Abstract
RNA editing by adenosine deamination has been shown to generate multiple isoforms of several neural receptors, often with profound effects on receptor function. However, little is known about the regulation of editing activity during development. We have developed a large-scale RNA sequencing protocol to determine adenosine-to-inosine (A-to-I) editing frequencies in the coding region of genes in the mammalian brain. Using the 454 Life Sciences (Roche) Amplicon Sequencing technology, we were able to determine even low levels of editing with high accuracy. The efficiency of editing for 28 different sites was analyzed during the development of the mouse brain from embryogenesis to adulthood. We show that, with few exceptions, the editing efficiency is low during embryogenesis, increasing gradually at different rates up to the adult mouse. The variation in editing gave receptors like HTR2C and GABAA (gamma-aminobutyric acid type A) a different set of protein isoforms during development from those in the adult animal. Furthermore, we show that this regulation of editing activity cannot be explained by an altered expression of the ADAR proteins but, rather, by the presence of a regulatory network that controls the editing activity during development.
Adenosine to inosine (A-to-I) RNA editing is a co- or post-transcriptional event on double-stranded RNA. This activity plays an important role by increasing the diversity of the proteome when encoded sequence is targeted. Since inosine is interpreted as guanosine by the translation machinery, A-to-I RNA editing in coding sequences often results in amino acid changes that alter protein function. The enzymes that catalyze this type of editing are known as ADARs (adenosine deaminases that act on RNA) (Bass et al. 1997). There are three members of this group in vertebrates, ADAR, ADARB1, and ADARB2. ADAR and ADARB1 are the only enzymes with proven deaminase activity. They are found in most tissues with the highest expression in the brain (Melcher et al. 1996; Chen et al. 2000). In the mammalian brain, RNA editing fine-tunes the features of several proteins involved in neurotransmission, where single amino acid changes alter gating and kinetic properties of a number of neural receptors. The edited site that has been proven to be most important is situated in the transcript coding for subunit 2 of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole (AMPA) glutamate receptor (Gria2). This site (Q/R) encodes a glutamine (Q) in its nonedited form (CAG), while the edited form (CIG) encodes an arginine (R). Editing at this position is close to 100%, and it controls the calcium permeability, resulting in receptors impermeable to Ca2+ ions (Sommer et al. 1991). Genetically modified mice that render uneditable Gria2 transcripts die a few weeks after birth because of epileptic seizures, as a direct consequence of calcium influx (Brusa et al. 1995). The equivalent Q/R site is also found in the Grik1 and Grik2 subunits of the kainite receptor. Transgenic mice homozygote for a noneditable form of Grik2 are susceptible to kainite induced seizures, indicating that editing also at this site is important for regulation of calcium permeability (Vissel et al. 2001). Moreover, Grik2 is also edited at other positions within the encoded sequence (the I/V, Y/C, and M/V site) that could be involved in additional regulation of calcium permeability (Kohler et al. 1993). Another editing site termed the “R/G site” is found in the Gria2, Gria3, and Gria4 subunits of the glutamate receptor. Edited receptors where an arginine is substituted for a glycine have been shown to recover faster from desensitization (Lomeli et al. 1994). The serotonin receptor HTR2C is the only G-protein-coupled receptor known to be edited (Burns et al. 1997). Five sites (A, B, C′, C, and D) in close proximity to each other in the second intracellular domain of the receptor can be edited. Editing of the HTR2C receptor affects the affinity to the G-protein and decreases constitutive activity (Niswender et al. 1999; Wang et al. 2000). The two most recent additions of edited receptors in the CNS is the potassium voltage-gated ion channel KCNA1 and the GABAA (gamma-aminobutyric acid type A) receptor. Gabra3, coding for the alpha3 subunit of the GABAA receptor, is efficiently edited at one site, giving rise to an isoleucine-to-methionine change in the third transmembrane region of the subunit (Ohlson et al. 2007). Also, in the transcript of the human KCNA1 channel, an isoleucine codon is changed upon editing, but here the edited ITT codon is recognized as a valine (Bhalla et al. 2004). Editing of both Gabra3 and KCNA1 has been suggested to have functional consequences on the receptors (Bhalla et al. 2004; Rula et al. 2008). The editing enzyme ADARB1 edits its own pre-mRNA within intron 4 to induce alternative splicing (Rueter et al. 1999). Splicing at the alternative 3′ splice site alters the reading frame and results in diminished expression of the catalytically active protein. Auto-editing at this position has been proposed to be a mechanism for ADARB1 to balance its own expression (Feng et al. 2006).
Previous studies show that editing of some sites in the kainate and the AMPA glutamate receptor transcripts in rat are regulated during development (Lomeli et al. 1994; Paschen et al. 1994; Schmitt et al. 1996; Bernard et al. 1999). Owing to the large impact of A-to-I editing on several processes in the CNS, it is of interest to understand how editing is regulated and what impact an altered editing activity may have on the developing brain. In this study, we use the large-scale 454 Life Sciences (Roche) Amplicon Sequencing technique (Roche) to examine editing at a high resolution. We analyze the editing levels of known mammalian ADAR substrates at four different developmental stages of the mouse brain. To our knowledge, this is the first time deep sequencing has been used to investigate the frequencies of site-selective A-to-I RNA editing. This technique allows us to analyze the sequence of several thousand individual transcripts allowing quantification of low editing frequencies with high statistical significance. Interestingly, with few exceptions, our data reveal that A-to-I editing is low in early embryogenesis and increases during development. This implicates that regulated A-to-I RNA editing is important for the process of brain maturation. Furthermore, the protein levels of the ADAR enzymes remain constant during development and cannot explain the increase in editing activity. This indicates that the editing activity of ADAR and ADARB1 is regulated in an age-dependent manner to acquire a precise balance between nonedited and edited isoforms.
Results
Site-selective editing increases during brain development
We wanted to investigate if the editing pattern of known selectively edited sites changes during the development of the brain. To analyze editing frequencies, total RNA was isolated from mouse brain at different developmental stages from embryonic to young adult mice. RNAs known to be targets for A-to-I editing were amplified by reverse transcriptase followed by polymerase chain reaction (RT-PCR). The products were subsequently sequenced according to the 454 Amplicon Sequencing protocol. This technique allowed us to analyze the sequence of several thousands of individual transcripts. All sequences corresponding to the A-to-I editing substrates were aligned to the genomic sequence and analyzed for A-to-G changes. The extent of editing in total brain extracts was determined at embryonic days 15 and 19 (E15 and E19) as well as postnatal days 2 and 21 (P2 and P21).
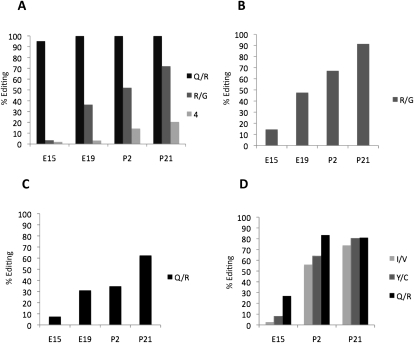
We discovered a general trend that the editing efficiency in the substrates shows a developmental regulation with very low levels of editing during embryogenesis that increase over time. Although most sites follow this pattern, there is a variation in how gradual the increase in editing efficiency is. As previously shown (Seeburg et al. 1998), the Q/R site of Gria2 is quickly saturated to the adult editing levels close to 100% already at E19 (Fig. 1A; Table 1). To our surprise, a site only 4 nucleotides (nt) downstream from the Q/R site is developmentally regulated, showing only 5% editing at E15 increasing to 20% in P21. Even though the effect of editing at this site is silent, giving no change of the translational code, the regulation of the editing efficiency is evident and cannot be explained by accessibility to the editing enzyme. Editing at the R/G site of Gria2 has been shown to increase during development (Lomeli et al. 1994). We confirmed this result and determined the editing at stage E15 as 4%, gradually increasing to 72% at P21 (Fig. 1A; Table 1). A similar pattern is seen for the R/G site of Gria3 with a somewhat higher frequency of editing at E15 (15%) that increases to 91% at P21 (Fig. 1B).
Figure 1.
Editing levels of the glutamate receptor transcripts at four different developmental stages; E15, E19, P2, and P21 of the mouse brain determined by 454 Amplicon Sequencing after RT-PCR. (A) The percentage of transcripts edited at Q/R site, the nearby +4 site, and the R/G site in Gria2. (B) Editing frequency at the R/G position of the Gria3 transcripts. (C) The level of editing at the Q/R site of the Grik1 transcripts. (D) Editing frequency at the Q/R, Y/C and I/V sites of the Grik2 transcripts.
Table 1.
Editing frequencies from embryonic days 15 and 19 (E15 and E19) as well as postnatal days 2 and 21 (P2 and P21)
aThe upper and lower limits of a confidence interval of 95%.
bThe number of transcripts sequenced.
cA novel site of editing in the Adarb1 transcript termed according to earlier positions (Dawson et al. 2004).
The transcripts of Grik1 and Grik2 show elevated editing levels during development at all sites (Fig. 1C,D). The Q/R site of Grik1 is inefficiently edited at E15 (8%) and increases to 62% at P21. In Grik2, all sites—Q/R, Y/C, and I/V—are edited between 3% and 27% at the early E15 stage and increase to 74%–81% editing at stage P21 (Fig. 1D). These results reveal that editing of the glutamate receptors is inefficient during embryogenesis with one exception, the Q/R site of Gria2.
Novel sites of A-to-I editing are also regulated during development
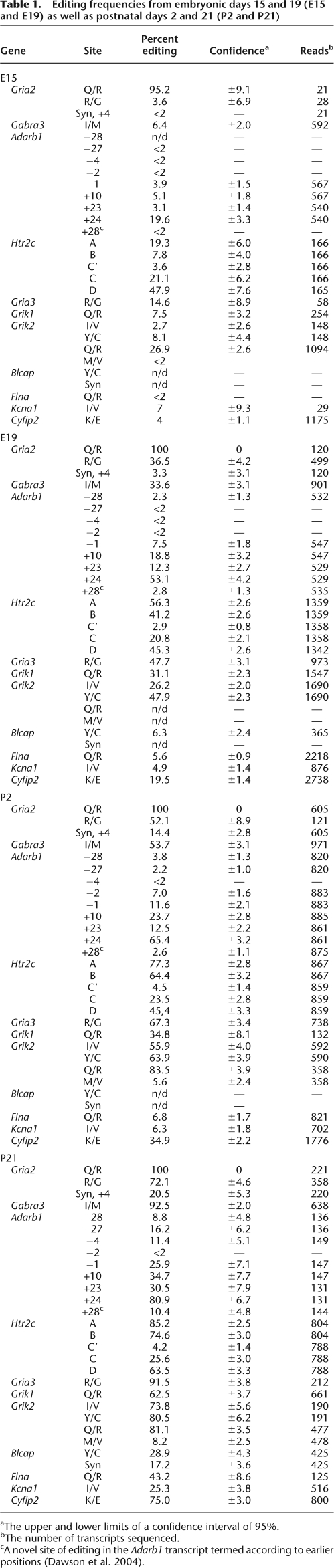
The Gabra3 transcript coding for the alpha3 subunit of the GABAA receptor was recently found by us to be edited at one site (I/M) by both ADAR and ADARB1 (Ohlson et al. 2007). Alpha3 is the only subunit of the GABAA receptor known to be edited. A dramatic increase in editing during development is seen at the I/M site of Gabra3 (Fig. 2A). Only 6% of the transcripts are edited at E15, whereas 54% of the transcripts are edited at day 2 of the newly born mice. At the later P21 stage, the editing frequency reaches 93%. We have previously analyzed the editing efficiency at the I/M site of Gabra3 during the postnatal stages 2, 7, 12, and 21 using the conventional cloning and sequencing method described previously (Ohlson et al. 2007). The results from the 454 Amplicon Sequencing are in good correlation to these data. To further investigate the biological variation between individuals, we used direct sequencing of PCR products from three different brains at four developmental days (E15–P21) (Supplemental Fig. S1A). From these data, we can conclude that the individual differences in editing efficiency are minor.
Figure 2.
Efficiency of editing in the transcripts of Gabra3, Cyfip2, Flna, and Kcna1 at four developmental stages, E15, E19, P2, and P21 of the mouse brain determined by 454 Amplicon Sequencing. (A) Editing frequency at the I/M site of Gabra3 transcripts. (B) Editing frequency at the K/E site of Cyfip2 transcripts. (C) Editing frequency at the Q/R site of Flna transcripts. (D) Editing levels at the I/V site of the Kcna1 transcripts.
Also, editing of the recently reported A-to-I editing sites in the cytoplasmic Fragile X mental retardation interacting protein (Cyfip2) and filamin A protein (Flna) mRNAs (Levanon et al. 2005) is regulated during development. The K/E position of Cyfip2 shows a gradual increase in the editing pattern through development with only 4% editing at E15 and 75% editing at the P21 stage (Fig. 2B). This position is mainly targeted by ADARB1, as was shown in two recent studies by Nishimoto et al. (2008) and Riedmann et al. (2008).
The Q/R site of the Flna transcript shows an interesting editing pattern during the maturation of the brain. The transcripts are inefficiently edited to <7% at E15, E19, and P2, while at P21 the editing frequency drastically increases to 43% (Fig. 2C). A similar pattern is seen for the I/V site of Kcna1. Here inefficient editing between 5% and 7% is seen until P2, with an increase to 25% at P21 (Fig. 2D). This is a surprisingly low level of constitutive editing compared to what previously has been determined (Bhalla et al. 2004). The stepwise increase in editing of these two substrates indicates that they are regulated in a different way from the other targets.
Editing changes the panel of serotonin receptor 2c isoforms expressed during development
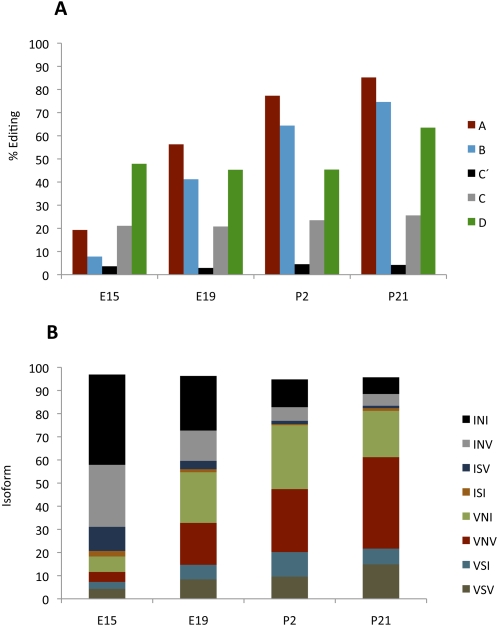
The serotonin receptor transcript Htr2c can be edited at five sites (A, B, C′, C, and D) in its coding sequence (Burns et al. 1997). Various combinations of editing at the A–D sites can, in theory, give rise to 24 different protein isoforms since seven different amino acids are possible at these sites depending on the editing pattern. The frequency of editing the A and B sites follows the same trend observed on most substrates analyzed during development—low embryonic editing levels that steadily increase over time (Fig. 3A). The C′ site is not efficiently edited at any time during development, and the C site shows low editing levels, whereas editing at the D site remains constant at an intermediate level (Fig. 3A). To determine the distribution of possible isoforms, we analyzed editing at all five positions within single transcripts. We find that the transcripts with no editing, giving rise to the INI (Ile-Asn-Ile) isoform, are the most common ones during early embryogenesis; at E15, 39% of the transcripts escaped editing (Fig. 3B). This isoform decreases during development, and only 7% of the transcripts are nonedited at P21 (Fig. 3B). The second most common isoform during embryogenesis is the INV (Ile-Asn-Val) isoform, derived from transcripts edited at the D site. At E15, 27% of the transcripts are edited only at the D position, whereas only 5% are edited at P21 (Fig. 3B). The A and B sites are the most highly edited, with 85% and 75% editing at P21 (Fig. 3A). At the later postnatal stages, these two sites are often edited in concert with editing at the D site (63% at P21). As a result of this coupling, the VNV (Val-Asn-Val) isoform is the most common form in the adult brain. During development, there is an increase of this isoform from 4% at E15 to 40% at P21 (Fig. 3B). The editing frequency at the C′ site was low all through development with only 3%–4% editing, frequently not accompanied by editing at other sites. This result shows that the fully edited isoform VGV (Val-Gly-Val) is very rare even in the adult animal, where <1% of the total transcripts of the serotonin receptor are edited at all sites (Supplemental Table S1).
Figure 3.
Editing of the serotonin receptor at the four developmental stages E15, E19, P2, and P21. (A) Editing levels at the A, B, C′, C, and D sites of the Htr2c transcripts was determined by 454 Amplicon Sequencing after RT-PCR. (B) The distribution of the HTR2C protein isoforms during development as a consequence of the editing events.
In summary, our results indicate that there is a shift in the representation of different isoforms between early and late developmental stages. During embryogenesis, the predominant isoforms are the nonedited INI (39%) and the INV (27%), while in the adult brain, the most common isoforms are VNV (40%), VNI (20%), and VSV (15%) (Fig. 3B).
To further investigate if there is a biological difference in editing between individuals, we analyzed three different brain samples at four developmental stages (E15–P21) using direct PCR sequencing of a region covering all five editing sites in the Htr2c transcript (Supplemental Fig. S1B). Since the editing efficiency between the different sites and developmental stages varies between 3% and 85%, this also gave us the opportunity to compare direct PCR sequencing with the 454 Amplicon Sequencing analyses. Together with the result from the Gabra3 direct sequencing (Supplemental Fig. S1), we concluded that: (1) The individual differences between brain samples at the same developmental stage are small, and the editing pattern within a sample can be superimposed on the other at the same stage; (2) less than 20% editing is not detectable by direct PCR sequencing (see C′ and C sites for Htr2c in Supplemental Fig. S1B); and (3) at 50% editing determined by 454 Amplicon Sequencing, the G peak is higher than the A peak in the chromatogram from direct PCR sequencing (see P2 for Gabra3 and D site for Htr2c in Supplemental Fig. S1). This indicates that the peak height of the sequencing chromatogram is not directly correlated to the level of editing.
Auto-editing and alternative splicing increase during development
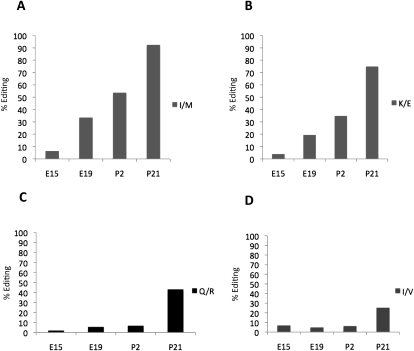
ADARB1 edits its own transcript at several sites within intron 4 (Rueter et al. 1999). There are eight reported editing sites in this region of ∼60 nt (Dawson et al. 2004). All of these sites show elevated editing during development (Table 1). Editing at one of these sites (−1) has been shown to induce alternative splicing that probably leads to a truncated, inactive ADARB1 protein if translated (Rueter et al. 1999). In our sequence analysis, we observed a developmental increase in editing also at this site (Fig. 4A; Table 1). At E15, 5% of the transcripts are edited at the −1 site, whereas 25% of the transcripts are edited at P21. Three sites (+10, +23, and +24) are more efficiently edited than the −1 site reaching 35%, 30%, and 81% editing at P21 (Fig. 4A). A novel editing site at +28 was found to be 10% edited at P21 (Table 1). Contradictory to recently published data in rat brain (Hang et al. 2008), we detected editing at sites −1, +10, +23, +24, and +28 in Adarb1 at the embryonic stages of the mouse brain.
Figure 4.
Editing of the Adarb1 transcripts during development and the ratio between normal and alternatively spliced Adarb1 transcripts. (A) Editing frequency at the −1, 10, 23, and 24 sites of Adarb1 transcripts was determined by 454 Amplicon Sequencing of RT-PCR products from four developmental stages, E15, E19, P2, and P21. (B) Detection of normal and alternatively spliced Adarb1 transcripts by RT-PCR during the six developmental stages, E15, E19, P2, P14, P21, and >P21. (C) Protein levels of ADAR and ADARB1 during development of the mouse brain. Western blot analysis showing the ADAR (110 kDa) and ADARB1 expression during the developmental stages, E15, E19, P2, P14, and P21 as indicated. An anti-actin antibody was used as a loading control.
To investigate if the increase in editing at the −1 site also influences the ratio between normal and alternative splicing, we used semiquantitative RT-PCR with a primer pair detecting both the normal and the alternative Adarb1 transcript. We find that the alternatively spliced transcript is made as a direct consequence of editing at the −1 site and therefore increases during development (Fig. 4B). In older animals, the alternatively spliced transcript is more abundant than the normally spliced transcript. This is not consistent with the relatively low editing frequency of 25% at this site at P21 (Fig. 4A). One explanation for this could be that a large fraction of the transcripts are rapidly spliced at the −1 site and therefore not detectable when the pre-mRNA is sequenced. Nevertheless, the functional mRNA appears to be expressed at a constant level during development (Fig. 4B).
The levels of ADAR proteins do not change during maturation of the brain
We wanted to investigate if the increase in editing frequency in the developing brain is correlated with increased expression levels of the ADAR proteins. Consequently, lysates from mouse brain at E15, E19, P2, P7, P21, and adult >P21 were used for Western blot to detect the protein levels of ADAR and ADARB1. We detect a constant protein expression of the 110-kDa ADAR protein from the early E15 stage to the later >P21 stage (Fig. 4C). A similar pattern is observed for ADARB1, showing no significant difference in protein levels between the early embryonic stages and the adult brain. Thus, our data reveal that most sites of selective editing in the mammalian brain are regulated during brain development but the regulation cannot be explained simply by an altered expression of the ADAR enzymes.
Discussion
We have used the 454 Amplicon Sequencing protocol to evaluate A-to-I editing within single transcripts of the developing brain. The editing efficiency during the development of the mouse brain was examined for the known mammalian editing substrates at four different developmental stages, two embryonic and two postnatal. The 454 Amplicon Sequencing technique allowed us to detect even low levels of editing with high statistical significance. We see a general trend of editing being regulated during development with increased editing activity over time in most substrates. The only real exception to this trend is the Q/R site in Gria2, which is edited to a high level through development. This is also in line with previous results showing that the unedited Q form is not essential for brain development, while the edited R form is (Kask et al. 1998; Higuchi et al. 2000). We find this site to be edited in all except one transcript already at E15. However, it has previously been shown in rat that the equivalent Q/R site in Grik1 and Grik2 is developmentally regulated (Bernard and Khrestchatisky 1994; Paschen et al. 1994; Schmitt et al. 1996). Our data are in total agreement with these analyses, but in addition, we can see that the I/V and Y/C sites of Grik2 also are edited with a low efficiency at embryogenesis that increases during development. These results indicate that editing regulation is not limited to specific sites, but rather that the Q/R site of Gria2 is an exception to the rule. As previously shown, we also see that the R/G site of Gria2 is developmentally regulated (Lomeli et al. 1994). Indeed, our data show a global regulation of most of the substrates that is independent of substrate specificity. ADAR and ADARB1 have both overlapping and distinct specificities for editing of the target sites (Higuchi et al. 2000; Hartner et al. 2004; Wang et al. 2004). Our data indicate no consensus-editing pattern between any of these sites, implying that there is no general trend that separates ADAR from ADARB1 editing during development. For example, the ADARB1-specific sites—Y/C of Grik2, K/E of Cyfip2, and the −1 site of the Adarb1 transcript—increase during development. The same pattern can be seen for the ADAR-specific targets of the A and B sites in the serotonin receptor. Furthermore, sites that can be efficiently edited by both ADAR and ADARB1, like the Gria2 R/G site and the Gabra3 I/M site, are indistinguishable from regulation of editing of other sites. Our results from the 454 Amplicon Sequence analysis confirm what others and we previously have seen in the developmental regulation of Gabra3 mRNA editing using conventional cloning and sequencing. Taken together, this implies that 454 Amplicon Sequencing is a reliable method well suited for large-scale screening of RNA editing. In addition, this method is highly sensitive and allows detection of editing <20% not possible to detect by direct sequencing of PCR products.
Previous studies have demonstrated that ADARB1 can edit its own pre-mRNA at a site within intron 4 that creates an alternative 3′ splice site 47 nt upstream of the normal splice site (Rueter et al. 1999). Splicing at this alternative splice site (−1) results in a loss of the functional ADARB1 protein, suggesting that auto-editing functions as a regulatory feedback loop to control protein expression. When we analyzed the editing activity at the −1 site of the Adarb1 transcript, an increase in editing frequency was observed during development (Fig. 4A; Table 1). We next analyzed the ratio between the normal and the alternatively spliced transcript and could see an increase in the production of the alternative transcript over time, in line with an increased auto-editing. When the developmental expression of the Adarb1 transcript was analyzed in a previous study, no distinction between the normal and the alternatively spliced transcript was made (Paupard et al. 2000). In agreement with their results, we see an increase in the total expression of the Adarb1 mRNA (Fig. 4B). However, in our analysis, the normal transcript remained at what appears to be a constant level throughout development. This is also in line with expression of the ADAR enzymes, which remained at a constant level during development (Fig. 4C). This result indicates that the auto-editing of Adarb1 maintains a constant protein expression during development and does not account for the increased editing frequency observed. Therefore, other factors are likely to regulate the activity of ADAR and ADARB1.
One factor that is important for the activity of the ADAR enzymes is inositol hexakiphosphate (IP6). When the crystal structure of the catalytic domain of ADARB1 was determined, IP6 was discovered to be buried within the enzyme core contributing to protein folding. One possible explanation of why ADAR is less active during early embryogenesis is if the production of IP6 is limited at this stage. In neural-specific tissue in mice at E8.5, the expression of Ipk2 required for IP6 production increases significantly (Frederick et al. 2005; Verbsky et al. 2005). Mice deficient for Ipk2 die at E9.5 with abnormal folding of the neural tube, indicating that IP6 is important during early embryogenesis. This result implies that IP6 is normally present during development and not the limiting factor that decreases the activity of the ADAR enzymes during early development.
A more likely explanation for the regulation is a positive factor that induces ADAR activity. It was recently shown in Caenorhabditis elegans that the ADBP-1 protein (ADR-2 binding protein-1) interacts with the A-to-I editing enzyme ADR-2 and retains it in the nucleus (Ohta et al. 2008). In an adbp-1 mutant strain, A-to-I RNA substrates were not edited, indicating that ADBP-1 is required for efficient editing. It is possible that a similar protein regulates editing in the mammalian brain by inducing editing activity during the maturation of the brain. This would also explain how some substrates with an optimal sequence and structure, like the Q/R site in Gria2, do not require additional factors for efficient editing during early embryogenesis. No homolog to the ADBP-1 protein has been identified in mammalian cells, making it more likely that an unrelated protein would induce the activity in higher eukaryotes.
Another factor that has been found to modulate editing activity is SUMO1 (small ubiquitin-like modifier 1). The human ADAR1 protein can be modified by SUMO1 on lysine 418, thereby reducing the editing activity (Desterro et al. 2005). SUMO1 was also shown to colocalize with ADAR1 in the nucleolus. Therefore, upon conjugation, SUMO1 might transport ADAR1 to the nucleolus, thereby separating ADAR from its targets. SUMO1 expression has been detected in embryonic stem cells; it is therefore a possible candidate as a negative regulator during early development.
What are the consequences of the low embryonic editing activity compared to editing in adults? It is clear from our results that the ratio between nonedited and edited isoforms of several neural receptors is different during embryogenesis from that in the adult animal. Our results indicate that the nonedited isoforms are used during early development, whereas the edited isoforms are required for a functional adult brain. Perhaps the clearest example is seen in the serotonin receptor 2c. Editing at the five sites (A, B, C′, C, and D) in various combinations has the potential to produce 24 different isoforms of the receptor. However, only a fraction of these transcript isoforms are produced in a detectable amount in the brain. In all, 24 RNA isoforms were found, but 11 of these were expressed <1% at any time during development (Supplemental Table S1). Included in this latter group are the all-edited transcripts that produce the VGV (Val-Gly-Val) isoform. Interestingly, even though we have not determined the local variations in different brain regions, it is clear that the overall distribution of the isoforms changes during development. At embryogenesis, the predominant isoforms are the non-edited INI (Ile-Asn-Ile) and INV (Ile-Asn-Val) with only the D-site edited (Fig. 3B). The nonedited INI isoform gives a high degree of constitutive activity compared to the edited isoforms (Niswender et al. 1999). These receptors are used in the immature brain when the network between neurons is undeveloped. In the adult brain, the most common isoform is the partially edited VNV form followed by the VNI and VSV isoforms. Evidence from several studies shows that extended editing of Htr2c decreases the ability of the receptor to couple to the G-protein and thereby communicate with the internal cellular environment (for review, see Werry et al. 2008). This leads to dramatic changes in signaling between the embryonic and adult states that most likely are important for brain development. It is possible that even minor changes in editing pattern during development can have a large impact on the mature brain. The integrated neuronal circuits in the adult brain require fast transmission between the pre- and post-synaptic neuron. Editing at the R/G site in the subunits of the glutamate receptor and the I/V site of the voltage-gated potassium channel Kcna1 results in functions that influence the kinetics of the receptors that are suitable for the mature brain. It has recently been suggested that editing of the alpha3 transcript Gabra3 results in receptors with slower activation and faster deactivation rates (Rula et al. 2008). However, it is not known what effects this might have on the GABAA receptor in vivo. Unpublished data from our laboratory suggest that editing of alpha3 inhibits trafficking of the receptor to the cell surface (data not shown). Editing might therefore play a role in the switch between the alpha3-containing receptors used during early development and the alpha1-containing receptors that are most common in the mature brain (Laurie et al. 1992).
We show here how 454 Amplicon Sequencing can be used to determine editing at many sites simultaneously and analyze the changes in editing during development. Our conclusion is that there is an extensive restraint on editing during early development and that it most likely has functional consequences for brain maturation. This powerful method can be used to determine editing levels in diseased individuals that might be caused by diminutive changes in editing patterns but sufficient to cause neurological dysfunctions.
Methods
Tissue preparation
Total brain from NMRI mice was collected at four different developmental stages—E15, E19, P2, and P21. The tissue was quickly removed and snap-frozen in nitrogen oxygen and kept at −80°C until further use.
RNA extraction
Total RNA from the mouse brain was isolated with TRIzol reagent according to the manufacturer's instructions (Invitrogen). First-strand cDNA synthesis was made from 3 μg of total RNA using random hexamer deoxyoligonucleotides (Invitrogen) and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions.
454 Amplicon Sequencing
PCR products were amplified using primers specific for the known A-to-I editing substrates in Table 1 and the FastStart High Fidelity PCR System (Roche). Adaptor deoxyoligonucleotides of 18 bases were linked to the specific primers according to 454 Amplicon Sequencing instructions. The sequences of the primers can be provided by the authors upon request. PCR products were purified on a 1.5% agarose gel using nucleospin extract II (MACHEREY-NAGEL). Of the gel-purified products, 5 ng was used for 454 Amplicon Sequencing (Margulies et al. 2005). The sequence determination was made according to the instructions by the manufacturer (Roche).
Identification of edited transcripts from 454 Amplicon Sequencing
For the subsequent collection and compilation of the data from 454 Amplicon Sequencing, we used in-house scripts. The correct gene tag to a sequence was recognized by the gene-specific primer that initiated each sequence. We used DIALIGN 2 (Morgenstern 1999) to create multiple sequence alignments (MSAs). For each edited and sequenced region, we aligned the corresponding data and included genomic data in which the coordinates were known for the expected edited site. For all positions of the known editing events, we calculated the proportion between As and Gs. For the Htr2c and Adarb1 edited regions, we used a different approach, since multiple editing events in a limited region inferred low quality to the alignment in the same region. To ensure not to include mis-aligned As or Gs in the wrong category, we used a pattern-matching approach. In the sequence spanning the edited region, all positions containing an edited A were classified with a logical OR, that is, either an A OR G. Hence, the pattern sequence would match the correct 454 Amplicon Sequence regardless of A/G ambiguities. Inherent in Perl, all As and Gs could be directly calculated through the matching.
Determination of editing frequencies
The error estimation of editing frequencies is given in Table 1 and was calculated by
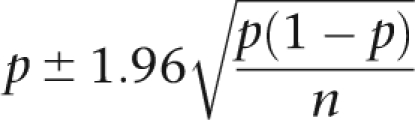 |
where p is the proportion of Gs (or editing frequency) and n is the sample size, in this case, the number of reads that were used.
PCR of alternatively spliced ADARB1
For the detection of alternatively spliced Adarb1 transcript, PCR reactions were made, following RNA extraction and cDNA synthesis as described above. Using a primer pair located in exon 4 and exon 5 allowed us to detect both the normal and alternatively spliced transcripts. The sequence of the primers can be provided by the authors upon request. To ensure linearity, the PCR reactions using Taq polymerase (Fermentas) were run for 25 cycles.
Western blot
Crude extract from mouse brain was obtained by homogenizing the brain with 3 volumes of Lysis-M buffer (Roche) according to the manufacturer's description. Lysates containing 30 μg of protein and 2× sample buffer (Bio-Rad) were boiled for 5 min, separated on a 10% polyacrylamide gel, and transferred to a polyvinylidine fluoride membrane (Bio-Rad). Following blocking in 10% nonfat milk in TBS-Tween, the membranes were incubated with antibodies against the ADAR proteins and actin as internal control. The ADAR-specific antibody was a kind gift from Dr. Brenda Bass (University of Utah). The ADARB1 antibody was raised in rabbit as previously described (Ohlson et al. 2005). The actin antibody was obtained from Sigma. A swine anti-rabbit coupled to horseradish peroxidase (DakoCytomation) was used as a secondary antibody that was visualized by the ECL-plus Western blot detection kit (GE healthcare).
Acknowledgments
We thank Christina Holmberg and the 454 Amplicon Sequence Facility at the KTH Genome Center, Stockholm. We thank Patrick Young for critically reading the paper. The Swedish Research Council funded this work.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study are available at http://www.molbio.su.se/Research/ohmanlab/454_database/ and have been deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA008179.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089409.108.
References
- Bass B.L., Nishikura K., Keller W., Seeburg P.H., Emeson R.B., O'Connell M.A., Samuel C.E., Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Khrestchatisky M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J. Neurochem. 1994;62:2057–2060. doi: 10.1046/j.1471-4159.1994.62052057.x. [DOI] [PubMed] [Google Scholar]
- Bernard A., Ferhat L., Dessi F., Charton G., Represa A., Ben-Ari Y., Khrestchatisky M. Q/R editing of the rat GluR5 and GluR6 kainate receptors in vivo and in vitro: Evidence for independent developmental, pathological and cellular regulation. Eur. J. Neurosci. 1999;11:604–616. doi: 10.1046/j.1460-9568.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Bhalla T., Rosenthal J.J., Holmgren M., Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Brusa R., Zimmermann F., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson T.R., Sansam C.L., Emeson R.B. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Keegan L.P., Jaffray E., Hay R.T., O'Connell M.A., Carmo-Fonseca M. SUMO1 modification alters ADAR1 editing activity. Mol. Biol. Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Sansam C.L., Singh M., Emeson R.B. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol. Cell. Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick J.P., Mattiske D., Wofford J.A., Megosh L.C., Drake L.Y., Chiou S.T., Hogan B.L., York J.D. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang P.N., Tohda M., Matsumoto K. Developmental changes in expression and self-editing of adenosine deaminase type 2 pre-mRNA and mRNA in rat brain and cultured cortical neurons. Neurosci. Res. 2008;61:398–403. doi: 10.1016/j.neures.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Hartner J.C., Schmittwolf C., Kispert A., Muller A.M., Higuchi M., Seeburg P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Kask K., Zamanillo D., Rozov A., Burnashev N., Sprengel R., Seeburg P.H. The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc. Natl. Acad. Sci. 1998;95:13777–13782. doi: 10.1073/pnas.95.23.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler M., Burnashev N., Sakmann B., Seeburg P.H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: Diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Laurie D.J., Wisden W., Seeburg P.H. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J. Neurosci. 1992;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon E.Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H., Mosbacher J., Melcher T., Hoger T., Geiger J.R., Kuner T., Monyer H., Higuchi M., Bach A., Seeburg P.H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher T., Maas S., Herb A., Sprengel R., Seeburg P.H., Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Nishimoto Y., Yamashita T., Hideyama T., Tsuji S., Suzuki N., Kwak S. Determination of editors at the novel A-to-I editing positions. Neurosci. Res. 2008;61:201–206. doi: 10.1016/j.neures.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Niswender C.M., Copeland S.C., Herrick-Davis K., Emeson R.B., Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- Ohlson J., Ensterö M., Sjöberg B.M., Öhman M. A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 2005;33:e167. doi: 10.1093/nar/gni169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson J., Pedersen J.S., Haussler D., Öhman M. Editing modifies the GABAA receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Fujiwara M., Ohshima Y., Ishihara T. ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans. Genetics. 2008;180:785–796. doi: 10.1534/genetics.108.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W., Dux E., Djuricic B. Developmental changes in the extent of RNA editing of glutamate receptor subunit GluR5 in rat brain. Neurosci. Lett. 1994;174:109–112. doi: 10.1016/0304-3940(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Paupard M.C., O'Connell M.A., Gerber A.P., Zukin R.S. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 2000;95:869–879. doi: 10.1016/s0306-4522(99)00431-5. [DOI] [PubMed] [Google Scholar]
- Riedmann E.M., Schopoff S., Hartner J.C., Jantsch M.F. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Rula E.Y., Lagrange A.H., Jacobs M.M., Hu N., Macdonald R.L., Emeson R.B. Developmental modulation of GABAA receptor function by RNA editing. J. Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J., Dux E., Gissel C., Paschen W. Regional analysis of developmental changes in the extent of GluR6 mRNA editing in rat brain. Brain Res. Dev. Brain Res. 1996;91:153–157. doi: 10.1016/0165-3806(95)00175-1. [DOI] [PubMed] [Google Scholar]
- Seeburg P.H., Higuchi M., Sprengel R. RNA editing of brain glutamate receptor channels: Mechanism and physiology. Brain Res. Brain Res. Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- Sommer B., Kohler M., Sprengel R., Seeburg P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Verbsky J., Lavine K., Majerus P.W. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc. Natl. Acad. Sci. 2005;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B., Royle G.A., Christie B.R., Schiffer H.H., Ghetti A., Tritto T., Perez-Otano I., Radcliffe R.A., Seamans J., Sejnowski T., et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron. 2001;29:217–227. doi: 10.1016/s0896-6273(01)00192-1. [DOI] [PubMed] [Google Scholar]
- Wang Q., O'Brien P.J., Chen C.X., Cho D.S., Murray J.M., Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin 2C receptors. J. Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D.L., Weiss M.J., Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Werry T.D., Loiacono R., Sexton P.M., Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol. Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]