Abstract
A proportion of the genetic variants underlying complex phenotypes do so through their effects on gene expression, so an important challenge in complex trait analysis is to discover the genetic basis for the variation in transcript abundance. So far, the potential of mapping both quantitative trait loci (QTLs) and expression quantitative trait loci (eQTLs) in rodents has been limited by the low mapping resolution inherent in crosses between inbred strains. We provide a megabase resolution map of thousands of eQTLs in hippocampus, lung, and liver samples from heterogeneous stock (HS) mice in which 843 QTLs have also been mapped at megabase resolution. We exploit dense mouse SNP data to show that artifacts due to allele-specific hybridization occur in ∼30% of the cis-acting eQTLs and, by comparison with exon expression data, we show that alternative splicing of the 3′ end of the genes accounts for <1% of cis-acting eQTLs. Approximately one third of cis-acting eQTLs and one half of trans-acting eQTLs are tissue specific. We have created an important systems biology resource for the genetic analysis of complex traits in a key model organism.
It is now well established that natural genetic variation contributes significantly to variation in gene expression, and the loci of regulatory variation for individual transcripts have been mapped in yeast (Brem and Kruglyak 2005), mice (Schadt et al. 2003), rats (Hubner et al. 2005), humans (Morley et al. 2004), maize (Shi et al. 2007), Eucalyptus (Kirst et al. 2005), and Arabidopsis (West et al. 2007). These studies show that variation in transcript abundance arises from expression quantitative trait loci (eQTL) that can be classified as either local (cis-eQTLs) or distant (trans-eQTLs) regulatory variants (Rockman and Kruglyak 2006). Typically, multiple eQTLs contribute to variation in the abundance of a single transcript.
In rodents, combining genome-wide expression analysis and QTL mapping has been shown in some cases to lead to the identification of genes involved in quantitative phenotypes (Karp et al. 2000; Bystrykh et al. 2005; Mehrabian et al. 2005; Schadt et al. 2005). However, for this approach to become widely applicable to the thousands of QTLs so far identified, it is necessary to perform the following:
-
Measure transcript abundance in multiple tissues in large populations. It is not clear how many tissues will need to be assayed, or at how many time points, to provide a sufficiently comprehensive coverage of transcript abundance to enable gene identification at the thousands of QTLs known in rodents (Flint et al. 2005). The equivalence of eQTLs across tissues is a key assumption if we are to use eQTLs identified in one tissue or time point as a surrogate. Estimates of the correspondence between tissues vary in the rat: Only 15% were found to be common to both fat and kidney (Hubner et al. 2005), although 63%–88% of cis-eQTLs were reported to overlap in a study of liver, adipose, muscle, and brain gene expression in a BXH F2 intercross (Meng et al. 2007).
The statistical power of many eQTL experiments may be too low, assuming that eQTLs may have effect sizes similar to QTLs contributing to other phenotypes (∼3% in an F2) (Flint et al. 2005). Schadt and colleagues have used two F2 populations consisting of 111 and 334 mice (Doss et al. 2005; Mehrabian et al. 2005; Schadt et al. 2005; GuhaThakurta et al. 2006); the larger of these is predicted to have ∼80% power to detect an eQTL explaining >8% of the variance (Lynch and Walsh 1998). The 35 recombinant inbred mouse (Chesler et al. 2005) and 30 rat (Hubner et al. 2005) strains in which eQTLs have been mapped have 80% power to detect effects of >20% (Belknap et al. 1996). This relative lack of power is also apparent in the difficulties in detecting trans-eQTL, for which a high genome-wide level of statistical significance is needed compared with that required to detect a cis-eQTL where the locus is known a priori. While most eQTLs mapped in yeast are trans-acting (Yvert et al. 2003), few have been discovered in human studies (Dixon et al. 2007) and the proportion in rodents is unknown (Rockman and Kruglyak 2006).
Map both eQTLs and QTLs at high resolution. Earlier studies that combine expression and phenotypic measurements on the same cohort have used crosses between inbred strains in which the mapping resolution is of the order of tens of megabases. Alternative mouse populations offer much higher mapping resolution, such as heterogeneous stocks (Valdar et al. 2006b), outbred animals (Yalcin et al. 2004), and the collaborative cross (Valdar et al. 2006a). Due to improvements in high-throughput genotyping and methods of analysis, these populations can now be used in whole genome association studies.
Exclude artifactual explanations for eQTLs. Single-nucleotide polymorphisms (SNPs) can have a significant impact on microarray-based assays of transcript abundance (Alberts et al. 2007; Walter et al. 2007; Benovoy et al. 2008). Analysis of data from Affymetrix arrays on 30 recombinant mouse inbred lines (Bystrykh et al. 2005) indicated that almost half of the reported 100 most significant cis-eQTLs could be attributed to sequence diversity in probe regions (Alberts et al. 2007). The recently released high density SNP data for mouse inbred strains (Frazer et al. 2007; Yang et al. 2007) now make it possible to identify potential false cis-eQTLs.
Here we provide a comprehensive resource for integrating genetics and transcriptional profiling by mapping transcriptional abundance in hippocampus, liver, and lung in genetically heterogeneous stock (HS) mice, descended from eight inbred progenitor strains (A/J, AKR/J, BALBc/J, CBA/J, C3H/HeJ, C57BL/6J, DBA/2J, and LP/J) (Valdar et al. 2006b). The same animals have been used to map 843 phenotypic QTLs for 97 phenotypes, mapped into an average 95% confidence interval of 2.8 Mb (Valdar et al. 2006b). Many of these QTLs contain a small number of genes and will provide a powerful resource for exploring the relationship between variation in gene expression and quantitative phenotypic variation. All analyses are made available by our genome browser http://gscan.well.ox.ac.uk.
Results
We analyzed gene expression in hippocampus, liver, and lung using Illumina arrays with 47,430 oligonucleotides that assess expression of 21,288 Ensembl annotated transcripts, representing 19,687 genes. For the hippocampus we used 460 HS brains and also measured expression in the eight HS founders with both Illumina arrays and Affymetrix exon arrays. For the liver and lung a subset of 260 animals and the HS founders were tested with Illumina arrays only. The HS tissues were taken from animals that had been previously genotyped with 13,459 markers and phenotyped for 97 traits (Valdar et al. 2006b).
Genome scans were performed to map the eQTLs for all transcripts and tissues on to build 37 of the mouse genome. Because a transcript generally has more than one eQTL, and because different combinations of eQTLs may explain the variation in a transcript equally well, we mapped eQTLs using resample-based model averaging (described in the Supplemental Methods). We measured the robustness of the support for an eQTL by the resample-based model inclusion probability (RMIP), a generalization of the bootstrap posterior probability (BPP) we have described earlier (Valdar et al. 2006b). This measures the fraction of times the eQTL is included in a multiple-eQTL model in repeated subsamples of the data (Valdar et al. 2006b). From simulation of QTLs explaining 5% of the phenotypic variance, a detected eQTL that exceeds a RMIP threshold of 0.5 will be true in 85% of cases, and in 70% of cases for a threshold of 0.25 (Valdar et al. 2006b). At a RMIP threshold of 0.25, about one false positive eQTL occurs every four genome scans; no false positives are detected at a RMIP of 1.0. Unless otherwise stated, we consider only eQTLs with RMIPs ≥0.25.
eQTLs were categorized as either cis or trans depending on whether an eQTL peak was either less or more than 2 Mb from the midpoint of its cognate transcript (in the HS, linkage disequilibrium measured by the mean r2 falls to <0.5 within 2 Mb and is <0.2 within 8 Mb) (Valdar et al. 2006b). The relationship between the number of eQTLs detected and distance from the cognate transcript is shown in Supplemental Figure 1. The number of eQTLs detected depends on the RMIP threshold applied. In the hippocampus, at the most stringent RMIP value of 1.0, we detected 2732 cis-eQTLs and 205 trans-eQTLs; at the lower RMIP value of 0.25 there are 3961 cis-eQTLs and 4586 trans-eQTLs (Fig. 1).
Figure 1.
(A) The number of cis-eQTLs for each of three tissues at different RMIP thresholds (vertical axis). Numbers are shown for cis-eQTLs containing no annotated SNPs (crosses) and for all cis-eQTLs (diamonds). Lung data are shown in blue, liver in green, and hippocampus in red (260 animals) and black (460 animals). (B) The number of trans-eQTLs for each of three tissues at different RMIP thresholds (vertical axis). Lung data are shown in blue, liver in green, and hippocampus in red (260 animals) and black (460 animals).
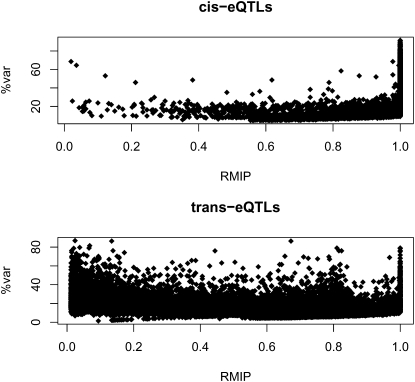
It is important to appreciate that, while the RMIP measures the robustness of the eQTL detection, it is not a measure of the effect size of the eQTL. This is shown by the lack of correlation with the percentage of variance explained (which is itself highly correlated with the ANOVA –log10 of the P-value [logP] [r = 0.974]; Fig. 2). The median effect size of all cis-eQTLs with RMIP ≥0.25 is 20.83% (mean 26.35%) and 15.19% (mean 14.86%) for trans-eQTLs; 43 trans-eQTLs and 775 cis-eQTLs had effect sizes in excess of 50%.
Figure 2.
Relationship between the RMIP and effect size (expressed as percentage of the variation in transcript level [%var]) for cis- and trans-eQTLs.
Many large effect cis-eQTLs are due to SNPs within the target sequence
The large effect of many cis-eQTLs suggests that they may be due to highly penetrant mutations, which might in turn aid the discovery of the causative sequence variant. However, it is also possible that, as others have observed, sequence polymorphisms within the probe sequences are causing artifactual cis-eQTLs (Alberts et al. 2007; Walter et al. 2007). The extensive SNP data sets available for the mouse make it possible to investigate the contribution of SNPs to cis-eQTLs (Frazer et al. 2007; Yang et al. 2007).
In the hippocampus, 694 cis-eQTLs with RMIP ≥0.25 contain an annotated SNP (17% of the total). There is a highly significant enrichment of SNPs in large effect cis-eQTLs: Figure 3 shows the distribution of logPs for cis-eQTLs in the hippocampus detected by probes containing annotated SNPs, compared to cis-eQTLs with no known SNP. The presence of SNPs correlates significantly with logP values (P < 10−16). About 20% of cis-eQTLs with a logP between 10 and 20 (which constitute 8.3% of all cis-eQTLs) contain an annotated central SNP; more than half of cis-eQTLs with a logP >50 do so (2.9% of all cis-eQTLs). This suggests that many of the cis-eQTLs we have detected may be in part due to differences in hybridization efficiency.
Figure 3.
Effect of sequence variants on cis-eQTL detection. The frequency (vertical axis) of logP (negative logarithm of the P-value) scores (horizontal axis) for cis-acting eQTLs is shown. Data for cis-eQTLs with no annotated variant (solid line) are compared with data for cis-eQTLs with terminal variants (a SNP in the first two or last base pairs of the probe sequence) and with central variants (defined as any SNP that is not a terminal variant).
An alternative explanation is that loci dense in SNPs are also dense in cis-eQTLs, and so a SNP within a probe reflects the presence of other SNPs nearby, some of which affect gene expression. However, Figure 3 shows that there is an important difference between eQTLs with SNPs in the center of the probe (and which are more likely to affect hybridization) and probes with SNPs in the terminal regions (within the last two base pairs at either end of the probe). A χ2 test in which eQTLs containing SNPs were classified by logP (low: logP < 10; medium: 10 < logP < 20; high: logP > 20) and by SNP (central or terminal) was significant at P < 4 × 10−5. On average, while every central SNP adds 7.5 units (s.e. ± 0.5) to the logP, every terminal SNP increases it by 0.4 (s.e. ± 1.4; not significant). This suggests that SNPs are causing the variation in signal, because we would not expect probes with centrally located SNPs to be distributed differently across the genome from probes with terminally located SNPs.
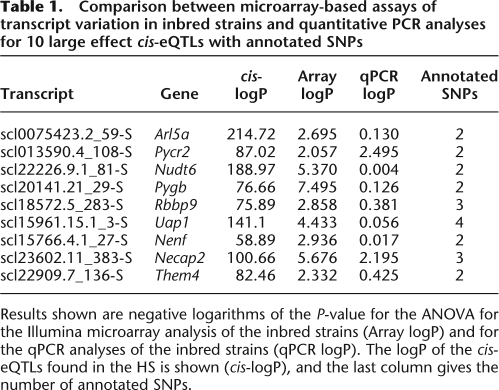
We confirmed experimentally whether SNPs within probes were contributing to differential hybridization by performing quantitative PCR (qPCR). We chose 10 transcripts that contained two or more central annotated SNPs and for which mapping indicated the sole source of variation was attributable to a large cis-eQTL (percent variation >50). We designed primers to amplify the same exon as that detected by the Illumina array, using primers that hybridized to DNA without annotated SNPs. We then measured transcript abundance in the eight inbred progenitor strains by qPCR and by using Illumina arrays. The results are given in Table 1. Only two of the 10 transcripts show significant strain variation by qPCR.
Table 1.
Comparison between microarray-based assays of transcript variation in inbred strains and quantitative PCR analyses for 10 large effect cis-eQTLs with annotated SNPs
Results shown are negative logarithms of the P-value for the ANOVA for the Illumina microarray analysis of the inbred strains (Array logP) and for the qPCR analyses of the inbred strains (qPCR logP). The logP of the cis-eQTLs found in the HS is shown (cis-logP), and the last column gives the number of annotated SNPs.
We estimated the fraction of unannotated SNPs in probe sets by sequencing 45 probes (without annotated SNPs), randomly selected from three ranges of logPs. We found six central SNPs in 17 probes with logPs >20, three in 12 probes with cis-eQTLs with logPs between 10 and 20 and no variants in 16 probes with cis-eQTLs with logPs <10. Logistic regression shows the relationship between logP and the presence of a novel SNP is significant (P < 0.05) and that every increase of 10 logP units ups the log odds of a SNP by 0.13 (±0.07). Applying this model to the logPs of the remaining unannotated cis-eQTLs, we estimate that a further 16% (95% confidence interval: 9%–31%) would contain SNPs. Including both known and unknown SNPs, ∼30% of cis-eQTLs are expected to contain a SNP.
Since some of the cis-eQTLs at transcripts containing annotated SNPs reflect true variation in transcript abundance, we incorporated all cis-eQTL results into our web-based interface to the phenotypic QTLs (http://gscan.well.ox.ac.uk). We indicate whether the probe contains an annotated SNP or not by a difference in color (red for SNPs, black for no known SNPs).
Few cis-eQTLs are due to splicing variation
The existence of many large effect cis-eQTLs, where some alleles correspond to very low or no expression and other alleles produce high levels of expression, was surprising. For example, in the hippocampus, after leaving out cis-eQTLs with annotated SNPs there are 288 with effects >50%, including 20 with effects >80%. Since we measured gene expression using 3′-based expression arrays, there is a possibility that some of the large effect cis-eQTLs may be attributable to allele-specific alternative splicing of the 3′ end of the genes (Kwan et al. 2008), while the variation in expression of other parts of the genes are low.
To estimate the contribution of splicing variation to all cis-eQTLs (not just the largest), we took advantage of the derivation of the HS animals from inbred progenitor strains. We first obtained a conservative estimate of the extent (though not the frequency) of isoform variation in the HS from that measured in the eight progenitors in three replicate experiments. Using Affymetrix exon arrays consisting of 216,129 probe sets, (each probe set representing a single exon, and in total representing 15,620 unique genes) we looked for exons with patterns of strain-specific expression in the hippocampus that differed significantly from the overall pattern of strain expression for the gene. After removing 23,499 probe sets with annotated SNPs (Frazer et al. 2007), there was evidence for isoform variation at 1985 genes at a false discovery rate of 0.05.
Since a proportion of the isoform variation detected by the array is likely to be due to unannotated polymorphisms in the probe-target sequences (Benovoy et al. 2008), we looked for differences in the hybridization of individual oligonucleotides within a probe set that do not follow the overall pattern of strain differences for that probe set (this is equivalent to looking for an interaction between oligonucleotide and strain in an ANOVA of the probe set). However, a significant interaction can be due to anything that differentially affects hybridization of one oligonucleotide in the probe set, not just a SNP, so this is a conservative way of assessing the contribution of sequence variants.
We identified 234 genes in which the probe sets' ANOVA interaction test was not significant (P-value of >0.05) and that were also predicted to show splicing variation between strains. These splice isoforms could not be explained as artifacts of annotated SNPs. Out of the 3225 cis-eQTLs in the hippocampus with RMIP >0.25 and without annotated SNPs, 47 intersected with this set. However, the Illumina probe, used to detect the cis-eQTL, overlapped the exon probe set in only 20 of those cases, meaning that in 27 cases the alternative splicing event could not be causing the expression difference detected by the Illumina assay. This limits the fraction of alternative splicing events causing expression eQTLs to 20/3225, or 0.6%.
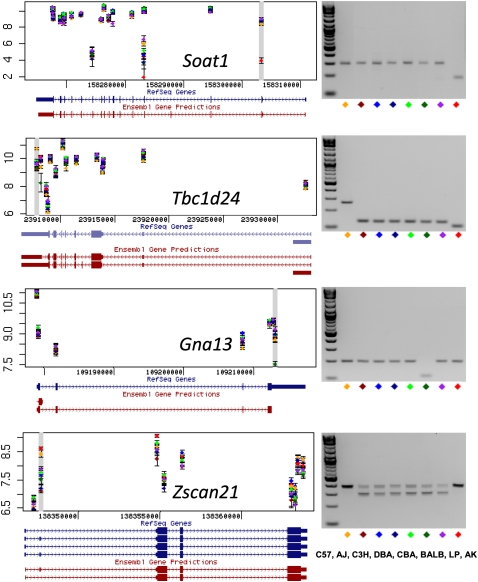
To test these cases we designed specific primers for the 20 candidate splicing variants coinciding with eQTLs. Using quantitative real-time PCR we detected a significant interaction between exon and strain in eight cases. In two genes (Gna13 and Tbc1d24) we observed two products in the melt curves from RNA isolated from different inbred strains, suggesting that there was an additional intron not annotated in Ensembl, an interpretation supported by finding different PCR product sizes when we used primers to amplify the spanning exons of predicted splice isoforms (Fig. 4).
Figure 4.
Splicing variants. Predicted transcript isoforms for four genes (Soat1, Tbc1d24, Gna13, and Zscan21) are shown on the left, based on the exon-array data. Each dot is the average of three measurements of the probe set signal intensity for one of the eight inbred strains. A gray horizontal bar identifies probe sets judged to show significant variation between the strains. RefSeq and Ensembl predicted gene structure are taken from the UCSC Genome Browser (http://www.genome.ucsc.edu). Exon PCR results are shown on the right. In two cases (Gna13 and Tbc1d24) PCR primers amplify across a single terminal exon. For Soat1 and Zscan21 primers were designed that amplify flanking exons. Each mouse inbred strain is indicated by a different color. Strain names are as follows: C57, C57BL/6J; AJ, A/J; C3H, C3H/HeJ; DBA, DBA/2J; CBA, CBA/J; BALB, BALBc/J; LP, LP/J; AK, AKR/J. The PCR results suggest the following as an explanation for the inbred strains differences in exon-specific hybridization signal: (1) exon skipping of the second exon of Soat1 in AKR/J (AK, red diamond); (2) newly identified spliced introns within 5′ UTR of Tbc1d24 in all strains except for C57BL/6J (C57, yellow); (3) newly identified spliced intron within the 3′ UTR of Gna13 in BALBc/J (BALB, dark green); (4) alternative splicing by exon skipping of the second exon of Zscan21 in all strains except for C57BL/6J and AKR/J (C57; yellow; AK, red).
In summary, our results indicate that large variation in alternative splicing may explain only a minority of cis-eQTLs that we have detected (e.g., Gna13 and Tbc1d24, Fig. 4). However, our data also show that large variation in alternative splicing, which could contribute to the phenotypic QTL, could be missed by 3′-based expression arrays (e.g., Soat1 and Zscan21; Fig. 4).
Tissue specificity of eQTLs
We compared gene expression in the hippocampus, liver, and lung in 260 mice, excluding the additional 200 hippocampus samples. The number of transcripts with expression levels significantly above negative control probes differed between the three tissues: 12,966 for liver, 19,243 for lung, and 20,743 for hippocampus. These numbers were not reflected in the proportion of heritable transcripts in the three tissues: 4198 transcripts had heritabilities >10% in liver, 7990 in lung, and 2661 in the hippocampus. Thus, while there are many more detectable transcripts in the hippocampus, heritable variation is most common in the lung.
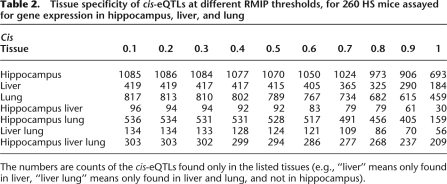
As expected from tissue differences in transcript heritability, there is only modest correlation between the eQTLs in different tissues. Figure 1 shows the number of eQTLs detected at RMIPs from 0.1 to 1 after removing probes with annotated SNPs. In order to provide estimates of tissue specificity in eQTL detection, in Table 2 we show the number of eQTLs (without annotated SNPs) detected at all RMIP thresholds from 0.1 to 1. For a given RMIP threshold, an eQTL is said to be tissue specific if the RMIP exceeds that threshold in one tissue and is zero in other tissues. By this definition approximately one third of cis-acting eQTLs and one half of trans-acting eQTLs are tissue specific. For cis-eQTLs that are expressed in two tissues, the mean difference between the effect size (expressed as percentage of the variation explained) is 3.3% (standard deviation of 15.7).
Table 2.
Tissue specificity of cis-eQTLs at different RMIP thresholds, for 260 HS mice assayed for gene expression in hippocampus, liver, and lung
The numbers are counts of the cis-eQTLs found only in the listed tissues (e.g., “liver” means only found in liver, “liver lung” means only found in liver and lung, and not in hippocampus).
Tissues differ in the number of eQTLs: most were detected in the lung, which at an RMIP of ≥0.1 contains >11,000 compared with 8770 in the hippocampus and 8400 in the liver (Fig. 1). However, this summary obscures a difference in the prevalence of cis- and trans-eQTLs. At RMIPs between 0.1 and 0.7 every tissue has an excess of trans-eQTLs, but the ratio of cis to trans varies significantly between tissues: In the hippocampus trans-eQTLs are twice as common as cis-, and more than fives times more common in the liver (the lung is intermediate).
cis-eQTLs that contain SNPs (not included in Table 2) are less tissue specific than cis-eQTL without SNPs, suggesting that tissue-specific gene expression has been underreported. Table 3 shows that eQTLs not containing SNPs are three times more likely to be found in a single tissue rather than in all three, while the numbers of eQTLs with SNPs are approximately equal, whether they occur in one or all three tissues (ratio 0.85).
Table 3.
Tissue specificity of cis-eQTLs according to the presence or absence of annotated SNPs
Numbers in the first column refer to the number of tissues in which a cis-eQTL is found. cis-eQTLs that contain SNPs are less tissue-specific than cis-eQTLs without SNPs.
Mapping resolution
The mean 95% confidence intervals for cis-eQTLs is 2.45 Mb (s.d. 1.50) and for trans-eQTLs 3.75 (s.d. 1.31). Figure 5 shows the relationship between resolution and logP: For every increase in 10 logP units resolution decreases by −0.76 Mb for cis-eQTLs and by −0.70 Mb for trans-eQTLs. The mean number of genes in the cis-eQTL and trans-eQTLs 95% intervals is 40.7 and 47.1, respectively. In Supplemental Table 1 we provide a list of genes that lie within eQTLs containing five or fewer genes.
Figure 5.
Bootstrap derived 95% confidence of cis- and trans-eQTLs plotted against their respective logP. The size of the interval is shown on the vertical axis in megabases, and the ANOVA logP on the horizontal axis. Trans-eQTLs are shown as open circles, cis-eQTLs as filled circles. A regression curve fitted to the square root of the confidence interval is shown for trans-eQTLs (dotted) and cis-eQTLS (continuous).
Discussion
We have provided a high resolution map of thousands of expression QTLs in HS mice; on average cis-eQTLs are mapped into 2.45 Mb 95% confidence intervals and trans-eQTLs into 3.75 Mb intervals. We observed a remarkable degree of tissue specificity: Approximately one third of cis-acting eQTLs and one half of trans-acting eQTLs are tissue specific (Table 2). We find that trans-eQTLs outnumber cis-eQTLs.
To interpret our results, it is important to appreciate the way we analyze the HS mice, a structured population which contains animals of differing degrees of genetic relatedness. We report evidence for the existence of an eQTL using a resample-based model inclusion probability, the RMIP. The ANOVA logPs from the association analysis, which in other designs would provide a robust measure of statistical significance, do not distinguish ghost peaks (that arise from genotype correlations) from real peaks. If we relied solely on logPs and single locus associations, we would detect many apparently large effect trans-eQTL that were in fact ghosts of the true cis-effects.
For phenotypes where there is a single true peak, the RMIP is more likely to be correlated with the logP value (and effect size). We found more single large effect (high logP) cis- than trans-eQTL and as a result RMIPs close or equal to one are more common among the cis-eQTLs. This explains the pattern in Figure 1, where there are fewer cis-eQTLs than trans-eQTLs with RMIPs <0.6.
Our results raise a number of cautions about using eQTL data. The first concerns the validity of cis-eQTLs. We were able to use high density SNP data for mouse inbred strains (Frazer et al. 2007; Yang et al. 2007) to identify potential false cis-eQTLs. We found that, among cis-eQTLs with a logP >20, ∼40% contain a known polymorphism in the target sequence. Furthermore, from sequencing we estimated the frequency of unannotated SNPs and conclude that overall ∼30% of cis-eQTLs contain a SNP.
For a number of reasons, it is unlikely that the increased variation in expression for probes containing SNPs can be attributed to additional nearby SNPs lying within functional elements (GuhaThakurta et al. 2006). Critically, we find that SNPs lying within terminal regions of the probes do not have a significant effect on logPs, while SNPs lying within central regions of the oligonucleotide do; furthermore, the more variants within a probe, the higher the logP (Fig. 3). Testing large effect eQTLs that contain annotated SNPs failed to confirm expression variation in eight out of 10 cases. We also found that cis-eQTLs containing SNPs are less tissue specific than cis-eQTLs without SNPs, suggesting that tissue-specific gene expression has been underreported. cis-eQTLs that occur in multiple tissues and contain SNPs are extremely likely to be due, at least in part, to hybridization artifacts.
We find that splicing variation is not an important contributor to cis-eQTLs identified by 3′-based expression arrays, but we note that possible important variation in splicing patterns might be missed. Less than 1% of the cis-eQTLs in the hippocampus coincided with a gene predicted to have isoform differences between strains. Furthermore, where the Illumina and Affymetrix exon array probe sets interrogated the same exon, we validated less than half of the predicted isoforms. While we cannot rule out the possibility that alternative splicing occurs in some of those genes excluded because their probe sets contained SNPs, we would expect it to occur at a similar rate to that observed in the genes we did analyze.
The second issue concerns the usefulness of eQTLs for identifying quantitative trait genes (Bystrykh et al. 2005; Mehrabian et al. 2005; Schadt et al. 2005). In its simplest formulation, the causal link between sequence variant, gene expression, and phenotype arises because the sequence variant is responsible for both a cis-eQTL and the QTL contributing to a quantitative phenotype. Consequently the two loci must coincide and, among the many genes that lie within the confidence interval of the phenotypic QTL, those involved in the phenotype should have transcripts whose abundance is in part under the control of a cis-eQTL. Since there are >2000 known QTLs in mice, but less than a hundred genes have been robustly identified (Flint et al. 2005), methods that accelerate gene discovery are needed. We have identified 799 phenotypic QTLs that contain at least one eQTL within their 95% confidence intervals (Valdar et al. 2006b). Our results, freely available at http://gscan.well.ox.ac.uk, could in principle lead to the identification of genes at 95% of QTLs in the HS.
By combining analysis of networks of gene expression covariance (Chen et al. 2008) and haplotype structure (Yalcin et al. 2005), our data will be useful for determining the extent to which, through their control over gene expression, eQTLs may reveal the presence of genes involved in quantitative phenotypes (Bystrykh et al. 2005; Mehrabian et al. 2005; Schadt et al. 2005; Dixon et al. 2007; Goring et al. 2007). We expect that our resource will be an important starting point for identifying genes in many complex traits.
Methods
Tissue preparation and RNA extraction
Tissue from HS animals was snap-frozen in liquid nitrogen. Whole brains were cut in half sagittally and the left hemisphere soaked into RNAlater-Ice (Ambion) for 20 h at −20°C. The hippocampus was dissected for RNA extraction by using TissueLyser and RNeasy Lipid Tissue Kit (Qiagen). Tissue samples (25 mg for liver and lung down to a few milligrams for hippocampus) were homogenized using 5-mm stainless steel beads (Qiagen) on a Tissue Lyser (Retsch MM300 Mixer Mill) for 10 min at 25 Hz. Total RNA was then extracted using the RNeasy 96 Universal Tissue Kit (Qiagen) for liver and lung and the RNeasy Lipid Tissue Kit (Qiagen) for hippocampus according to the manufacturer's instructions. RNA quantity and integrity were assessed using a NanoDrop ND-1000 Spectrophotometer and an Agilent 2100 Bioanalyser. Total RNA samples with RNA Integrity Number >9 were used for messenger RNA amplification. For the Affymetrix mouse exon array, hippocampus was isolated from the eight inbred mice and stored in −80 (four animals of each strain were used). Total RNA was extracted as described above.
Messenger RNA amplification and labeling
Messenger RNA molecules were amplified using the MessageAmp II-96 Kit (Ambion) according to the manufacturer's instructions. First strand cDNA was synthesized using 300 ng of total RNA and oligo(dT) primers. In vitro transcription was carried out at 37°C for 14 h during which biotinylated UTPs (75 mM Biotin-16-UTP, Ambion) were used for RNA labeling. Messenger RNA quantity and size were determined in the same way as total RNA using a NanoDrop ND-1000 Spectrophotometer and an Agilent 2100 Bioanalyser. Size ranged from 250 to 5500 nucleotides (nt) with a peak centered at 1000–1500 nt.
Arrays hybridization, washing, and scanning
For the Illumina arrays, 1.5 mg of labeled messenger RNA was hybridized to Illumina Sentrix mouse-6 expression beadchips. After 17-h hybridization at 55°C, beadchips were washed according to recommended protocols from Illumina. FluoroLink Cy3-labeled Streptavidin (Amersham Biosciences) was used to detect expression signals. The BeadStation 500 G system was then used to scan the beadchips. For the exon arrays a GeneChip Fluidics Station 450/250 was used to wash and stain the Mouse Exon 1.0 ST Array. Quality control was carried out using Affymetrix Expression Console Software Version 1.0.
Microarray expression data handling
The Illumina Mouse WG-6 v1 BeadArray contains 47,429 unique probe sequences. These were mapped to Build 37 (mm9) using the alignment tool BLAT (Kent 2002) and perfect and unique matches were retained. This resulted in 30,029 probe sequences matched to a genomic location pertaining to 19,688 and 21,289 unique Ensembl gene and transcript identifiers, respectively.
Data generated from scanning were imported into Illumina BeadStudio version 3.0 to generate background subtracted signal values for each bead type. Positive signals were defined to be probes with signals in excess of the mean of the negative control probes +2 standard deviations, giving the thresholds of hippocampus 3.58, liver 3.77, lung 3.92. Processed data were then exported to the R statistical computing language, transformed, and normalized using the BioConductor package, vsn (Huber et al. 2002), and analyzed for differential expression using the limma package (Smyth 2004). This vsn package incorporates a variance stabilizing data transformation and normalization that generates generalized log2 values. The limma package identifies differential expression through linear modeling and an empirical Bayesian approach to estimate the test statistic. Differential transcripts were identified as those with P-values ≤0.01 and fold changes of ≥1.5.
Genetic mapping of expression QTLs
The genetic mapping of expression QTLs (eQTLs) proceeded by first identifying a suitable linear model formulation for the expression phenotype; second, performing genome scans that assess the effects of single loci; third, establishing significance thresholds based on null simulations; fourth, performing multiple QTL modeling of transcripts that have significant loci. We used a modification of the resample-based methods described in Valdar et al. (2006b). A complete description of our approach is provided as Supplemental material.
Alternative splicing analysis
Exon expression data from Affymetrix arrays measured in triplicate on the eight HS founders were analyzed for evidence of alternative splicing and for evidence of SNP artifact. Each exon was represented on the microarray by a probe set containing four nonoverlapping 25-mer probes.
Signal estimates were derived from the CEL files by quantile sketch normalization using the PLIER algorithm for probe set (exon-level) intensities and IterPLIER for gene-level intensities using the Expression Console software (Affymetrix). Only “core” level probe sets (probe sets assigned to the highest confidence level) were used in the analysis. Gene-level iterPLIER estimates are derived by combining correlated probe sets, predicted to map into the same transcript cluster (according to the meta-probe set list). The iterPLIER algorithm iteratively discards probes that do not correlate well with the overall gene-level signal and then recalculates the signal estimate to derive a robust estimation of the gene expression value.
Candidate exons for alternative splicing were detected by testing for significant differences in probe set signal between different strains after controlling for the gene-level differences. This was done with ANOVA to compare between two linear models that predict the observed expression level of each probe set by the gene level or by both the gene level and the strain type. An estimated local false discovery rate value (as implemented in R in the “fdrtool” package) was assigned to each probe set, expressing the probability of not being differentially expressed.
To test for SNP effects, ANOVA was performed separately within each probe set, modeling the intensity yrks observed in replicate r, oligonucleotide k (k = 1…4) and strain s as yrks = μ + αr + βk + γs + θks + erks. Evidence for a hybridization artifact affecting a particular combination of oligonucleotide probe and strain is indicated by the contribution of the interaction term θks being significant in an ANOVA.
Acknowledgments
This work was supported by the Wellcome Trust. M.J. was funded by an intra European Marie Curie fellowship under the Framework 6 Programme. W.V. was funded by a grant from the European Union Framework 6 Programme, contract number LHSG-CT-2003-503265 and by the Medical Research Council, UK.
Footnotes
[Supplemental material is available online at www.genome.org and at http://gscan.well.ox.ac.uk The expression data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. E-MTAB-86 and E-MTAB-88.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.088120.108.
References
- Alberts R., Terpstra P., Li Y., Breitling R., Nap J.P., Jansen R.C. Sequence polymorphisms cause many false cis eQTLs. PLoS One. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap J.K., Mitchel S.R., Crabbe J.C. Type I and II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains: Computer simulation and empirical results. Behav. Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- Benovoy D., Kwan T., Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008;36:4417–4423. doi: 10.1093/nar/gkn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R.B., Kruglyak L. The landscape of genetic complexity across 5700 gene expression traits in yeast. Proc. Natl. Acad. Sci. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrykh L., Weersing E., Dontje B., Sutton S., Pletcher M.T., Wiltshire T., Su A.I., Vellenga E., Wang J., Manly K.F., et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics.”. Nat. Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., MacNeil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E.J., Lu L., Shou S., Qu Y., Gu J., Wang J., Hsu H.C., Mountz J.D., Baldwin N.E., Langston M.A., et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Doss S., Schadt E.E., Drake T.A., Lusis A.J. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Valdar W., Shifman S., Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- GuhaThakurta D., Xie T., Anand M., Edwards S.W., Li G., Wang S.S., Schadt E.E. Cis-regulatory variations: A study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics. 2006;7:235. doi: 10.1186/1471-2164-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl. 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Hubner N., Wallace C.A., Zimdahl H., Petretto E., Schulz H., Maciver F., Mueller M., Hummel O., Monti J., Zidek V., et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- Karp C.L., Grupe A., Schadt E., Ewart S.L., Keane-Moore M., Cuomo P.J., Kohl J., Wahl L., Kuperman D., Germer S., et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 2000;1:221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M., Basten C.J., Myburg A.A., Zeng Z.B., Sederoff R.R. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics. 2005;169:2295–2303. doi: 10.1534/genetics.104.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B. Genetics and analysis of quantitative traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- Mehrabian M., Allayee H., Stockton J., Lum P.Y., Drake T.A., Castellani L.W., Suh M., Armour C., Edwards S., Lamb J., et al. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat. Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- Meng H., Vera I., Che N., Wang X., Wang S.S., Ingram-Drake L., Schadt E.E., Drake T.A., Lusis A.J. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl. Acad. Sci. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M.V., Kruglyak L. Genetics of global gene expression. Nat. Rev. Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Lamb J., Yang X., Zhu J., Edwards S., Guhathakurta D., Sieberts S.K., Monks S., Reitman M., Zhang C., et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Uzarowska A., Ouzunova M., Landbeck M., Wenzel G., Lubberstedt T. Identification of candidate genes associated with cell wall digestibility and eQTL (expression quantitative trait loci) analysis in a Flint × Flint maize recombinant inbred line population. BMC Genomics. 2007;8:22. doi: 10.1186/1471-2164-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R. Simulating the collaborative cross: Power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006a;172:1783–1797. doi: 10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Solberg L.C., Gauguier D., Burnett S., Klenerman P., Cookson W.O., Taylor M.S., Rawlins J.N., Mott R., Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006b;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Walter N.A., McWeeney S.K., Peters S.T., Belknap J.K., Hitzemann R., Buck K.J. SNPs matter: Impact on detection of differential expression. Nat. Methods. 2007;4:679–680. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M.A., Kim K., Kliebenstein D.J., van Leeuwen H., Michelmore R.W., Doerge R.W., St Clair D.A. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Willis-Owen S.A., Fullerton J., Meesaq A., Deacon R.M., Rawlins J.N., Copley R.R., Morris A.P., Flint J., Mott R. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat. Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- Yalcin B., Flint J., Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 2005;171:673–681. doi: 10.1534/genetics.104.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bell T.A., Churchill G.A., Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yvert G., Brem R.B., Whittle J., Akey J.M., Foss E., Smith E.N., Mackelprang R., Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]