Lerosey et al, 2009).
Tumour cell ploidy
The majority of tumours have a hyperdiploid (near-triploid or penta/hexaploid) DNA content, whereas less than half of neuroblastic tumours are diploid (Look et al, 1984; Kaneko et al, 1987). Locoregional tumours are commonly hyperdiploid; diploidy is more common in advanced-stage tumours. A number of studies have shown that in patients <18 months of age with metastatic disease, hyperdiploidy in combination with a non-amplified MYCN gene and the lack of specific segmental chromosome aberrations (such as 11q deletion) are predictive of a favourable outcome (George et al, 2005; Schleiermacher et al, 2007). However, in patients <18 months of age with metastatic disease with diploid, MYCN non-amplified tumours have a statistically significantly worse outcome.
Evaluation of genomic markers in the INRG analytic cohort
Relationship of genetic aberrations
In the INRG analytic cohort (n=8800; see Cohn et al (2009) for details), 29% were diploid, 16% were MYCN amplified, 21% had an 11q aberration, 23% had a 1p aberration, and 48% had a 17q gain. Statistically significant associations of genetic factors were identified (Table 1). 11q aberration was associated with 17q gain (P<0.0001) and inversely associated with MYCN amplification (P=0.0006), but 11q aberration was not associated with diploidy or 1p aberration. Diploidy and 17q gain were not associated, but all other pair-wise comparisons were highly statistically significant (P<0.01).
Table 1. Tests of association of genetic factors.
| MYCN amplification | 1p aberration | 11q aberration | 17q gain | |
|---|---|---|---|---|
| Diploidy | P<0.0001 | P<0.0001 | P=0.1242 | P=0.3613 |
| MNA | P<0.0001 | P=0.0006* | P=0.0096 | |
| 1p aberration | P=0.0613 | P<0.0001 | ||
| 11q aberration | P<0.0001 |
*Inversely associated. MNA MYCN amplification
Outcome by clinical and genetic subgroups
In the survival-tree regression approach of Cohn and Pearson et al (2009), patients were clustered into meaningful pretreatment groups that were homogeneous in terms of outcome and prognostic factors (clinical and biological). For the descriptive purposes of this manuscript, the INRG analytic cohort was further subdivided. Outcome by genetic factors vs INSS stage, age, and primary tumour site is shown in Tables 2a, and 2b, respectively. Within each genetic factor subgroup, the patterns of outcome differences that have been observed in the overall population prevailed: (a) EFS and overall survival (OS) decreased with increasing stage (Table 2a); (b) older patients had worse outcome than younger patients (Table 2b) and (c) patients with adrenal primary tumour site had worse outcome than those with non-adrenal tumour (Table 2b).
Table 2a. Event-free and overall survival (EFS and OS) of genetic factors by INSS stage.
|
INSS stage 1
|
INSS stage 2
|
INSS stage 3
|
INSS stage 4s
|
INSS Stage 1,2,3,4s
|
INSS Stage 4
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | |
| Hyper diploid (n=2611) | 719 (28) | 93±2 | 99±1 | 415 (16) | 88±3 | 96±2 | 452 (17) | 82±3 | 86±3 | 232 (9) | 86±3 | 92±3 | 1818 (70) | 88±1 | 94±1 | 728 (28) | 47±3 | 53±3 |
| Diploid n=1086) | 196 (18) | 90±4 | 96±2 | 119 (11) | 84±5 | 90±5 | 152 (14) | 65±6 | 66±6 | 62 (6) | 77±9 | 77±9 | 529 (49) | 80±3 | 84±3 | 527 (50) | 31±3 | 36±3 |
| MYCN, not amplified (n=5947) | 1626 (27) | 93±1 | 98±1 | 947 (16) | 85±2 | 97±1 | 1013 (17) | 81±2 | 89±1 | 481 (8) | 82±2 | 91±2 | 4067 (68) | 87±1 | 95±1 | 1798 (31) | 45±2 | 54±2 |
| MYCN, amplified (n=1155) | 48 (4) | 50±12 | 76±9 | 39 (3) | 57±12 | 67±11 | 217 (19) | 45±4 | 48±4 | 47 (4) | 41±9 | 45±9 | 351 (30) | 46±4 | 53±4 | 787 (69) | 21±2 | 25±2 |
| 1p normal (n=1659) | 489 (29) | 93±2 | 98±1 | 264 (16) | 83±3 | 98±1 | 313 (19) | 77±3 | 88±3 | 152 (9) | 86±4 | 95±3 | 1218 (73) | 86±1 | 95±1 | 430 (26) | 42±3 | 50±3 |
| 1p aberration (n=493) | 48 (10) | 76±9 | 100 | 29 (6) | 63±11 | 81±9 | 90 (18) | 52±7 | 61±7 | 29 (6) | 60±13 | 67±12 | 196 (40) | 60±5 | 74±4 | 280 (57) | 24±4 | 32±4 |
| 11q normal (n=844) | 227 (27) | 90±3 | 99±1 | 141 (17) | 71±7 | 95±3 | 152 (18) | 77±5 | 85±4 | 69 (8) | 86±7 | 95±5 | 589 (70) | 82±3 | 94±2 | 233 (28) | 42±5 | 50±5 |
| 11q aberration (n=220) | 11 (5) | 73±22 | 91±14 | 11 (5) | 45±34 | 91±16 | 35 (16) | 51±11 | 75±11 | 8 (4) | 38±30 | 63±38 | 65 (30) | 51±10 | 79±8 | 153 (70) | 28±6 | 47±7 |
| 17q normal (n=187) | 36 (19) | 78±9 | 97±4 | 22 (12) | 77±12 | 100 | 47 (25) | 74±7 | 85±6 | 15 (8) | 92±11 | 92±11 | 120 (64) | 78±5 | 92±3 | 60 (32) | 40±7 | 46±7 |
| 17q gain (n=175) | 20 (11) | 95±9 | 100 | 19 (11) | 51±18 | 84±14 | 16 (9) | 75±13 | 94±7 | 4 (2) | 100 | 100 | 59 (34) | 76±8 | 93±5 | 104 (59) | 24±6 | 38±7 |
Table 2b. Event-free and overall survival (EFS and OS) of genetic factors, by age and primary tumour site at diagnosis.
|
Age <547 days
|
Age > 547 days
|
Adrenal primary tumour site
|
Non-adrenal primary tumour site
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | n (%) | 5-year EFS±s.e. | 5-year OS±s.e. | |
| Hyper diploid (n=2611) | 1816 (70) | 88±1 | 95±1 | 795 (30) | 48±3 | 54±3 | 1023 (39) | 73±2 | 78±2 | 1473 (56) | 78±2 | 85±1 |
| Diploid (n=1086) | 425 (39) | 70±4 | 75±4 | 661 (61) | 46±3 | 50±3 | 489 (45) | 49±4 | 54±3 | 556 (51) | 60±3 | 64±3 |
| MYCN, not amplified (n=5947) | 3657 (61) | 88±1 | 95±1 | 2290 (39) | 51±1 | 60±1 | 2439 (41) | 69±1 | 76±1 | 3265 (55) | 77±1 | 86±1 |
| MYCN, amplified (n=1155) | 404 (35) | 36±3 | 41±3 | 751 (65) | 26±2 | 30±2 | 732 (63) | 27±2 | 33±2 | 384 (33) | 33±4 | 37±4 |
| 1p normal (n=1659) | 1095 (66) | 88±1 | 96±1 | 564 (34) | 49±3 | 59±3 | 751 (45) | 71±2 | 78±2 | 881 (53) | 77±2 | 88±2 |
| 1p aberration (n=493) | 185 (38) | 57±5 | 65±5 | 308 (62) | 26±3 | 38±4 | 323 (66) | 33±4 | 44±4 | 157 (32) | 48±5 | 58±5 |
| 11q normal (n=844) | 510 (60) | 83±3 | 93±2 | 334 (40) | 46±4 | 57±4 | 359 (43) | 64±4 | 72±4 | 451 (53) | 73±3 | 85±3 |
| 11q aberration (n=220) | 63 (29) | 61±11 | 85±8 | 157 (71) | 26±5 | 47±6 | 126 (57) | 34±6 | 52±7 | 93 (42) | 35±9 | 63±9 |
| 17q normal (n=187) | 104 (56) | 80±5 | 92±3 | 83 (44) | 44±6 | 53±6 | 74 (40) | 60±7 | 65±7 | 105 (56) | 67±6 | 82±5 |
| 17q gain (n=175) | 65 (37) | 68±9 | 83±7 | 110 (63) | 27±5 | 41±6 | 116 (66) | 37±7 | 51±7 | 53 (30) | 53±9 | 67±9 |
However, as shown in Table 2a, only a limited number of data on the prognostic impact of some segmental aberrations are available, impeding reliable interpretation of some of the data (e.g., 17q gain in stage 1 and 4 s tumours). Thus, larger data sets on segmental aberrations are needed to allow final statements on their prognostic impact (Table 3) .
Table 3. Consensus of genetic markers currently used for therapy stratification and proposed for future analyses.
| Genetic INRG risk classification markers | Techniques recommended/accepted | DNA probes recommended and comments |
|---|---|---|
| Obligatory markers | ||
| MYCN | I-FISH PCR, aCGH, MLPA | Two colour I-FISH: BAC or other large DNA-insert clone for the MYCN gene and a clone of similar size for a gene/locus on the long arm of chromosome 2 (e.g., LAF at 2q11); commercially available probes should be used whenever possible |
| 11q23 | I-FISH, PCR; pan-/multigenomic techniques: aCGH (oligo, clone or SNP based); MLPA | |
| Ploidy | Flow or static cytometry | Normal cells from the same patient should be used as a diploid standard |
| Genetic markers to be analysed prospectively | ||
| 1p | aCGH, SNP arrays, MLPA etc. | Commercially available platform preferred |
| 2p | ||
| DDX1 | ||
| NAG | ||
| ALK | ||
| 3p | ||
| 4p | ||
| 7q | ||
| 9p | ||
| 12p | ||
| 14q | ||
| 17q | ||
| Others | aCGH, SNP arrays |
aCGH=array-based comparative genomic hybridisation; BAC=bacterial artificial chromosomes; I-FISH=interphase fluorescence in situ hybridisation; INRG=International Neuroblastoma Risk Group; MLPA=multiplex ligation-dependent probe amplification; PCR=polymerase chain reaction; SNP=single nucleotide polymorphism.
Statistical considerations
Tests of association were carried out using a χ2-test. P-values <0.05 were considered statistically significant. For event-free survival (EFS) analysis, time to event was defined as the time from diagnosis until the time of first occurrence of relapse, progression, secondary malignancy, or death, or until time of last contact if no event occurred. For OS, time to event was defined as time until death, or until last contact if the patient was alive. The methods of Kaplan and Meier (1958) were used to calculate EFS and OS estimates, with s.e. according to Peto and Peto (1972).
International consensus by the INRG biology committee
Because of the relative rarity of neuroblastomas, and the uniqueness of many of the molecular diagnostic factors, the INRG Biology Committee recommended that the genetic studies required for INRG classification be carried out in experienced laboratories, typically central reference laboratories for the cooperative groups (see Table 4), to guarantee high consistency and quality of results (Ambros et al, 2003).
Table 4. Laboratories responsible for Neuroblastom Genetics.
| National Group | Name | Institution | Town | Country | |
|---|---|---|---|---|---|
| Australia | Michelle | Haber, PhD | Children's Cancer Institute Australia and COG | Sydney, NSW | Australia |
| COG | Julie | Gastier-Foster, PhD | Nationwide Children's Hospital | Columbus, OH | USA |
| Michael | Hogarty, MD | Children's Hospital of Philadelphia | Philadelphia, PA | USA | |
| Rochelle | Bagatell, MD | Children's Hospital of Philadelphia | Philadelphia, PA | USA | |
| Germany | Frank | Berthold, MD | Department of Pediatric Hematology and Oncology, University of Cologne | Cologne | Germany |
| Jessica | Theißen, PhD | Department of Pediatric Hematology and Oncology, University of Cologne | Cologne | Germany | |
| Manfred | Schwab, PhD | Division of Cytogenetics, German Cancer Research Center | Heidelberg | Germany | |
| Frank | Westermann, MD | Division of Cytogenetics, German Cancer Research Center | Heidelberg | Germany | |
| Freimut H | Schilling, MD | Department of Pediatric Oncology and Hematology, Olgahospital | Stuttgart | Germany | |
| Sabine | Stegmaier, PhD | Department of Pediatric Oncology and Hematology, Olgahospital | Stuttgart | Germany | |
| Niggli | Felix, MD | Department of Pediatric Oncology and Hematology | Zurich | Switzerland | |
| Japan | Akira | Nakagawara, MD | Chiba Cancer Center Research Institute | Chiba | Japan |
| Miki | Ohira, MD | Chiba Cancer Center Research Institute | Chiba | Japan | |
| Yasuhiko | Kaneko, MD | Saitama Cancer Center Research Institute | Saitama | Japan | |
| Junko | Takita, MD | Department of Pediatrics, University of Tokyo | Tokyo | Japan | |
| Hajime | Ohkita, MD | National Center for Child Health and Development | Tokyo | Japan | |
| SIOPEN Biology | Peter F | Ambros, PhD | CCRI, Children's Cancer Research Institute | Vienna | Austria |
| Inge M | Ambros, MD | CCRI, Children's Cancer Research Institute | Vienna | Austria | |
| Frank | Spelemann, PhD | Center for Medical Genetics-OK5, University of Ghent | Ghent | Belgium | |
| Nadine | Van Roy, PhD | Center for Medical Genetics-OK5, University of Ghent | Ghent | Belgium | |
| Ales | Vicha, MD | Department of Pediatric Hematology and Oncology, Motol | Prague | Czech Republic | |
| Jean | Bénard, MD | Lab De Pharmacologie Clinique et Moleculaire, Instiut Gustav Roussy | Villejuife-Cedex | France | |
| Alexander | Valent, PhD | Lab De Pharmacologie Clinique et Moleculaire, Instiut Gustav Roussy | Villejuife-Cedex | France | |
| Jérome | Couturier, MD | Unité de Cytogénétique oncologique, Institut Curie – Section Médicale | Paris | France | |
| Olivier | Delattre, PhD | Unité de Cytogénétique oncologique, Institut Curie – Section Médicale | Paris | France | |
| Gudrun | Schleiermacher, MD | Unité de Cytogénétique oncologique, Institut Curie, Section Médicale | Paris | France | |
| Marta | Jeison, PhD | Pediatric Hematology Oncology Department, Schneider Children's Medical Center of Israel | Petah Tikva | Israel | |
| Gian Paolo | Tonini, PhD | Translational Paediatric Oncology, National Cancer Research Institute (IST) | Genoa | Italy | |
| Raffaella | Defferrari, PhD | Translational Paediatric Oncology, National Cancer Research Institute (IST) | Genoa | Italy | |
| Katia | Mazzocco, PhD | Translational Paediatric Oncology, National Cancer Research Institute (IST) | Genoa | Italy | |
| Klaus | Beiske, MD | Department of Pathology, Rikshospitalet | Oslo | Norway | |
| Barbara | Marques, PhD | Centro de Genética Humana, Instituto Nacinal de Saúde | Lisbon | Portugal | |
| Nicole | Gross Ph.D. | Recherche en Oncologie Pédiatrique | Lausanne | Switzerland | |
| Rosa | Noguera, MD | Facultad de Medicina | Valencia | Spain | |
| Tommy | Martinsson, PhD | Gahlgrenska Univ. Hospital/East, Gothenburg University | Gothenburg | Sweden | |
| Deborah A. | Tweddle, MD | Department of Health | Newcastle | UK | |
| John | Lunec, PhD | Northern Institute for Cancer Research | Newcastle | UK | |
| Nick | Bown, PhD | Institute of Human Genetics | Newcastle | UK |
Consensus on the genetic markers to be currently used in the INRG risk classification system
As reported in the study by Cohn and Pearson et al (2009) survival-tree regression analyses of the INRG database confirmed the prognostic significance of MYCN amplification, 11q aberrations, and ploidy, in addition to age and stage, in different subgroups of the INRG cohort. Table 3 includes these three genetic markers and also those, which should be analysed prospectively.
Tumour sampling/storing procedure and indication of the tumour cell content
An adequate amount of tumour material (i.e., 107 tumour cells) from at least two different regions of the tumour should be obtained. In collaboration with the institutional pathologist, the tumour cell content must be determined and recorded. The latter is an indispensable prerequisite to avoid false results and has to be carried out by the pathologist. A section (cryo- or paraffin) has to be kept to each tumour piece used for genetic or expression studies. In certain cases, interphase fluorescence in situ hybridisation (I-FISH) can be used to decide on the tumour cell number. A tumour cell content of more than 60% is required for most molecular studies and of more than 20% for ploidy measurement. Differentiating/maturing tumours can have fewer tumour cells, and therefore, the interpretation of these data need to de done with caution. For I-FISH, a low tumour cell number can also be sufficient, in case of numeric/structural chromosome aberrations.
Molecular studies on maturing/mature tumours, such as ganglioneuroblastomas or ganglioneuromas, (Ambros et al, 1996; Brodeur, 2003) require meticulous identification of tumour and Schwannian stromal cells, and can thus only be undertaken when using an appropriate system (e.g., on ganglionic cells or by microdissection, ideally on paraffin sections). In case of needle-core biopsies, for obtaining tumour material from different areas, an exact determination of the tumour cell content is crucial again (Frostad et al, 1999). Interphase fluorescence in situ hybridisation results of disseminated tumour cells in the bone marrow (Ambros et al, 2001) can be given only if MYCN amplification is present due to the higher error-proneness if segmental/numeric aberrations are evaluated in mixtures of normal and tumour cells. In case of immunological preselection and automatic relocation of DTCs (Mehes et al, 2001), information on genetic aberrations other than MYCN amplification can be given as well.
On account of the advent of new techniques enabling large retrospective studies, it is essential to store frozen tumour samples and/or extracted DNA, RNA, or protein (at −80°C or in liquid nitrogen) with exactly determined tumour cell content. Non-neoplastic reference cells from the same patient should be stored as well. The INRG Biology Subcommittee further emphasises the clear need for biobanking of high quality biological materials from neuroblastoma patients, and this must be central to the SOP of any cooperative group for the collection of diagnostic material (Qualman et al, 2005). For further details and recommendations, see Figure 2 and the guidelines indicated by Ambros and Ambros (2001).
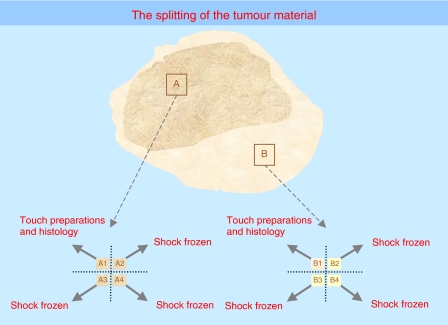
Figure 2.
Recommendations concerning the splitting of the tumour material for resected tumours or surgical biopsies. All specimens should be transported to the pathology laboratory as quickly as possible. From there, the snap frozen and/or OCT embedded material should then be transported immediately to the biology lab (can be used for any type of DNA, RNA or protein work). Normal reference cells (e.g., peripheral blood) should be sent to the reference laboratories.
Evaluation and the reporting of the MYCN gene copy status
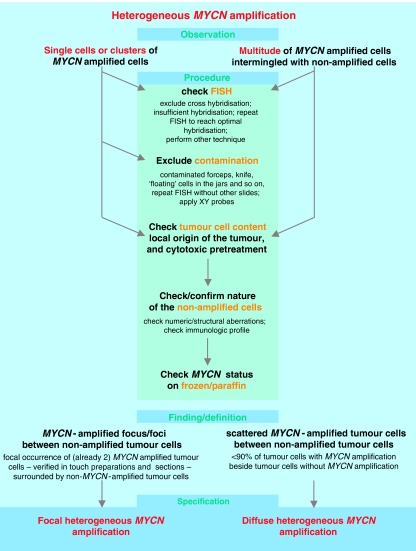
The INRG Biology Committee agreed that MYCN status should be evaluated in every resected neuroblastic tumour, including the Schwann cell stroma-rich categories (Shimada et al, 1999). Recommended and accepted techniques to detect structural and/or segmental aberrations are summarised in Table 3, and recommendations concerning the evaluation and terminology are given in Tables 5, 6, 7 and 8. Interphase fluorescence in situ hybridisation is preferred because it has a number of advantages compared with the other techniques, most important of which is direct quality control of the hybridization quality by the microscopist. Heterogeneous MYCN amplification requires meticulous analysis of the tumour specimen (Figure 3). At the present time, the prognostic significance of heterogeneously MYCN amplified neuroblastomas is not known, but amplification of a substantial number of tumour cells in a specimen is still considered an ominous sign.
Table 5. MYCN terms and definitions of I-FISH results.
|
MYCN status analysed by I-FISH
| ||
|---|---|---|
| Terms | Description | Comments |
| MYCN not amplified (normal MYCN status) | A balanced ratio between the MYCN-specific signals and the signal number of the reference probe on the chromosome 2q arm | Neuroblastic tumours have a propensity to polyploidisation, especially but not exclusively after therapy with the occurrence of giant polyploid nuclei. The number of MYCN signals must not exceed the number of reference signals on 2q (caveat: the centromeric probe is unqualified as reference probe because of frequent centromeric associations in such tumours). The number of other chromosomes or the DNA index should be checked as well. |
| 2p24 gain: | The term 2p24 gain is suggested as a descriptive generic term for MYCN signal numbers exceeding up to 4-fold the number of reference signals on chromosomal arm 2q. This pattern could reflect either: | The use of a 2p specific probe in addition to 2q (optionally a centromeric probe) is recommended to clarify the presence of a chromosome 2p gain vs a restricted MYCN gain. The discrimination could be important since a MYCN gain could indicate an ‘incipient’ MYCN amplification. |
| (a) 2p gain | (a) a gain of short arms of chromosome 2, for example, from unbalanced translocations involving 2p; or | Equal number of MYCN and 2p signals, exceeding 2q signals (mostly a consistent excess of one or two MYCN and 2p signals). |
| (b) MYCN gain | (b) a gain of the MYCN gene either chromosomally integrated (such as duplications) or extrachromosomally | An inconsistent MYCN excess with a varying number of excess signals is much more likely in line with MYCN gain as extrachromosomally elements. The latter can also be found in tumours with heterogeneous MYCN amplification. |
| MYCN amplification, ‘homogeneous’ | More than 4-fold increase in the MYCN signal number compared with the reference probe located on the chromosome 2q | Details on the amplification grade (a) >4–10 times; (b) >10 times; (c) >30 times amplification, and also on the type of amplification either double minutes (dmin) or homogeneously staining regions (hsr) should be given in the report. Besides tumour cells with amplification defined as such, a proportion of tumour cells can also show ‘MYCN gain’. However, tumour cells without MYCN gain or amplification are extremely rare. |
| MYCN amplification, ‘heterogeneous’ | Generic term for the coexistence of amplified as well as non-amplified tumour cells in the same tumour | Single cells (at least two) or cell clusters or a multitude of cells with MNA besides proven tumour cells without MNA. The terms ‘focal’ and ‘diffuse MNA’ are specifications, which, however, can only be attributed after evaluation of the tissue sections. In case amplified tumour cells are found in addition to non-amplified cells, a very meticulous procedure is strongly recommended to exclude false-positive or false negative results (see Figure 3). |
| (a) Focal heterogeneous MYCN amplification | Defined as the more or less focal (one focus or several foci – multifocal) occurrence of MYCN-amplified cells surrounded by non-amplified tumour cells. | |
| (b) Diffuse heterogeneous MYCN amplification | Designates a tumour, which contains the MYCN amplified cells in a scattered pattern besides non-MYCN amplified tumour cells. | |
| No result | Should be specified: unclear or not interpretable; | |
| inadequate tumour cell content; no tumour; not carried out | ||
I-FISH=interphase fluorescence in situ hybridisation.
Table 6. Terms and definitions for segmental chromosome aberrations of I-FISH results.
|
Segmental chromosome aberrations analysed by I-FISH
| ||
|---|---|---|
| Terms | Description | Comments |
| Normal status | Balanced ratio between the signal numbers of the chromosomal region of interest and the reference signals on the opposite arm of the chromosome | In the case of a 2/2 ratio, an isodisomy with a complete LOH of all loci located on the investigated chromosome cannot be excluded. However, uniparental isodisomies are extremely rare in NB tumours. |
| I-FISH imbalance (inconclusive with regard to LOH) | Unbalanced ratio between the signal numbers of the chromosomal region of interest and the reference signals with more than one signals of the chromosomal region of interest | Does not necessarily correspond to an LOH, and is therefore called inconclusive. PCR should be performed to clarify this result. In NB tumours, imbalances frequently, but not always, reflect LOH. Fluorescence in situ hybridisation imbalances can also occur focally. Interphase fluorescence in situ hybridisation imbalances should only be diagnosed if hybridisation failure can be excluded. At least 200 cells should be evaluated. |
| Deletion | Unbalanced ratio between the signal numbers of the chromosomal region of interest and the reference signals with only one signal of the chromosomal region of interest | This hybridisation pattern corresponds to an LOH. Deletions can also occur focally. Interphase fluorescence in situ hybridisation deletion should only be diagnosed if hybridisation failure can be excluded. At least 200 cells should be evaluated. |
| Gain | Up to 4-fold excess of signal numbers of the chromosomal region of interest compared with the reference signals | |
| No result | Should be specified: unclear or not interpretable; inadequate tumour cell content; no tumour; Not carried out | |
I-FISH=interphase fluorescence in situ hybridisation.
Table 7. Terms and definitions of segmental chromosome aberrations analysed by PCR.
|
Losses of chromosomal parts analysed by PCR
| ||
|---|---|---|
| Terms | Description | Comments |
| Normal status | Similar band intensities | |
| Allelic imbalance (inconclusive) | Different band intensities; The result can mean either allele disequilibrium or LOH | One band is relatively weaker, when compared with the ratio of constitutional DNA controls. Needs further clarification by FISH (copy number of the respective chromosome has to be known) and re-evaluation of the tumour cell content (an LOH could be masked by a high number of normal cells). Alternatively, different PCR probes, located on both arms (p and q) can circumvent the need for the use of another technique. Allele disequilibrium means an imbalance between the number of maternal and paternal alleles (in the case of odd numbers of the respective chromosome), but not a physical loss of alleles |
| Allelic loss | Loss of one band | One band has completely, or almost completely disappeared |
| No result | Should be specified: unclear or not interpretable; inadequate tumour cell content (<60%); constitutional homozygosity; no DNA; no tumour; not carried out | |
FISH=fluorescence in situ hybridisation.
Table 8. Terms and definitions of chromosome aberrations analysed by MLPA.
|
Chromosome and gene status analysed by MLPA
| ||
|---|---|---|
| Terms | Description | Comments |
| Normal status | Balanced ratio between the majority of signals (signal intensity is visualised in the graphic representation of the MLPA results as height of the bars) of both chromosomal arms | In the case of a balanced ratio, a uniparental isodisomy (a uniparental isodisomy means that both chromosomes are derived from one parent. Thus, the two chromosomes are not homologous but identical. However, uniparental isodisomies are rare in NB tumours) with a complete LOH of all loci located on the investigated chromosome cannot be excluded |
| Segmental chromosome aberration | ||
| Loss | Unbalanced ratio between the signals of the chromosomal region of interest (at least two adjacent probes) and the reference signals (at least two probes) signals | This result could correspond to a FISH deletion that reflects an LOH, or a FISH imbalance, which does not necessarily indicate an LOH. |
| Gain | Unbalanced ratio (low signal excess) between the signals of the chromosomal region of interest (at least two adjacent probes) and the reference signals (at least two) of the chromosomal region of interest | |
| The threshold between gain and amplification needs to be determined for the experimental system with the help of other techniques,for example, I-FISH | ||
| Amplification | Unbalanced ratio (high signal excess) between the signals of a gene and all other probes located on the same chromosome | The threshold between gain and amplification needs to be determined for the experimental system with the help of other techniques, for example, I-FISH |
| No result | Should be specified: unclear or not interpretable; inadequate tumour cell content; no tumour; not carried out | |
FISH=fluorescence in situ hybridisation; MLPA=multiplex ligation-dependent probe amplification; I-FISH=interphase fluorescence in situ hybridisation.
Figure 3.
Heterogeneous MYCN amplification and recommended procedure for the clarification of the underlying genetic pattern.
Evaluation and reporting of segmental genetic aberrations: gains and losses of chromosome parts
The term ‘structural’ aberrations (also known as structural chromosomal instability (s)-CIN) designates any kind of chromosomal alterations including gene amplification. To produce precise definitions, consensus was reached to introduce the term ‘segmental’ aberrations for gains and losses of chromosome parts. In neuroblastomas, most segmental chromosome aberrations are unbalanced, that is, they are associated with regional losses or gains of chromosome parts. Balanced aberrations, that is, reciprocal translocations without losses of genetic material, are thought to be relatively uncommon in neuroblastomas. Segmental aberrations (such as 11q23 and 1p36.3 deletions) can be identified by a number of techniques shown in Table 3. Currently, either I-FISH or PCR are carried out to detect segmental aberrations, but the INRG Biology Committee recommends array-based methods, multiplex ligation-dependent probe amplification (MLPA) or similar techniques in the future. The latter techniques are currently being validated (for nomenclature (I-FISH, PCR, and MLPA), see Tables 5, 6, 7 and 8; part of the nomenclature was developed by the SIOPEN Biology Group (for group members, see Table 4 and Ambros et al, 2003)).
Tumour cell DNA content (ploidy)
Ploidy was analysed in the INRG cohort using the definitions diploid (i.e., DI ⩽1.0) vs hyperdiploid (DI >1.0) as published in Look et al (1991). More recent data suggest that prognostic classification may be further refined using specific ranges of DI The INRG Biology Committee recommends recording the exact numerical DI value for each tumour, so that clinically relevant cutoffs for DI can be determined (for techniques see Table 3). Any other method can be applied as long as it allows discrimination of the diploid DNA peak. However, an unknown number of normal cells contained in the tumour specimen under investigation, including tumour-derived, but non-neoplastic diploid Schwann cells are an important source of false data interpretation. Thus, the DNA content of maturing/mature tumours, which mostly develop from near-triploid neuroblastomas is easily misdiagnosed as diploid.
Future prospects: new techniques
There is clear consensus that the use of genetic data derived from diagnostic neuroblastoma tumours will remain central to patient treatment planning and gain even more power. The INRG Biology Committee addressed the issues of ongoing and future work directed at prognostic biomarker discovery and validation, as well as how these data sets can be leveraged to identify molecular targets for novel therapeutics. In Table 3, 12 genetic markers to be analysed prospectively are mentioned. Pan-genomic investigations may help to identify additional genomic areas of interest.
DNA-based biomarkers
A number of studies suggest that the pattern of DNA-based genomic changes is prognostic in neuroblastoma, and that whole genome analysis should be carried out rather than a series of individual locus-specific assays. Therefore, the INRG Biology Subcommittee suggests using pan- or multigenomic techniques enabling an analysis of all relevant genomic loci.
Multiplex ligation-dependent probe amplification is a PCR-based technique that detects a large variety of segmental aberrations and gene amplifications in a robust manner (for technical details, see (Schouten et al, 2002). Quantification of losses, gains, and amplifications in small amounts of DNA can be determined, and simultaneous investigation of a large number of loci, covering all currently known important aberrant regions found in neuroblastomas, can be carried out in a single assay (Villamon et al, 2008). The robust nature of the results and the relatively low cost of the MLPA kits make this technique attractive for routine neuroblastoma analysis.
Chip-based technologies have also been used to molecularly classify neuroblastoma tumours (Bilke et al, 2005; George et al, 2005; Selzer et al, 2005; Spitz et al, 2006; Stallings et al, 2006; Lastowska et al, 2007; Michels et al, 2007; Mosse et al, 2007; Tomioka et al, 2008). Comparative genomic hybridisation has reached a high coverage of the target sequences and become more widely used. The advantage of single nucleotide polymorphism (SNP)-based platforms is the simultaneous detection of physical gains and losses and the detection of copy neutral LOH (George et al, 2005). Still, these techniques cannot be considered as routine techniques but as excellent tools to identify so far undetected genomic regions of prognostic impact. The INRG Biology Committee considers it a priority to work towards a single diagnostic tool that will reliably and accurately detect allelic deletions and gains, as well as MYCN amplification, and estimate overall DNA content (ploidy), on a single platform. This has been achieved using in-house arrays (Maris et al, 2005; Janoueix-Lerosey et al, 2009), but a commercially available platform will be needed to implement this or a similar technology (e.g., MLPA) uniformly in clinical laboratories across various groups and countries.
RNA-based biomarkers
Several groups have recently shown that genome-wide gene expression profiling can identify differentially expressed transcripts that provide reliable prognostic information (Wei et al, 2004; Ohira et al, 2005; Schramm et al, 2005; Asgharzadeh et al, 2006; Oberthuer et al, 2006; Lastowska et al, 2007; Tomioka et al, 2008). Although these studies are of outstanding quality, they each suffer from relatively small sample sizes, necessitating preliminary validation of proposed gene sets. In addition, there was very little overlap in the gene sets that were identified as prognostic by each group, so a consensus gene set, that is, predictive of outcome has yet to be identified. International collaboration will be required to unequivocally determine whether mRNA expression profiles, likely focused on a representative set of genes, are a sensitive and specific enough molecular assay to be used in the clinic, and/or if these data are synergistic with, or override the information derived from the assays of DNA alterations.
Molecular targets
Neuroblastoma treatment will continue to rely on risk grouping based on tumour genomic features. Increasing attention is now focused on utilising these data sets to discover therapeutic targets. It stands to reason that any prognostically relevant genomic aberration might also signal a molecular aberration that is critical to the maintenance of the malignant phenotype, and thus can be targeted for therapy. Regions of DNA copy number gain that result in the overexpression of a protein that is druggable in patients succumbing to the disease can theoretically be identified in carefully annotated data sets in which both high quality DNA- and RNA-based microarray data are available. Thus, a parallel focus on therapeutic target discovery and validation will further increase the significance of genomic efforts used in the discovery, validation, or even clinical application phases of implementation.
Conclusion
Currently, neuroblastoma treatment planning is not possible without detailed knowledge of tumour cell genomics. International efforts were and are needed not only to identify the most significant genetic markers but also to identify and coordinate particularly suitable techniques for their evaluation in all patients. For future studies, new technologies, which provide a comprehensive picture of the tumour cell genome are recommended. Moreover, the INRG Biology Committee achieved consensus on the nomenclature of genetic aberrations and developed definitions of the terms to be used. The INRG Biology Committee is dedicated to providing the highest possible reproducibility and reliability of genetic markers enabling a uniform INRG classification and forming the basis for international clinical and translational studies. Finally, many of these recommendations apply not only to neuroblastoma tumours but also to any tumour entity for which genetic factors are essential for therapy decisions.
Acknowledgments
This study was supported by the William Guy Forbeck Research Foundation, USA; the Little Heroes Cancer Research Fund, USA; the St Anna Kinderkrebsforschung, Vienna, Austria (PFA and IMA); Cancer Research UK (ADJP); and the Audrey Evans Endowed Chair (GMB), the Giulio D’Angio Endowed Chair (JMM), and the National Health and Medical Research Council and Cancer Institute NSW, Australia (MH).
Footnotes
The study was presented in part at the SIOP Meeting 2007.
References
- Ambros IM, Benard J, Boavida M, Bown N, Caron H, Combaret V, Couturier J, Darnfors C, Delattre O, Freeman-Edward J, Gambini C, Gross N, Hattinger CM, Luegmayr A, Lunec J, Martinsson T, Mazzocco K, Navarro S, Noguera R, O’Neill S, Potschger U, Rumpler S, Speleman F, Tonini GP, Valent A, Van Roy N, Amann G, De Bernardi B, Kogner P, Ladenstein R, Michon J, Pearson AD, Ambros PF (2003) Quality assessment of genetic markers used for therapy stratification. J Clin Oncol 21: 2077–2084 [DOI] [PubMed] [Google Scholar]
- Ambros IM, Zellner A, Roald B, Amann G, Ladenstein R, Printz D, Gadner H, Ambros PF (1996) Role of ploidy, chromosome 1p, and Schwann cells in the maturation of neuroblastoma [see comments]. N Engl J Med 334: 1505–1511 [DOI] [PubMed] [Google Scholar]
- Ambros PF, Ambros IM (2001) Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 37: 492–504 [DOI] [PubMed] [Google Scholar]
- Ambros PF, Ambros IM, Strehl S, Bauer S, Luegmayr A, Kovar H, Ladenstein R, Fink FM, Horcher E, Printz G, Mutz I, Schilling F, Urban C, Gadner H (1995) Regression and progression in neuroblastoma. Does genetics predict tumour behaviour? Eur J Cancer 31A: 510–515 [DOI] [PubMed] [Google Scholar]
- Ambros PF, Mehes G, Hattinger C, Ambros IM, Luegmayr A, Ladenstein R, Gadner H (2001) Unequivocal identification of disseminated tumor cells in the bone marrow by combining immunological and genetic approaches—functional and prognostic information. Leukemia 15: 275–277 [DOI] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A, Seeger RC (2006) Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst 98: 1193–1203 [DOI] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, Brodeur GM, Cohn SL, Matthay KK, Maris JM (2005) Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med 353: 2243–2253 [DOI] [PubMed] [Google Scholar]
- Bilke S, Chen QR, Whiteford CC, Khan J (2005) Detection of low level genomic alterations by comparative genomic hybridization based on cDNA micro-arrays. Bioinformatics 21: 1138–1145 [DOI] [PubMed] [Google Scholar]
- Bourdeaut F, Ribeiro A, Paris R, Pierron G, Couturier J, Peuchmaur M, Delattre O (2008) In neuroblastic tumours, Schwann cells do not harbour the genetic alterations of neuroblasts but may nevertheless share the same clonal origin. Oncogene 27: 3066–3071 [DOI] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Lastowska M, O′Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, Laureys G, Speleman F (1999) Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma (see comments). N Engl J Med 340: 1954–1961 [DOI] [PubMed] [Google Scholar]
- Brodeur GM (2003) Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 3: 203–216 [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM (1984) Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Caron H (1995) Allelic loss of chromosome 1 and additional chromosome 17 material are both unfavourable prognostic markers in neuroblastoma. Med Pediatr Oncol 24: 215–221 [DOI] [PubMed] [Google Scholar]
- Caron H, van SP, Buschman R, Pereira-do TR, Maes P, Beks L, de KJ, Voute PA, Vergnaud G, Westerveld A, Slater R, Versteeg R (1996a) Allelic loss of the short arm of chromosome 4 in neuroblastoma suggests a novel tumour suppressor gene locus. Hum Genet 97: 834–837 [DOI] [PubMed] [Google Scholar]
- Caron H, van SP, de KJ, Bokkerink J, Egeler M, Laureys G, Slater R, Westerveld A, Voute PA, Versteeg R (1996b) Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma (see comments). N Engl J Med 334: 225–230 [DOI] [PubMed] [Google Scholar]
- Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, Mosseri V, Simon T, Garaventa A, Castel V, Matthay KK (2009) The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D′Angio GJ, Evans AE, Koop CE (1971) Special pattern of widespread neuroblastoma with a favourable prognosis. Lancet 1: 1046–1049 [DOI] [PubMed] [Google Scholar]
- De Preter K, Speleman F, Combaret V, Lunec J, Laureys G, Eussen BH, Francotte N, Board J, Pearson AD, De Paepe A, Van Roy N, Vandesompele J (2002) Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod Pathol 15: 159–166 [DOI] [PubMed] [Google Scholar]
- Evans AE, Gerson J, Schnaufer L (1976) Spontaneous regression of neuroblastoma. Natl Cancer Inst Monogr 44: 49–54 [PubMed] [Google Scholar]
- Frostad B, Martinsson T, Tani E, Falkmer U, Darnfors C, Skoog L, Kogner P (1999) The use of fine-needle aspiration cytology in the molecular characterization of neuroblastoma in children. Cancer 87: 60–68 [PubMed] [Google Scholar]
- George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, Look AT, Maris JM (2007) Genome-Wide Analysis of Neuroblastomas using High-Density Single Nucleotide Polymorphism Arrays. PLoS ONE 2: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Kenyon R, McGuckin AG, Kohl N, Kogner P, Christiansen H, Pearson AD, Lunec J (1997) Analysis of candidate gene co-amplification with MYCN in neuroblastoma. Eur J Cancer 33: 2037–2042 [DOI] [PubMed] [Google Scholar]
- George RE, London WB, Cohn SL, Maris JM, Kretschmar C, Diller L, Brodeur GM, Castleberry RP, Look AT (2005) Hyperdiploidy plus nonamplified MYCN confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol 23: 6466–6473 [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A, Plantaz D, Rubie H, Valteau-Couanet D, Thomas C, Combaret V, Rousseau R, Eggert A, Michon J, Speleman F, Delattre O (2009) Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol 27(7): 1026–1033 [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Kanda N, Maseki N, Sakurai M, Tsuchida Y, Takeda T, Okabe I (1987) Different karyotypic patterns in early and advanced stage neuroblastomas. Cancer Res 47: 311–318 [PubMed] [Google Scholar]
- Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53: 457–481 [Google Scholar]
- Ladenstein R, Ambros IM, Potschger U, Amann G, Urban C, Fink FM, Schmitt K, Jones R, Slociak M, Schilling F, Ritter J, Berthold F, Gadner H, Ambros PF (2001) Prognostic significance of DNA di-tetraploidy in neuroblastoma. Med Pediatr Oncol 36: 83–92 [DOI] [PubMed] [Google Scholar]
- Lastowska M, Viprey V, Santibanez-Koref M, Wappler I, Peters H, Cullinane C, Roberts P, Hall AG, Tweddle DA, Pearson AD, Lewis I, Burchill SA, Jackson MS (2007) Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene 26: 7432–7444 [DOI] [PubMed] [Google Scholar]
- Look AT, Hayes FA, Nitschke R, McWilliams NB, Green AA (1984) Cellular DNA content as a predictor of response to chemotherapy in infants with unresectable neuroblastoma. N Engl J Med 311: 231–235 [DOI] [PubMed] [Google Scholar]
- Look AT, Hayes FA, Shuster JJ, Douglass EC, Castleberry RP, Bowman LC, Smith EI, Brodeur GM (1991) Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol 9: 581–591 [DOI] [PubMed] [Google Scholar]
- Manohar CF, Salwen HR, Brodeur GM, Cohn SL (1995) Co-amplification and concomitant high levels of expression of a DEAD box gene with MYCN in human neuroblastoma. Genes Chromosomes Cancer 14: 196–203 [DOI] [PubMed] [Google Scholar]
- Maris JM, Hii G, Gelfand CA, Varde S, White PS, Rappaport E, Surrey S, Fortina P (2005) Region-specific detection of neuroblastoma loss of heterozygosity at multiple loci simultaneously using a SNP-based tag-array platform. Genome Res 15: 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, White PS, Beltinger CP, Sulman EP, Castleberry RP, Shuster JJ, Look AT, Brodeur GM (1995) Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res 55: 4664–4669 [PubMed] [Google Scholar]
- Mehes G, Luegmayr A, Ambros IM, Ladenstein R, Ambros PF (2001) Combined automatic immunological and molecular cytogenetic analysis allows exact identification and quantification of tumor cells in the bone marrow. Clin.Cancer Res. 7: 1969–1975 [PubMed] [Google Scholar]
- Michels E, Vandesompele J, De Preter K, Hoebeeck J, Vermeulen J, Schramm A, Molenaar JJ, Menten B, Marques B, Stallings RL, Combaret V, Devalck C, De Paepe A, Versteeg R, Eggert A, Laureys G, Van Roy N, Speleman F (2007) ArrayCGH-based classification of neuroblastoma into genomic subgroups. Genes Chromosomes Cancer 46: 1098–1108 [DOI] [PubMed] [Google Scholar]
- Mosse YP, Diskin SJ, Wasserman N, Rinaldi K, Attiyeh EF, Cole K, Jagannathan J, Bhambhani K, Winter C, Maris JM (2007) Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer 46: 936–949 [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, Ernestus K, Konig R, Haas S, Eils R, Schwab M, Brors B, Westermann F, Fischer M (2006) Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol 24: 5070–5078 [DOI] [PubMed] [Google Scholar]
- Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, Hirata T, Kubo H, Goto T, Yamada S, Yoshida Y, Fuchioka M, Ishii S, Nakagawara A (2005) Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell 7: 337–350 [DOI] [PubMed] [Google Scholar]
- Perez CA, Matthay KK, Atkinson JB, Seeger RC, Shimada H, Haase GM, Stram DO, Gerbing RB, Lukens JN (2000) Biologic variables in the outcome of stages I and II neuroblastoma treated with surgery as primary therapy: a children′s cancer group study. J Clin Oncol 18: 18–26 [DOI] [PubMed] [Google Scholar]
- Peto R, Peto J (1972) Asymptotically efficient rank invariant test procedures. J R Stat Soc A 135: 185–198 [Google Scholar]
- Qualman SJ, Bowen J, Fitzgibbons PL, Cohn SL, Shimada H (2005) Protocol for the examination of specimens from patients with neuroblastoma and related neuroblastic tumors. Arch Pathol Lab Med 129: 874–883 [DOI] [PubMed] [Google Scholar]
- Schleiermacher G, Michon J, Huon I, d′Enghien CD, Klijanienko J, Brisse H, Ribeiro A, Mosseri V, Rubie H, Munzer C, Thomas C, Valteau-Couanet D, Auvrignon A, Plantaz D, Delattre O, Couturier J (2007) Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer 97: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, Schulte JH, Klein-Hitpass L, Havers W, Sieverts H, Berwanger B, Christiansen H, Warnat P, Brors B, Eils J, Eils R, Eggert A (2005) Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene 24: 7902–7912 [DOI] [PubMed] [Google Scholar]
- Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J (1983) Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305: 245–248 [DOI] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D (1985) Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 313: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL (2005) Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 44: 305–319 [DOI] [PubMed] [Google Scholar]
- Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, Castleberry RP (1999) The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 86: 364–372 [PubMed] [Google Scholar]
- Spitz R, Hero B, Ernestus K, Berthold F (2003) Gain of distal chromosome arm 17q is not associated with poor prognosis in neuroblastoma. Clin Cancer Res 9: 4835–4840 [PubMed] [Google Scholar]
- Spitz R, Oberthuer A, Zapatka M, Brors B, Hero B, Ernestus K, Oestreich J, Fischer M, Simon T, Berthold F (2006) Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer 45: 1130–1142 [DOI] [PubMed] [Google Scholar]
- Squire JA, Thorner PS, Weitzman S, Maggi JD, Dirks P, Doyle J, Hale M, Godbout R (1995) Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene 10: 1417–1422 [PubMed] [Google Scholar]
- Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, O′Meara A, Breatnach F (2006) High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res 66: 3673–3680 [DOI] [PubMed] [Google Scholar]
- Thompson PM, Maris JM, Hogarty MD, Seeger RC, Reynolds CP, Brodeur GM, White PS (2001) Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res 61: 679–686 [PubMed] [Google Scholar]
- Tomioka N, Oba S, Ohira M, Misra A, Fridlyand J, Ishii S, Nakamura Y, Isogai E, Hirata T, Yoshida Y, Todo S, Kaneko Y, Albertson DG, Pinkel D, Feuerstein BG, Nakagawara A (2008) Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene 27: 441–449 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Baudis M, De Preter K, Van Roy N, Ambros P, Bown N, Brinkschmidt C, Christiansen H, Combaret V, Lastowska M, Nicholson J, O’Meara A, Plantaz D, Stallings R, Brichard B, Van den Broecke C, De Bie S, De Paepe A, Laureys G, Speleman F (2005) Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol 23: 2280–2299 [DOI] [PubMed] [Google Scholar]
- Villamon E, Piqueras M, Mackintosh C, Alonso J, de Alava E, Navarro S, Noguera R (2008) Comparison of different techniques for the detection of genetic risk-identifying chromosomal gains and losses in neuroblastoma. Virchows Arch 453: 47–55 [DOI] [PubMed] [Google Scholar]
- Weber A, Imisch P, Bergmann E, Christiansen H (2004) Coamplification of DDX1 correlates with an improved survival probability in children with MYCN-amplified human neuroblastoma. J Clin Oncol 22: 2681–2690 [DOI] [PubMed] [Google Scholar]
- Wei JS, Greer BT, Westermann F, Steinberg SM, Son CG, Chen QR, Whiteford CC, Bilke S, Krasnoselsky AL, Cenacchi N, Catchpoole D, Berthold F, Schwab M, Khan J (2004) Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res 64: 6883–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]