Abstract
Temporal and spatial variations in the concentrations of signaling intermediates in a living cell are important for signaling in complex networks because they modulate the probabilities that signaling intermediates will interact with each other. We have studied 30 signaling sensors, ranging from receptors to transcription factors, in the physiological activation of murine ex vivo T cells by antigen-presenting cells. Spatiotemporal patterning of these molecules was highly diverse and varied with specific T cell receptors and T cell activation conditions. The diversity and variability observed suggest that spatiotemporal patterning controls signaling interactions during T cell activation in a physiologically important and discriminating manner. In support of this, the effective clustering of a group of ligand-engaged receptors and signaling intermediates in a joint pattern consistently correlated with efficient T cell activation at the level of the whole cell.
INTRODUCTION
Cellular activation requires the transmission of signals through complex signaling networks. The ability of signaling intermediates such as receptors, enzymes, or adaptor proteins to interact with each other is encoded by their biochemical properties (1–3). Large-scale protein interaction maps reveal possible interactions within signaling networks (4). In addition, and much less well understood, the concentrations of signaling intermediates vary greatly in time and space inside living cells (5–11). We refer to these variations as “spatiotemporal patterns.” These patterns are likely critical for how signaling is regulated in living cells because enrichment of two proteins at the same location and at the same time increases their probability of interaction. Within this context, small variations in protein concentrations can have important functional consequences. For example, the twofold reduction in protein abundance achieved genetically by loss of a single allele often substantially modulates cellular function (12–14).
As a regulator of protein interaction probabilities, spatiotemporal patterning needs to be studied at the systems level. The spatiotemporal patterning of an individual signaling intermediate only becomes important when it can be compared to the patterns of multiple other components of the same network. Here, we have executed such a systems analysis of the spatiotemporal patterning of T cell activation.
Physiological T cell activation requires a cellular interaction with an antigen-presenting cell (APC) and the engagement of multiple receptors. The T cell receptor (TCR) on CD4+ T cells recognizes antigenic peptide presented by major histocompatability complex (MHC) class II on the surface of the APC (15). Costimulatory receptors, particularly CD28 and the integrin lymphocyte function-associated antigen–1 (LFA-1), enhance T cell activation (16, 17). The ligands for these costimulatory receptors, B7 and intercellular adhesion molecule–1 (ICAM-1), are also found on the surface of the APC. For a number of reasons, T cells are an excellent model in which to study spatiotemporal patterning. Signaling is restricted in time by cell coupling (18–21); receptor-ligand engagement is restricted in space to the T cell–APC interface; and additional patterning occurs within the interface. By studying receptors and signaling intermediates one at a time, numerous investigators have found that some signaling intermediates preferentially accumulate at the center of the T cell–APC interface, whereas other intermediates prefer the edge of the interface (22–34). However, a systems overview that relates all of these individually determined patterns to each other has not emerged from these studies. An important open question for such an overview is how diverse is the spatiotemporal organization of T cell activation.
If the spatiotemporal patterning of different signaling intermediates differed dramatically, then the likelihood of these molecules interacting with each other would vary substantially in time and space during T cell activation. Only then would spatiotemporal patterning be a substantial regulator of T cell activation. We found this to be the case here, where we studied the spatiotemporal patterns of 30 molecules, including receptors, kinases, adaptor proteins, small guanosine triphosphatases (GTPases), and cytoskeletal components, in murine ex vivo (primary) T cells activated by APCs and antigenic peptides. Our data set thus constitutes an important resource for the further investigation of the regulation of T cell signaling. As a first such investigation, we used cluster analysis to identify a group of ligand-engaged receptors and signaling intermediates that exhibited similar patterning. Comparing different T cells and T cell activation conditions, we showed that the joint clustering of these molecules was tightly linked to the efficiency of T cell signaling as determined biochemically at the cellular level. These data thus establish spatiotemporal patterning as an important regulator of the efficiency of T cell signaling during the physiological activation of primary T cells. A second conclusion that emerges from the comparison of different T cell activation conditions is that each condition generated a distinct spatiotemporal signature. Thus, spatiotemporal patterning is a highly discriminating characteristic of the T cell signaling state. Given the universality of spatiotemporal patterning, our findings are likely to apply to numerous other signaling systems.
RESULTS
An experimental system to study spatiotemporal patterning in T cell activation at the systems scale
To investigate spatiotemporal patterning during T cell activation in a physiologically relevant context, we activated murine primary TCR transgenic T cells with professional APCs and antigenic peptide and cultured them briefly in the presence of the T cell growth factor interleukin-2 (IL-2). We then studied the interaction of such “primed” T cells with professional APCs in vitro. Performing experiments in T cells expressing a TCR transgene enabled the engagement of the TCR by its natural ligand peptide–MHC complex, whereas the use of professional APCs provided ligands to as many costimulatory receptors as possible. The signaling intermediates of T cell activation that we chose to image represent central elements of T cell biology (fig. S1). These included receptors, such as TCR, CD28, CD2, LFA-1, Ly108-1, and the CXC chemokine receptor CXCR4; receptor-associated kinases, including ζ chain–associated protein kinase of 70 kD (ZAP-70), phosphatidylinositol 3-kinase (PI3K), and tyrosine kinase expressed in hepatocellular carcinoma (Tec); and proximal adaptor molecules, such as linker of activated T cells (LAT). In addition, we visualized distal signaling intermediates, such as phospholipase C–γ (PLC-γ), that link receptor engagement to key second messengers, as well as molecules linked to receptor internalization [Cbl-interacting protein of 85 kD (Cin85) and CD2-associated protein (CD2AP)], the activation of transcription factors [protein kinase C θ (PKCθ)], and the regulation of the cytoskeleton (the small GTPases Cdc42, Rac, and Rho). We also studied the lipids phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PIP3) and enzymes involved in their turnover phosphatidylinositol phosphate 5-kinase (PIP5K); the transcription factor nuclear factor of activated T cells (NFAT); and components of the T cell cytoskeleton, such as actin and tubulin.
To visualize the patterns of these elements of T cell activation, various types of sensors were used or generated (Table 1). Receptor-ligand pairs were imaged by visualizing the fusion protein of the receptor or its ligand fused to green fluorescent protein (GFP), which yielded data on the distribution of the entire receptor population or of its ligand-bound component, respectively (35). For cytoplasmic proteins, we directly labeled the entire protein or constructed “recruitment sensors” or “binding sensors.” To construct a recruitment sensor, which was done for ZAP-70, PI3K, Tec, Itk (inducible T cell kinase), and PLC-γ, GFP was fused to the domains of the signaling intermediate that mediate binding to its upstream activators. Thus, total cellular enzymatic activity was not increased by abundance of these sensors. A recruitment sensor reports on the availability of binding sites for the signaling intermediate. To construct a binding sensor, which was done for PIP2, PIP3, Cdc42, Rac1, RhoA, and Arf6 (adenosine 5′-diphosphate ribosylation factor 6), GFP was fused to the domain of a downstream effector protein of the signaling intermediate that the effector uses to bind to that signaling intermediate. The binding sensor thus reports on the spatiotemporal patterns of the active form of the signaling intermediate. Although the design of each sensor was undertaken to reflect endogenous signaling activity, this has not been verified for all sensors.
Table 1.
Sensors used for the analysis of signaling intermediates. The table lists all of the sensors used in the first column, grouped by type of molecule, with their domain structure in the middle column and the relevant supplementary movie file listed in the right column.
| Signaling protein | Design | Movie or reference |
|---|---|---|
| Receptors | ||
| TCR | TCRζ-GFP (53 ), MHC-GFP (35 ) | Movies S3, S4 |
| CD3ε-GFP* (N.S.C.v.O.) | Movie S6 | |
| TCRζ 1,2 | TCRζ 1,2-GFP* (N.S.C.v.O.) | |
| TCRζ 1–6 | TCRζ 1-6-GFP* (N.S.C.v.O.) | Movie S5 |
| CD28 | B7-2-GFP (30) | (30) |
| CD2 | CD48-GFP (43) | (43) |
| LFA-1 | ICAM-1-GFP (54) | Movie S7 |
| Ly108-1 | LY108-1-GFP* | Movies S8, S9 |
| CXCR4-GFP | CXCR4-GFP* (N.S.C.v.O.) | Movie S10 |
| Cytoskeleton | ||
| Actin | Actin-GFP (55) | (56) |
| Microtubule organizing center (MTOC) | Tubulin-GFP (55) | (56) |
| Signaling intermediates | ||
| ZAP-70 | ZAP-70 tandem Src homology 2 (SH2)-GFP* | Movie S11 |
| PI3K | PI3K SH2-interSH2-SH2-GFP* | Movie S12 |
| SLAM-associated protein (SAP) | SAP-GFP* | Movie S13 |
| Tec | Tec pleckstrin homology (PH)-Tec homology (TH)-SH3-GFP* | Movie S14 |
| Itk | Itk PH-TH-SH3-SH2-GFP* | Movie S15 |
| PLC-γ | PLC-γ PH-SH2-SH2-GFP* | Movie S16 |
| PKCθ | PKCθ-GFP (A. Altman, personal communication) | Movie S2 |
| PKCη | PKCη-GFP* (G.F. and N.R.J.G.) | Movie S17 |
| LAT | LAT-GFP* | Movie S1 |
| Cin85 | GFP-Cin85* | Movie S18 |
| CD2AP | GFP-CD2AP* | Movie S19 |
| PIP5K γ87 | GFP-PIP5K γ87 (H. Yin, personal communication) | Movie S20 |
| PIP2 | GFP-PLC-δ PH (57) | Movie S21 |
| PIP3 | GFP-cytohesin PH* | Movie S22 |
| Cdc42 | Wiskott-Aldrich syndrome protein (WASP) GTPase protein binding domain (GBD)-GFP-Caax (52) | (52) |
| Rac | Plenty of SH3 (POSH) GBD-GFP-Caax* | Movie S23 |
| Rho | Rhotekin GBD-GFP-Caax* | Movie S24 |
| Arf6 | Myr-GFP-Golgi–localizing, γ-adaptin ear homology (GAEH) domain, ADP ribosylation factor (ARF)–binding protein (GGA) 3 GBD* | Movie S25 |
| WASP-family verprolin homologous protein 2 (WAVE2) | WAVE2-GFP (58) | Movie S26 |
| Transcription factors | ||
| NFAT | NFAT-GFP* | Movie S27 |
Previously unpublished sensor.
The various sensors were expressed in primed, primary TCR-transgenic T cells by retroviral transduction (36). These T cells were then sorted by flow cytometry (fig. S2) such that only those cells within a fivefold range of low abundance of GFP, which matched the high end of microscope sensitivity, were collected. As determined by quantitative Western blotting against purified GFP (fig. S3), this corresponded to (0.63 ± 0.12) × 106 molecules per T cell, a concentration of 4.8 ± 0.9 μM. With PKCθ as an example, we have related the sensor abundance to endogenous protein abundance. With quantitative immunoblotting against purified PKCθ (fig. S3), we determined an endogenous concentration of 2.5 ± 0.5 μM. Because the abundance of the sensor can be comparable to that of the endogenous protein, use of the lowest feasible concentration of sensor is critical to minimize interference of T cell activation by the abundance of the sensor. As a sensitive assay for this type of interference, we always determined the frequency of primary T cells that coupled to an APC upon contact. Fairly modest interference with T cell activation, caused by reductions in the concentration of agonist peptide or blockade of ligands for individual costimulatory receptors, affects T cell–APC coupling (37). Notably, the abundance in T cells of our sensors did not influence coupling.
To image the spatiotemporal patterns of the sensors, resting, transduced T cells were reactivated by incubation with APCs that had been exposed to the appropriate antigenic peptide. The distributions of sensors were determined in three dimensions as a function of time (image acquisition every 20 s for 15 min) (Fig. 1 and supplementary methods). This time range coincides with the peak of biochemically detectable signaling (18, 19, 38, 39). This was also the time frame of the translocation of the critical transcription factor NFAT to the nucleus (fig. S4), thus linking proximal signaling to the initiation of a transcriptional response. The data set contains 2945 cell couples, with an average of 51 T cell–APC couples for each experimental condition. For the determination of spatiotemporal patterns as described in detail in the supplementary methods, any accumulation of sensor at ≥135% of the background cellular fluorescence was classified into six spatial patterns (Fig. 2) at 12 time points (Fig. 1). The five mutually exclusive interface patterns were accumulation at the center of the T cell–APC interface (central), accumulation at the periphery of the interface (peripheral), accumulation that covered central and peripheral regions (diffuse), accumulation in a large T cell invagination (invagination), and accumulation in a broad interface lamellum (lamellum). In addition, accumulation at the T cell distal pole (distal) was scored separately (Fig. 2).
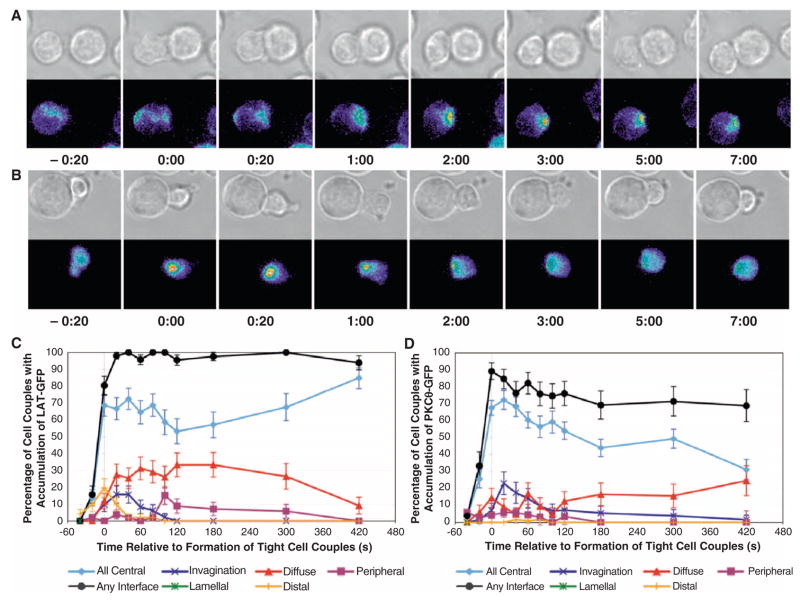
Fig. 1.
T cell–APC coupling triggers the rapid accumulation of sensors at the cellular interface in distinct patterns. Representative interactions of LAT-GFP–transduced (A) and PKCθ-GFP–transduced (B) 5C.C7 T cells with CH27 B cell lymphoma APCs in the presence of 10 μM MCC antigenic peptide (full stimulus) are shown at the indicated time points (in minutes) relative to the time of formation of a tight cell couple. Differential interference contrast (DIC) images are shown in the top rows, with top-down, maximum projections of three-dimensional LAT-GFP or PKCθ-GFP fluorescence data in the bottom rows. The LAT-GFP or PKCθ-GFP fluorescence intensities are displayed in a rainbow-like false-color scale (increasing from blue to red). Movies covering the entire time frame are available as movies S1 and S2, respectively. (C and D) The graphs display the percentage of cell couples that displayed accumulation of signaling intermediates with the indicated patterns (Fig. 2) relative to tight cell couple formation for T cells transduced with LAT-GFP (C) or PKCθ-GFP (D). Fifty-one cell couples were analyzed for the accumulation of LAT-GFP, whereas 111 cell couples were analyzed for the accumulation of PKCθ-GFP.
Fig. 2.
Spatiotemporal patterns of accumulation of sensors. Definitions of patterns, schematic representations, representative images, and associated supplementary movie files that prominently display the patterns are given. The en face view looks at the T cell–APC interface in the same way that an APC would. In the schemes, the outer gray circle delineates the entire interface, whereas the inner circle denotes its center. The top-down view looks down on the T cell from the top. In the schemes, the APC is not shown and would be on top of the T cell forming the flat T cell–APC interface.
As an example of how to determine a quantitative understanding of spatiotemporal patterning, we imaged the accumulation of PKCθ-GFP at the T cell–APC interface by spinning disk confocal microscopy, and we quantified the extent of translocation of this sensor during the formation of spatiotemporal patterns in three dimensions. At 1 min after the formation of a tight T cell–APC couple, the peak time of accumulation of PKCθ-GFP (Fig. 1D), the average volume of accumulation was 5.5 ± 0.8% of the entire volume of the T cell. Of the total cellular PKCθ-GFP, 11 ± 1.5% moved into this space, which led to a 3.0 ± 0.1-fold enrichment of this molecule in the accumulation volume and a reduction in the concentration of PKCθ-GFP in the remainder of the cell by only 12%. This enrichment of PKCθ-GFP is comparable to that determined for the TCR in fixed T cell–APC couples (29). Thus, spatiotemporal patterning can substantially enrich a protein in its area of accumulation without leading to substantial depletion of the protein elsewhere. The probability of interaction of two simultaneously accumulated signaling intermediates would thus increase, whereas that of two intermediates with divergent patterns would remain essentially unchanged.
Spatiotemporal patterning in T cell activation is highly diverse and dynamic
We determined the spatiotemporal patterns of 30 signaling components of T cell activation (Table 1) in 5C.C7 TCR transgenic T cells. The 5C.C7 TCR recognizes the moth cytochrome c (MCC) peptide (amino acid residues 89 to 103) as presented to the T cell by the MHC II allele I-Ek on the APC (40). We used CH27 B cell lymphoma APCs in the presence of a high concentration (10 μM) of MCC peptide. With PKCθ as an example, we showed that when primary, bone marrow–derived dendritic cells were used as APCs instead of CH27 cells, we obtained comparable results (fig. S5). Each sensor showed a unique spatiotemporal pattern (Fig. 3 and fig. S6), thus establishing dramatic diversity in the spatiotemporal organization of T cell signaling. Further illustrating this diversity, proteins that are biochemically highly related showed distinct differences in their patterning. For example, accumulation of the tyrosine kinase Itk was well sustained and showed a pronounced preference for the center of the interface in the first 2 min of cell coupling, with additional accumulation covering the entire interface afterwards (fig. S6). Its close homolog Tec, which shares with Itk an identical domain structure, 71% amino acid homology, and 55% amino acid identity, displayed only transient accumulation within the first minute of cell coupling, virtually exclusively at the center of the interface (fig. S6). Similar differences in patterning were observed for CD2AP and Cin85 (fig. S6), PKCθ, and PKCη (fig. S6), and previously for ezrin and moesin (41). Thus, spatiotemporal patterning might be particularly important in enhancing the functional distinction between two proteins when their biochemical properties are highly similar. Spatiotemporal patterns were also dynamic; for PKCθ, active Cdc42, and active Rac1, 9 to 74% of central patterns lasted for ≤1 min at any given time point (fig. S7). This indicates the high rate of turnover within the areas of accumulation and thus the substantial potential for dynamic regulation of the likelihood with which signaling intermediates would interact with each other.
Fig. 3.
Spatiotemporal patterning in T cell activation is highly diverse. In the interaction of primary 5C.C7 T cells with CH27 APCs in the presence of 10 μM MCC antigenic peptide the percentage occurrence of each of the six accumulation patterns (as defined in Fig. 2) was determined for 12 time points (Fig. 1) for 30 molecules involved in T cell activation (Table 1). Raw pattern analysis data are given in fig. S6. Here, patterns are graphically represented at 8 of the 12 time points analyzed as indicated at the right side of the panels in minutes. Gray denotes the outline of the T cell; the APC is not shown, but would be on top of the T cell. A large T cell invagination (43) is shown at early time points. Each signaling component of T cell activation is denoted by a unique, random color. To avoid crowding of panels, the 28 signaling components displayed were split into six panels at each time point. At each time point, the dominant accumulation pattern of a sensor was determined as described in the supplementary methods and is displayed in light or dark shades of that sensor’s color according to its dominance. If more than one sensor displays the same dominant pattern in a given panel, sensors are displayed as concentric pattern symbols in random order. An average of 53 cell couples (from a total of 1650 cell couples) were analyzed for each sensor (table S2).
With its diversity in time and space, spatiotemporal patterning will regulate the probabilities of interactions between signaling intermediates and thus the efficiency of T cell signaling itself. Our systems-level description of spatiotemporal patterning in T cell activation thus constitutes an important resource for the further investigation of T cell signaling. To allow investigators to mine this resource, the raw data and our analysis thereof will be made available upon request. To illustrate the usefulness of the data set, we addressed three prototypical questions here. First, can our systems data contribute to understanding the functions of molecular motifs? Varying the conditions of T cell activation leads to the differential phosphorylation of tyrosine residues in the ζ chain of the TCR-CD3 complex (42). Using mutants of some of these tyrosine residues, we could show that they served to control the targeting of the TCR to areas of varying signaling activity (supplementary text and fig. S8). Second, can our systems-level data contribute to understanding cellular structures? We have recently described a large T cell invagination that occurs immediately after formation of the T cell–APC couple (43). Selective enrichment of active Rho in the invagination, as discovered in our systems analysis (fig. S9), led us to establish that Rho was required for the dissolution of the T cell invagination (supporting text and fig. S10). Third, can our data help us to identify groups of signaling intermediates whose joint patterning is critical for the regulation of T cell signaling? We will address this question in the remainder of the manuscript.
Formation of the cSMAC signaling complex is related to efficient T cell signaling
The frequency of occurrence of each pattern at each time point (fig. S6) was used in a cluster analysis to determine which sensors showed the greatest similarity in their patterning (Fig. 4 and fig. S11). This analysis identified a group of closely related signaling intermediates, including ZAP-70, LAT, PKCθ, Itk, PLC-γ, Rac, and Rho, that associated with ligand-bound receptors, a complex that we call the “cSMAC signaling complex.” cSMAC stands for central supramolecular activation cluster (22, 29). The existence of this complex identifies the center of the T cell–APC interface as an area of active signaling on the time scale studied and for the activation of primed, primary T cells by professional APCs (see Discussion).
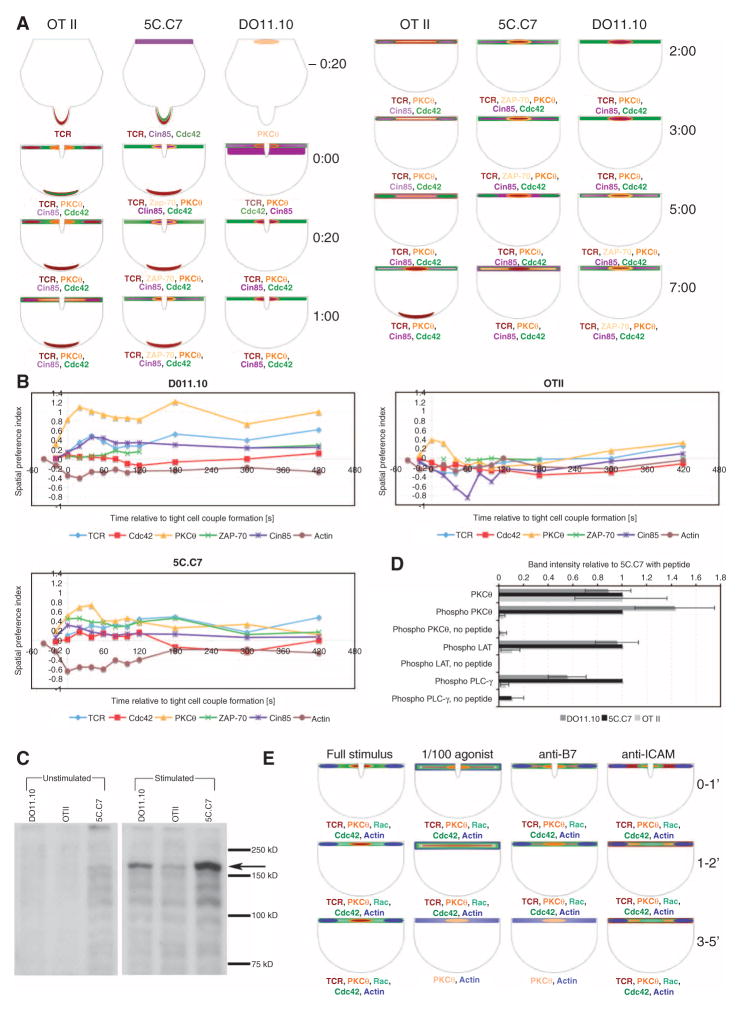
Fig. 4.
Cluster analysis of spatiotemporal patterning. Cluster analysis of the data presented in Fig. 3 and fig. S6 based on the five mutually exclusive interface patterns [central (C), invagination (Inv), diffuse (D), peripheral (P), and lamellum (L); Fig. 2] is given. The percentage occurrence of each pattern is given in shades of red from C-40 to L420 in the top part of the figure. In addition, to address the rate of pattern change, the percentage change per 20-s interval was tabulated (C-40 to L300 in the bottom part of the figure). Red indicates an increase and green a decrease in the percentage occurrence of a pattern relative to the previous time point. Cluster analysis based on Pearson’s correlation was performed on the entire data set and the cluster tree is given in pink. To assess reliability in the determination of pattern similarities, TCR clustering was determined with three sensors. ‘TCR’ refers to TCRζ-GFP, ‘TCR eng’ to ligand-engaged TCR, as determined by MHC II–GFP binding, and CD3e to CD3ε-GFP. The spatiotemporal patterns of these three TCR sensors were closely related, indicating high reliability in pattern determination. Relationships between signaling intermediates were not substantially changed by either including distal accumulation patterns or removing rates of pattern change (fig. S11), thus supporting the robust nature of the cluster analysis.
To assess the generality of the spatiotemporal patterning of signaling molecules during T cell signaling and that of the formation of the cSMAC signaling complex in particular, we determined the patterning of several sensors in the activation of primary T cells from two other TCR transgenic mouse models. The DO11.10 TCR recognizes the ovalbumin peptide (residues 324 to 340) presented by MHC II allele I-Ad (44), whereas the OTII TCR recognizes the same ovalbumin peptide when presented by MHC II allele I-Ab (45). As before, primary, in vitro primed T cells were retrovirally transduced, and transduced T cells that were sorted by fluorescence-activated cell sorting (FACS) were reactivated by professional APCs in the presence of a high concentration (10 μM) of antigenic peptide. A20 B cell lymphoma cells were used to activate the DO11.10 T cells, and interferon-γ (IFN-γ)–activated DC2.4 dendritic cells were used to activate the OTII T cells (46).
Comparison of the spatiotemporal patterning of signaling intermediates in the activation of 5C.C7, DO11.10, and OTII T cells revealed both similarities and differences (Fig. 5A and fig. S12). One similarity was the relative preference of a biosensor for the center or periphery of the T cell–APC interface. For example, PKCθ was always the most central sensor (Fig. 5B and fig. S12); however, the extent of segregation of signaling intermediates between the center and periphery of the T cell–APC interface differed dramatically between T cell types. With DO11.10 T cells, virtually all of the sensors showed a strong preference for recruitment to either the center or the periphery of the T cell–APC interface. In contrast, in OT II T cells only modest preferences were found; most sensors simultaneously covered parts of the center and the periphery of the interface at many time points (Fig. 5B and fig. S12). 5C.C7 T cells displayed an intermediate type of patterning, although this was closer to the DO11.10 phenotype than to that of the OT II cells. These data suggest that although signaling intermediates are sorted with similar spatial preferences, that is, closer to the center or periphery in all T cells, the extent of spatial segregation within the interface, including the formation of the cSMAC signaling complex, varied dramatically.
Fig. 5.
Formation of the cSMAC signaling complex is related to the efficiency of T cell signaling. (A) Spatiotemporal patterning of a subset of indicated sensors in the interaction of primary DO11.10, OTII, and 5C.C7 T cells with professional APCs in the presence of a 10 μM antigenic peptide is displayed similarly to that in Fig. 3. Raw pattern analysis data and data statistics are given in fig. S12. (B) The spatial preference index as defined in the supplementary methods is given for the data in (A) to illustrate differential centrality of patterning. (C and D) DO11.10, OTII, and 5C.C7 T cells, as indicated, were stimulated by professional APCs as described above in the presence of 10 μM antigenic peptide in a cell pellet for 5 min. As a control, antigenic peptide was omitted (“unstimulated”). T cell–APC extracts were analyzed by Western blotting for the presence of LAT-Y191, PLC-γ-Y783, and PKCθ-T538, as indicated. In (C), a representative blot for PLC-γ-Y783 (arrow) is shown. Both parts of the panel are derived from the same blot; with only the intermittent, unrelated lanes removed. In (D), band intensities relative to the 5C.C7 band for all blots are given with their standard errors. (E) Spatiotemporal patterning of a subset of indicated sensors in the interaction of primary 5C.C7 T cells with professional CH27 APCs under varying conditions of T cell activation conditions, as noted, is displayed similarly to that in Fig. 3. Raw pattern analysis data for hitherto unpublished data and data statistics are given in fig. S14.
Beyond its inherent interest (see Discussion), this variability in spatiotemporal patterning allowed us to address the importance of the cSMAC signaling complex by examining whether its formation was related to the efficiency of signaling at the level of the whole cell. We found that it was. First, we assessed phosphorylation of PKCθ, PLC-γ, and LAT as markers of their activation in T cell–APC extracts (Fig. 5, C and D). To standardize cell-couple formation, we forced contact between T cells and APCs by pelleting T cells with an excess of APCs during T cell activation. DO11.10 and 5C.C7 T cells showed substantial peptide-induced phosphorylation of PKCθ, PLC-γ, and LAT, whereas OTII T cells did not (Fig. 5, C and D). Interestingly, these pronounced differences in proximal T cell signaling did not necessarily extend to transcriptional regulation. Translocation of NFAT to the nucleus upon T cell activation exhibited similar kinetics in 5C.C7 and OTII T cells (fig. S4), as discussed below.
Second, we found that the ability of a T cell to couple with an APC upon transient contact varied in parallel with the later ability of the T cell to form a cSMAC signaling complex. Upon transient contact, 78.5 ± 2% of DO11.10 T cells coupled with peptide-incubated APCs, whereas 64 ± 3% of 5C.C7 T cells and only 42.5 ± 2.5% of OTII T cells did. Thus, the first stages of the segregation of signaling intermediates at the small initial contact point between the T cell and the APC may already regulate the efficiency of T cell signaling.
Third, we took the same DO11.10 TCR transgenic T cells and either differentiated them in vitro into cells with a T helper type 2 (TH2) effector phenotype or left them under neutral conditions (TH0). The reduced signaling activity of TH2 cells upon reactivation compared to TH1 and TH0 cells is already established (47). In addition, phosphorylation of LAT and PLC-γ in response to peptide-loaded APCs was reduced by 20% in TH2 T cells compared to that in TH0 cells. Through imaging of TH2 and TH0 DO11.10 T cells under identical conditions (activation by A20 B cell lymphoma cells in the presence of 10 μM Ova peptide), we determined the patterning of a critical component of the cSMAC signaling complex, the TCR (fig. S13). Although the percentage of cell couples that showed accumulation of the TCR at the T cell–APC interface did not differ, accumulation at the center of the interface was substantially reduced in the TH2 cells. The consistent relationship between the formation of the cSMAC signaling complex and efficient T cell signaling at the level of the whole cell strongly suggests that the cSMAC signaling complex is a critical regulator of the efficacy of T cell signaling. Moreover, the variability of spatiotemporal patterning establishes it as a discriminating descriptor of the state of T cell signaling.
TCR signal strength and costimulation selectively regulate spatiotemporal patterning during T cell activation
Next, we tested whether physiological changes in the stimulus of the T cell yielded substantial changes in patterning (Fig. 5E and fig. S14). Upon reactivation of sensor-expressing primary 5C.C7 T cells, we reduced the strength of the T cell stimulus by either decreasing the concentration of antigenic peptide 100-fold or by blocking ligands of either of two central costimulatory receptors, ICAM-1, the ligand for LFA-1, and B7-1 and B7-2, the ligands for CD28. Through the use of several sensors, we found distinct spatiotemporal signatures for all three conditions of T cell activation. Blocking B7-1 and B7-2 dramatically impaired sustained (>2 min) accumulation of sensors while leaving initial patterning intact (Fig. 5E and fig. S14). Reducing the concentration of antigenic peptide 100-fold interfered with the initial segregation of sensors into either central or peripheral patterns and impaired sustained (>2 min) accumulation of sensors (Fig. 5E and fig. S14). Blocking ICAM-1 did not affect the overall accumulation of sensors with any given pattern (with the exception of active Cdc42), yet it impaired the accumulation of sensors at the center of the T cell–APC interface (with the exception of active Rac) (Fig. 5E and fig. S14). These dramatic and distinct pattern changes upon physiological variation of the T cell stimulus corroborate an important and discriminating role of spatiotemporal patterning in the regulation of T cell signaling.
DISCUSSION
Spatiotemporal patterning in T cell activation is highly diverse
We performed a systems-level investigation of spatiotemporal patterning in the physiological activation of primary T cells by professional APCs. Our principal finding is that spatiotemporal patterning was highly diverse. The probabilities of interaction of any two signaling intermediates thus varied dramatically in time and space; as such, spatiotemporal patterning will likely critically regulate T cell signaling. Our data thus constitute an important resource for the further investigation of T cell signaling. The extent of protein redistribution within the T cell was modest; only about 10% of each sensor was moved to the area of accumulation. Although this movement led to a substantial, about threefold enrichment, in the concentration of the sensor in the area of accumulation, little change in the concentration of signaling intermediates occurred in the remainder of the cell. Spatiotemporal patterning thus is not likely to determine whether a signaling interaction occurs; instead, it may govern how likely the interaction is to occur. Cellular function will therefore be modulated rather than enabled or disabled as a consequence of changes in spatiotemporal patterning. The consistent relationship between the occurrence of a specific pattern in T cell activation (the cSMAC signaling complex) and the efficiency of T cell signaling as determined biochemically at the level of the whole cell strongly suggests that such modulation is physiologically important. Modulation of cellular function in general is critical under various physiological circumstances, for example, in cancer, metabolic regulation, or in finding a balance between an effective immune response to infection and autoimmunity. Understanding the regulation of highly diverse spatiotemporal patterning thus should affect numerous major areas of current biology.
Spatiotemporal patterning in T cell activation is variable
By investigating spatiotemporal patterning with a subset of sensors under various conditions, we found that spatiotemporal patterning was highly variable. This variability establishes spatiotemporal patterning as a highly discriminating characteristic of the cellular signaling state. This is of practical relevance in comparing data obtained from different experimental systems. We identified two major areas of variability. First, T cells expressing different TCRs segregated signaling intermediates within the T cell–APC interface in a highly variable manner even though all T cells were activated with a saturating stimulus, namely, professional APCs incubated with a high concentration of antigenic peptide. It will be of interest to understand how differential segregation of signaling intermediates is used as a mechanism to control T cell reactivity. In practical terms, spatiotemporal patterns of individual signaling intermediates will have to be interpreted within the systems context of the same TCR transgenic experimental system. Second, spatiotemporal patterning varied strongly as a function of the T cell stimulus. Reducing the concentration of the antigenic peptide or blocking individual costimulatory ligands each yielded distinct changes in spatiotemporal patterning. Great care has to be taken to make T cell activation conditions as similar as possible when comparing different experimental systems.
Two additional areas of variability seem likely. Naïve T cells may display spatiotemporal patterning that is different from those of the primed T cells used here, because they are more dependent on costimulation for efficient activation and because they are only about half the size of primed T cells. The common use of supported lipid bilayers as APC substitutes may alter spatiotemporal patterning, because the rigidity of the supported lipid bilayer likely interferes with T cell membrane dynamics at the nanometer scale in the segregation of receptor-ligand couples by size (35) and at the micrometer scale in macro-endocytic events, for example, the large T cell invagination that occurs immediately after formation of the T cell–APC couple (43). Thus, although the great variability in spatiotemporal patterning constitutes a rich source for improved understanding of signal transduction at the systems level, it also necessitates paying close attention to the specific cellular activation conditions used when comparing patterning data across experimental systems.
The center of the T cell–APC interface as a site of active signaling
Cluster analysis of spatiotemporal patterning in T cell activation identified the cSMAC signaling complex, thus characterizing the center of the T cell–APC interface as an area of active signaling. More importantly, formation of the cSMAC signaling complex was consistently related to efficient T cell signaling at the whole-cell level, implying that this relationship is likely to be physiologically relevant.
The suggestion of the center of the T cell–APC interface as a site of active signaling confirms the original hypothesis (29); however, it might seem at odds with recent data that suggest that TCR signaling occurs predominantly at the periphery and not at the center of the T cell–APC interface. We suggest that these results are not necessarily in conflict. When T cell activation is triggered by supported lipid bilayers and imaged selectively at the bilayer-T cell interface (33, 48), TCR engagement is initiated in microclusters at the periphery of the interface. These clusters then move toward the center of the interface where TCRs eventually accumulate, leaving relatively few proximal signaling intermediates. With the comparatively lower concentrations of antigenic peptide and the 3D imaging that we have used here, it seems unlikely that we could consistently observe peripheral microclusters. We found, however, that the TCR was relatively slow in accumulating at the center of the T cell–APC interface (fig. S15), consistent with initial engagement of the TCR in the periphery. In addition, the simultaneous occurrence of removal of the TCR from the distal pole and its increased accumulation at the center of the T cell–APC interface (fig. S6) suggests that substantial amounts of TCR could travel through the periphery of the interface on their way from the distal pole to the center. Moreover, although accumulation of the TCR at the center of the T cell–APC interface increased over time, accumulation of the associated signaling intermediates ZAP-70 and PKCθ declined (fig. S15). The intensity of signaling at the center of the T cell–APC interface thus diminished; however, it did not terminate. We suggest that the flexibility of the APC membrane allows continuous internalization of spent receptor-ligand couples at the center of the T cell–APC interface, thus limiting the accumulation of inert complexes and retaining the cSMAC as a site of active signaling. In support of this, central accumulation of Cin85 and CD2AP, the critical adaptors that link ligand-engaged TCR to the endocytic machinery, was seen at all time points (fig. S6), supporting earlier mathematical models of T cell signaling (49). Thus, whereas signaling certainly occurred at all locations within the T cell–APC interface, signaling at the center of the interface was most prominent.
The relationship between central patterning and efficient proximal signaling raises two questions: What are the consequences for distal signaling? What are the consequences for T cell function? Modest alterations in proximal signaling as regulated by spatiotemporal patterning do not necessarily need to result in changes in distal signaling: The dynamics of the translocation of NFAT to the nucleus were comparable in T cells from two TCR transgenic mice with divergent overall spatiotemporal organization, OTII and 5C.C7 (Fig. 5 and fig. S4). We suggest that such behavior occurs when a distal signaling event is threshold-dependent. If the proximal signaling strength is well above the threshold to trigger a distal signaling response, for example, with a strong stimulus as in the current study, then modest alterations in proximal signaling will likely remain inconsequential. If the proximal signaling strength is close to the threshold, however, then alterations in distal signaling will likely occur. To test this hypothesis, we addressed elevations in the concentration of intracellular Ca2+ as the immediate prerequisite for the nuclear translocation of NFAT in TH0 as compared to TH2 T cells. Whereas TH0 cells displayed effective formation of the cSMAC signaling complex, TH2 cells were less impressive (fig. S13). Further reduction of signaling centrality by a deficiency in Itk remained without substantial consequences for Ca2+ signaling in TH0 T cells. However, Ca2+ signaling was abrogated in Itk-deficient TH2 T cells.
A central element in the function of primed CD4+ T cells is to provide help to B cells and macrophages. This includes transcriptional responses, for example, sustained production of cytokines, and short-term cell biological responses, for example, directed cytokine secretion or effective cell coupling to enable receptor-ligand engagement at the resulting cellular interface. Spatiotemporal patterning is related to both types of processes. Primed Itk-deficient TH2 cells, in parallel with their severe impairment in establishing signaling centrality, cannot effectively induce the expression of messenger RNAs for the TH2-specific transcription factor GATA3 and various TH2-type cytokines, such as IL-4, upon restimulation (44). Similarly, effective receptor polarization is related to IL-2 production in rested as compared to anergic T cells and to the functional differentiation of naïve T cells into TH1 cells (50, 51). The actin dynamics that underlie effective cell coupling as a short-term T cell effector function depend on the effective accumulation of active Cdc42 at the center of the T cell–APC interface (52). Although only a systems-level analysis of relationships between spatiotemporal patterning and distal signaling events or T cell function can reliably address the importance of such relationships, the examples provided here strongly suggest that the effects of spatiotemporal patterning extend well beyond the regulation of the efficiency of proximal T cell signaling into various aspects of T cell function.
MATERIALS AND METHODS
Cells and reagents
In vitro primed, primary T cells from 5C.C7, OTII, and DO11.10 TCR transgenic mice (40, 44, 45) were generated as described (30, 43, 52). CH27 cells, IFN-γ–induced DC2.4 cells (46), and A20 cells were used as APCs as previously described (30, 43, 52). Sensors were generated with enhanced GFP (EGFP). Their domain structure is listed in Table 1. GFP was linked with 7 to 10 amino acid residues as encoded by the EGFP-expressing plasmid. Mature primary dendritic cells were prepared by culture of 5C.C7 bone marrow suspensions as described (43). Retroviral transduction was also performed as previously described (30, 43, 52). The concentrations of antigenic peptides were adjusted by dilution into non-antigenic peptide (32). Costimulation blockade with antibodies against ICAM-1 or B7-1 and B7-2 was performed as described (32). The following antibodies were used: antibodies against phospho-LAT Y191, PKCθ, phospho-PKCθ T538, phospho-PLC-γ Y783 (all obtained from Cell Signaling, Danvers, MA), and against GFP (Clontech, Mountain View, CA).
Image acquisition and image analysis
Details of the microscopy system and the techniques used in the acquisition and analysis of images are provided in the supplementary methods.
Biochemical assays
Phosphorylation states of LAT, PLC-γ, and PKCθ were determined by Western blotting analysis of cell extracts from T cell–APC couples that had interacted for 5 min, as previously described (30). The cellular concentrations of GFP fusion proteins and PKCθ were determined from cell extracts (30) generated from transduced and FACS-sorted primary 5C.C7 T cells (fig. S2). Purified GFP and PKCθ proteins were purchased from Biovision (Mountain View, CA) and Cell Signaling, respectively.
Supplementary Material
Fig. S1. Components of T cell activation examined in this study.
Fig. S2. FACS-based sorting of sensor-expressing T cells.
Fig. S3. Quantification of the abundance of biosensors and endogenous PKCθ.
Fig. S4. Rapid nuclear translocation of NFAT.
Fig. S5. Patterning of PKCθ in activated 5C.C7 T cells.
Fig. S6. Highly diverse spatiotemporal patterning during T cell activation.
Fig. S7. Dynamic nature of spatiotemporal patterns.
Fig. S8. Requirement for TCRζ ITAMs for effective recruitment of TCR to the cSMAC signaling complex.
Fig. S9. Enrichment of sensors in the large T cell invagination.
Fig. S10. Requirement for Rho activity for the resolution of the large T cell invagination.
Fig. S11. High diversity of spatiotemporal patterning during T cell activation.
Fig. S12. Variations in spatiotemporal patterning in activated T cells from different TCR transgenic mice.
Fig. S13. Reduced TCR clustering in TH2-polarized DO11.10 T cells.
Fig. S14. Regulation of spatiotemporal patterning by costimulation.
Fig. S15. Delayed central accumulation of the TCR compared to that of its dependent signaling intermediates.
Fig. S16. Similar dynamics of translocation of tubulin-GFP and the Arf6 sensor from the uropod to the interface upon formation of the T cell–APC interface.
Table S1. List of supplementary movies and a key that includes the sensor monitored, the time of cell coupling, and a description of the critical features observed.
Table S2. Numbers of cell couples analyzed.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 2.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 3.Feng S, Kasahara C, Rickles RJ, Schreiber SL. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.Alexander RT, Furuya W, Szászi K, Orlowski J, Grinstein S. Rho GTPases dictate the mobility of the Na/H exchanger NHE3 in epithelia: Role in apical retention and targeting. Proc Natl Acad Sci USA. 2005;102:12253–12258. doi: 10.1073/pnas.0409197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Van Keymeulen A, Wong K, Knight ZA, Govaerts C, Hahn KM, Shokat KM, Bourne HR. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J Cell Biol. 2006;174:437–445. doi: 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malliri A, Collard JG. Role of Rho-family proteins in cell adhesion and cancer. Curr Opin Cell Biol. 2003;15:583–589. doi: 10.1016/s0955-0674(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 12.Gong Q, Cheng AM, Akk AM, Alberola-Ila J, Gong G, Pawson T, Chan AC. Disruption of T cell signaling networks and development by Grb2 haploid insufficiency. Nat Immunol. 2001;2:29–36. doi: 10.1038/83134. [DOI] [PubMed] [Google Scholar]
- 13.Cornall RJ, Cyster JG, Hibbs ML, Dunn AR, Otipoby KL, Clark EA, Goodnow CC. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 14.Satterthwaite AB, Willis F, Kanchanastit P, Fruman D, Cantley LC, Helgason CD, Humphries RK, Lowell CA, Simon M, Leitges M, Tarakhovsky A, Tedder TF, Lesche R, Wu H, Witte ON. A sensitized genetic system for the analysis of murine B lymphocyte signal transduction pathways dependent on Bruton’s tyrosine kinase. Proc Natl Acad Sci USA. 2000;97:6687–6692. doi: 10.1073/pnas.110146697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by αβ T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 16.Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, van der Merwe PA. The nature of molecular recognition by T cells. Nat Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- 17.Sprent J. Presidential address to the American Association of Immunologists. Stimulating naive T cells. J Immunol. 1999;163:4629–4636. [PubMed] [Google Scholar]
- 18.Negulescu PA, Krasieva TB, Khan A, Kerschbaum HH, Cahalan MD. Polarity of T cell shape, motility, and sensitivity to antigen. Immunity. 1996;4:421–430. doi: 10.1016/s1074-7613(00)80409-4. [DOI] [PubMed] [Google Scholar]
- 19.Wei SH, Safrina O, Yu Y, Garrod KR, Cahalan MD, Parker I. Ca2+ signals in CD4+ T cells during early contacts with antigen-bearing dendritic cells in lymph node. J Immunol. 2007;179:1586–1594. doi: 10.4049/jimmunol.179.3.1586. [DOI] [PubMed] [Google Scholar]
- 20.Jacobelli J, Andres PG, Boisvert J, Krummel MF. New views of the immunological synapse: Variations in assembly and function. Curr Opin Immunol. 2004;16:345–352. doi: 10.1016/j.coi.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev. 2008;221:77–89. doi: 10.1111/j.1600-065X.2008.00589.x. [DOI] [PubMed] [Google Scholar]
- 22.Monks CR, Kupfer H, Tamir I, Barlow A, Kupfer A. Selective modulation of protein kinase C-θ during T-cell activation. Nature. 1997;385:83–86. doi: 10.1038/385083a0. [DOI] [PubMed] [Google Scholar]
- 23.Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egen JG, Allison JP. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 26.Freiberg BA, Kupfer H, Maslanik W, Delli J, Kappler J, Zaller DM, Kupfer A. Staging and resetting T cell activation in SMACs. Nat Immunol. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 27.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 28.Lee KH, Holdorf AD, Dustin ML, Chan AC, Allen PM, Shaw AS. T cell receptor signaling precedes immunological synapse formation. Science. 2002;295:1539–1542. doi: 10.1126/science.1067710. [DOI] [PubMed] [Google Scholar]
- 29.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 30.Purtic B, Pitcher LA, van Oers NS, Wülfing C. T cell receptor (TCR) clustering in the immunological synapse integrates TCR and costimulatory signaling in selected T cells. Proc Natl Acad Sci USA. 2005;102:2904–2909. doi: 10.1073/pnas.0406867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer BC, Kappler JW, Kupfer A, Marrack P. Complex and dynamic redistribution of NF-κB signaling intermediates in response to T cell receptor stimulation. Proc Natl Acad Sci USA. 2004;101:1004–1009. doi: 10.1073/pnas.0307858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wülfing C, Sumen C, Sjaastad MD, Wu LC, Dustin ML, Davis MM. Costimulation and endogenous MHC ligands contribute to T cell recognition. Nat Immunol. 2002;3:42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 33.Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, Dustin ML, Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 34.Zaru R, Cameron TO, Stern LJ, Müller S, Valitutti S. Cutting edge: TCR engagement and triggering in the absence of large-scale molecular segregation at the T cell-APC contact site. J Immunol. 2002;168:4287–4291. doi: 10.4049/jimmunol.168.9.4287. [DOI] [PubMed] [Google Scholar]
- 35.Wülfing C, Tskvitaria-Fuller I, Burroughs N, Sjaastad MD, Klem J, Schatzle JD. Interface accumulation of receptor/ligand couples in lymphocyte activation: Methods, mechanisms, and significance. Immunol Rev. 2002;189:64–83. doi: 10.1034/j.1600-065x.2002.18907.x. [DOI] [PubMed] [Google Scholar]
- 36.Costa GL, Benson JM, Seroogy CM, Achacoso P, Fathman CG, Nolan GP. Targeting rare populations of murine antigen-specific T lymphocytes by retroviral transduction for potential application in gene therapy for autoimmune disease. J Immunol. 2000;164:3581–3590. doi: 10.4049/jimmunol.164.7.3581. [DOI] [PubMed] [Google Scholar]
- 37.Tskvitaria-Fuller I, Mistry N, Sun S, Wülfing C. Protein transduction as a means of effective manipulation of Cdc42 activity in primary T cells. J Immunol Methods. 2007;319:64–78. doi: 10.1016/j.jim.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mustelin T, Taskén K. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J. 2003;371:15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wülfing C, Rabinowitz JD, Beeson C, Sjaastad MD, McConnell HM, Davis MM. Kinetics and extent of T cell activation as measured with the calcium signal. J Exp Med. 1997;185:1815–1825. doi: 10.1084/jem.185.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilani T, Khanna C, Zhou M, Veenstra TD, Bretscher A. Immune synapse formation requires ZAP-70 recruitment by ezrin and CD43 removal by moesin. J Cell Biol. 2007;179:733–746. doi: 10.1083/jcb.200707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Oers NS, Tohlen B, Malissen B, Moomaw CR, Afendis S, Slaughter CA. The 21- and 23-kD forms of TCRζ generated by specific ITAM phosphorylations. Nat Immunol. 2000;1:322–328. doi: 10.1038/79774. [DOI] [PubMed] [Google Scholar]
- 43.Singelton K, Parvaze N, Dama KR, Chen KS, Jennings P, Purtic B, Sjaastad MD, Gilpin C, Davis MM, Wülfing C. A large T cell invagination with CD2 enrichment resets receptor engagement in the immunological synapse. J Immunol. 2006;177:4402–4413. doi: 10.4049/jimmunol.177.7.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Au-Yeung BB, Katzman SD, Fowell DJ. Cutting edge: Itk-dependent signals required for CD4+ T cells to exert, but not gain, Th2 effector function. J Immunol. 2006;176:3895–3899. doi: 10.4049/jimmunol.176.7.3895. [DOI] [PubMed] [Google Scholar]
- 45.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 46.He T, Tang C, Xu S, Moyana T, Xiang J. Interferon γ stimulates cellular maturation of dendritic cell line DC2.4 leading to induction of efficient cytotoxic T cell responses and antitumor immunity. Cell Mol Immunol. 2007;4:105–111. [PubMed] [Google Scholar]
- 47.Weber KS, Miller MJ, Allen PM. Th17 cells exhibit a distinct calcium profile from Th1 and Th2 cells and have Th1-like motility and NF-AT nuclear localization. J Immunol. 2008;180:1442–1450. doi: 10.4049/jimmunol.180.3.1442. [DOI] [PubMed] [Google Scholar]
- 48.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supra-molecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J, Kanagawa O, Markiewicz M, Allen PM, Dustin ML, Chakraborty AK, Shaw AS. The immunological synapse balances T cell receptor signaling and degradation. Science. 2003;302:1218–1222. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- 50.Carlin LM, Yanagi K, Verhoef A, Nolte-’t Hoen EN, Yates J, Gardner L, Lamb J, Lombardi G, Dallman MJ, Davis DM. Secretion of IFN-γ and not IL-2 by anergic human T cells correlates with assembly of an immature immune synapse. Blood. 2005;106:3874–3879. doi: 10.1182/blood-2005-03-0996. [DOI] [PubMed] [Google Scholar]
- 51.Maldonado RA, Irvine DJ, Schreiber R, Glimcher LH. A role for the immunological synapse in lineage commitment of CD4 lymphocytes. Nature. 2004;431:527–532. doi: 10.1038/nature02916. [DOI] [PubMed] [Google Scholar]
- 52.Tskvitaria-Fuller I, Seth A, Mistry N, Gu H, Rosen MK, Wülfing C. Specific patterns of Cdc42 activity are related to distinct elements of T cell polarization. J Immunol. 2006;177:1708–1720. doi: 10.4049/jimmunol.177.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krummel MF, Sjaastad MD, Wülfing C, Davis MM. Differential clustering of CD4 and CD3ζ during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 54.Wülfing C, Sjaastad MD, Davis MM. Visualizing the dynamics of T cell activation: Intracellular adhesion molecule 1 migrates rapidly to the T cell/B cell interface and acts to sustain calcium levels. Proc Natl Acad Sci USA. 1998;95:6302–6307. doi: 10.1073/pnas.95.11.6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ballestrem C, Wehrle-Haller B, Hinz B, Imhof BA. Actin-dependent lamellipodia formation and microtubule-dependent tail retraction control-directed cell migration. Mol Biol Cell. 2000;11:2999–3012. doi: 10.1091/mbc.11.9.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tskvitaria-Fuller I, Rozelle AL, Yin HL, Wülfing C. Regulation of sustained actin dynamics by the TCR and costimulation as a mechanism of receptor localization. J Immunol. 2003;171:2287–2295. doi: 10.4049/jimmunol.171.5.2287. [DOI] [PubMed] [Google Scholar]
- 57.Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: Calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 59.We thank A. Hall, A. Altman, J. D. Schatzle, L. J. Berg, H. L. Yin, M. K. Rosen, and M. G. Roth for plasmids; D. J. Fowell for DO11.10 lymph node suspensions; and W. Woodward for statistical advice. This work was supported by the NIH. K.L.S. and K.T.R. were supported by the institutional training grant T32AI005284.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Components of T cell activation examined in this study.
Fig. S2. FACS-based sorting of sensor-expressing T cells.
Fig. S3. Quantification of the abundance of biosensors and endogenous PKCθ.
Fig. S4. Rapid nuclear translocation of NFAT.
Fig. S5. Patterning of PKCθ in activated 5C.C7 T cells.
Fig. S6. Highly diverse spatiotemporal patterning during T cell activation.
Fig. S7. Dynamic nature of spatiotemporal patterns.
Fig. S8. Requirement for TCRζ ITAMs for effective recruitment of TCR to the cSMAC signaling complex.
Fig. S9. Enrichment of sensors in the large T cell invagination.
Fig. S10. Requirement for Rho activity for the resolution of the large T cell invagination.
Fig. S11. High diversity of spatiotemporal patterning during T cell activation.
Fig. S12. Variations in spatiotemporal patterning in activated T cells from different TCR transgenic mice.
Fig. S13. Reduced TCR clustering in TH2-polarized DO11.10 T cells.
Fig. S14. Regulation of spatiotemporal patterning by costimulation.
Fig. S15. Delayed central accumulation of the TCR compared to that of its dependent signaling intermediates.
Fig. S16. Similar dynamics of translocation of tubulin-GFP and the Arf6 sensor from the uropod to the interface upon formation of the T cell–APC interface.
Table S1. List of supplementary movies and a key that includes the sensor monitored, the time of cell coupling, and a description of the critical features observed.
Table S2. Numbers of cell couples analyzed.