Abstract
Next-generation massively parallel sequencing technologies provide ultrahigh throughput at two orders of magnitude lower unit cost than capillary Sanger sequencing technology. One of the key applications of next-generation sequencing is studying genetic variation between individuals using whole-genome or target region resequencing. Here, we have developed a consensus-calling and SNP-detection method for sequencing-by-synthesis Illumina Genome Analyzer technology. We designed this method by carefully considering the data quality, alignment, and experimental errors common to this technology. All of this information was integrated into a single quality score for each base under Bayesian theory to measure the accuracy of consensus calling. We tested this methodology using a large-scale human resequencing data set of 36× coverage and assembled a high-quality nonrepetitive consensus sequence for 92.25% of the diploid autosomes and 88.07% of the haploid X chromosome. Comparison of the consensus sequence with Illumina human 1M BeadChip genotyped alleles from the same DNA sample showed that 98.6% of the 37,933 genotyped alleles on the X chromosome and 98% of 999,981 genotyped alleles on autosomes were covered at 99.97% and 99.84% consistency, respectively. At a low sequencing depth, we used prior probability of dbSNP alleles and were able to improve coverage of the dbSNP sites significantly as compared to that obtained using a nonimputation model. Our analyses demonstrate that our method has a very low false call rate at any sequencing depth and excellent genome coverage at a high sequencing depth.
Genetic polymorphisms contribute to variations in phenotypes, risk to certain diseases, and response to drugs and the environment. Genome-wide linkage analysis and positional cloning have been tremendously successful for mapping human disease genes that underlie monogenic Mendelian diseases (Jimenez-Sanchez et al. 2001). But most common diseases (such as diabetes, cardiovascular disease, and cancer) and clinically important quantitative traits have complex genetic architectures; a combination of multiple genes and interactions with environmental factors is believed to determine these phenotypes. Linkage analysis has significant limitations in its ability to identify common genetic variations that have modest effects on disease (Wang et al. 2005). In contrast, genome-wide association studies offer a promising approach for mapping associated loci. The completion of the human genome sequence (Lander et al. 2001; Venter et al. 2001) enabled the identification of millions of single nucleotide polymorphisms (SNPs) (Sachidanandam et al. 2001) and the construction of a high-density haplotype map (International HapMap Consortium 2005; International HapMap Consortium et al. 2007). These advances have set the stage for large-scale genome-wide SNP surveys for seeking genetic variations associated with or causative of a wide variety of human diseases.
For more than two decades, Sanger sequencing and fluorescence-based electrophoresis technologies have dominated the DNA sequencing field. And DNA sequencing is the method of choice for novel SNP detection, using either a random shotgun strategy or PCR amplification of regions of interest. Most of the SNPs deposited in dbSNP were identified by these methods (Sherry et al. 2001). A key advantage of the utility of traditional Sanger sequencing is the availability of the universal standard of phred scores (Ewing and Green 1998; Ewing et al. 1998) for defining SNP detection accuracy, in which the phred program assigns a score to each base of the raw sequence to estimate an error probability.
With high-throughput clone sequencing of shotgun libraries, a standard method for SNP detection (such as ssahaSNP; Ning et al. 2001) is to align the reads onto a reference genome and filter low-quality mismatches according to their phred score, known as the “neighborhood quality standard” (NQS) (Altshuler et al. 2000). With direct sequencing of PCR-amplified sequences from diploid samples, software, including SNPdetector (Zhang et al. 2005), novoSNP (Weckx et al. 2005), PolyPhred (Stephens et al. 2006), and PolyScan (Chen et al. 2007), has been developed to examine chromatogram files to detect heterozygous polymorphisms.
New DNA sequencing technologies, which have recently been developed and implemented, such as the Illumina Genome Analyzer (GA), Roche/454 FLX system, and AB SOLiD system, have significantly improved throughput and dramatically reduced the cost as compared to capillary-based electrophoresis systems (Shendure et al. 2004). In a single experiment using one Illumina GA, the sequence of approximately 100 million reads of up to 50 bases in length can be determined. This ultrahigh throughput makes next-generation sequencing technologies particularly suitable for carrying out genetic variation studies by using large-scale resequencing of sizeable cohorts of individuals with a known reference (Bentley 2006). Currently, using these technologies, three human individuals have been sequenced: James Watson's genome by 454 Life Sciences (Roche) FLX sequencing technology (Wheeler et al. 2008), an Asian genome (Wang et al. 2008), and an African genome (Bentley et al. 2008) sequenced by Illumina GA technology. Additionally, given such sequencing advances, an international research consortium has formed to sequence the genomes of at least 1000 individuals from around the world to create the most detailed human genetic variation map to date.
As noted, SNP detection methods for standard sequencing technologies are well developed; however, given distinct differences in the sequence data output from and analyses of next-generation sequencing, novel methods for accurate SNP detection are essential. To meet these needs, we have developed a method of consensus calling and SNP detection for the massively parallel Illumina GA technology. The Illumina platform uses a phred-like quality score system to measure the accuracy of each sequenced base pair. Using this, we calculated the likelihood of each genotype at each site based on the alignment of short reads to a reference genome together with the corresponding sequencing quality scores. We then inferred the genotype with highest posterior probability at each site using a Bayesian statistical method. The Bayesian method has been used for SNP calling for traditional Sanger sequencing technology (Marth et al. 1999) and has also been introduced for the analysis of next-generation sequencing data (Li et al. 2008a). In the method presented here, we have taken into account the intrinsic bias or errors that are common in Illumina GA sequencing data and recalibrated the quality values for use in inferring consensus sequence.
We evaluated this SNP detection method using the Asian genome sequence, which has 36× high-quality data (Wang et al. 2008). The evaluation demonstrated that our method has a very low false call rate at any sequencing depth, and excellent genome coverage in high-depth data, making it very useful for SNP detection in Illumina GA resequencing data at any sequencing depth. This methodology and the developed software described in this report have been integrated into the Short Oligonucleotide Alignment Program (SOAP) package (Li et al. 2008b) and named “SOAPsnp” to indicate its functionality for SNP detection using SOAP short read alignment results as input.
Results
System design for genotype calling
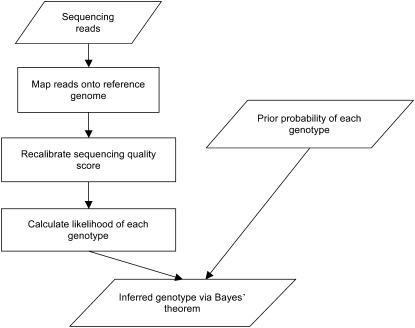
We used Bayes's theorem to infer the genotype given the observed allele types and quality scores at each chromosomal site. The steps for this method are depicted in Figure 1. For input data, the method used sequencing reads generated by the Illumina GA technology. These reads were then mapped onto a known reference genome (in this case, the genome sequence data from an Asian individual onto the build NCBI build 36.1 reference) using SOAP (Li et al. 2008b), and the alignment of uniquely mapped reads was used to build the consensus sequence for the sequenced genome. A sequencing quality score, which is an estimation of the sequencing error rate of each base, was recalibrated according to the observed mismatch rate of the read alignment onto the reference genome. Next, we calculated the likelihood of each observed genotype at each position on the genome. We then calculated the posterior probabilities of the genotypes using a Baysian formula that used the likelihood of observed genotypes and the estimated SNP rate between the sequenced sample and the reference genome as prior probability. The genotype assigned to each genomic location was the one with the highest probability, and that probability was transformed to a phred-like quality score to indicate the accuracy of the called genotype. Finally, a sum rank test was used to further eliminate any artificial heterozygous sites.
Figure 1.
Algorithmic overview of consensus calling for massively parallel resequencing. The program takes raw sequencing reads as input, maps them onto the reference genome, and calculates the likelihood of each possible genotype. It outputs the inferred genotype with highest posterior probability and its corresponding quality score.
Prior probability of each genotype
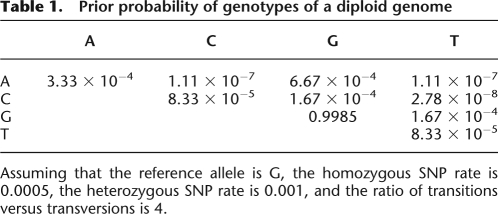
Given an available reference genome, the mutation rate of the newly sequenced individual and the reference genome can be estimated from known large SNP discovery studies. The estimated SNP rate between two human haploid chromosomes is about 0.001 (Sachidanandam et al. 2001). If we therefore assume that the human reference genome sequence has an error rate of 1 × 10−5 (Collins et al. 2004), each inferred haploid chromosome of a sequenced sample should have about one in a thousand bases different from the reference. Using these numbers, for diploid chromosomes, we set the homozygous SNP rate at 0.0005 and the heterozygous rate at 0.001.
According to a previous study on NCBI dbSNPs (Zhao and Boerwinkle 2002), transitions are four times more frequent than transversions among the substitution mutations, but there is little bias among each type of transition or transversion combination. Given this, these ratios were used in our SNP detection model. For example, assuming that the reference allele is G at a location, the prior probability of haploid genotypes is as follows: A and T are each 1.67 × 10−4; C is 6.67 × 10−4; and G is 0.999. The prior probability of the diploid genotypes GG is 0.9985; AA is 3.33 × 10−4; CC and TT are 8.33 × 10−5; AC and AT are 1.11 × 10−7; GC and GT are 1.67 × 10−4; AG is 6.67 × 10−4; and CT is 2.78 × 10−8 (Table 1).
Table 1.
Prior probability of genotypes of a diploid genome
Assuming that the reference allele is G, the homozygous SNP rate is 0.0005, the heterozygous SNP rate is 0.001, and the ratio of transitions versus transversions is 4.
Likelihood calculation using quality scores
The candidate allele types D at each location can be observed from the alignment of mapped reads on the reference genome. The likelihood of each assumed genotype Ti is P(D|Ti). All four attributes of each observed allele, including (1) allele type, (2) quality score, (3) coordinates on the read, and (4) t-th occurrence were integrated into our model of likelihood calculation to maximize information usage. (See Methods for details.)
Differences in bases between the reference and the new sequence can also be caused by errors during sequencing and by misalignment of short reads. Since the read length of next-generation sequencing is quite short, there is a higher possibility of reads from highly diverged genomic regions to be incorrectly mapped, thus creating an incorrect SNP call. We filtered most of these incorrect alleles by setting a frequency cutoff that we determined best to filter out these errors (discussed in the next section).
With regard to sequencing, errors are often not random; this is especially true for low quality bases and those near the 3′-end of reads. A ↔ C and G ↔ T substitution errors are significantly (P < 0.0001) overrepresented. Given these aspects, we used a multiple dimensional matrix to recalibrate the quality scores by taking into account read coordinates, the sequencing quality score, and substitution error bias. We then used these recalibrated quality scores to calculate the likelihood of the genotypes. Additionally, to avoid dependent errors, we reduced the sum quality of genotypes that may be due to the presence of duplicate clones of PCR amplification. Investigation of these problems and the means we devised to remedy them are discussed in the following sections.
Uniqueness and accuracy of read placement
To evaluate the uniqueness and accuracy of read mapping, we generated simulated short reads of different lengths from chromosome 12 of the NCBI human genome. The simulated reads contained a SNP rate of 0.001 against the reference, and mismatch sequencing errors were generated using a sampling of the quality scores from the Asian genome sequencing data. The simulated reads were realigned back to the whole human reference genome. We called a “best hit” for each simulated read or read-pair that mapped to a position on a chromosome with the lowest number of nucleotide differences between the read and the reference genome. A read having only a single best hit was considered uniquely aligned. Reads that had more than one “best hit” (meaning that they could be aligned to multiple positions with the same number of mismatches) were considered repeatedly aligned.
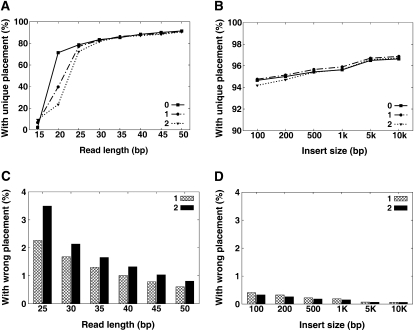
From these data, we calculated the percentage of reads that could be uniquely aligned. For single-end reads, the percent uniqueness increased sharply (10 times) for read lengths from 15 to 25 bp, but beyond this length, there was only a small change in percent uniqueness (Fig. 2A). For reads that were identical to the sequence at their mapped position, 78.6% of the 25-bp reads and 91.5% of the 50-bp reads were uniquely mapped. At a 35-bp length, which is the typical read length generated by Illumina GA technology, 85.4%, 86.3%, and 85.9% of the 0-, 1-, and 2-mismatch hits, respectively, are unique. The percent uniqueness in the simulation data was similar to that found from mapping the sequence reads generated from the Asian genome sequence. From our analysis, we also found that paired-end sequencing greatly improved the amount of uniquely mapped reads. We next fixed read length at 35 bp and simulated insert sizes from 100 bp to 10 kb with ±10% deviation and found that the percent uniqueness only slightly improved with increased insert size (Fig. 2B). At an insert size of 200 bp, 95.4% of the read pairs have unique placement.
Figure 2.
Uniqueness and accuracy of read placement. We produced a copy of human chromosome 12 with a 0.001 SNP rate to the NCBI reference, then simulated 36× reads for each given read length or paired-end insert size. The simulated error rate over all reads is ∼1%, and the standard deviation on paired-end insert size is 10%. (See Methods section for details.) All the read sequences were then aligned back to the reference genome, and then the uniqueness and accuracy of reads placement was evaluated: (A) unique placement of single-end reads; (B) unique placement of paired-end reads (read length: 35 bp); (C) wrong placement of single-end reads; (D) wrong placement of paired-end reads (read length, 35 bp).
When mapping using very short reads, it is likely that some reads containing true SNPs or reads containing sequencing errors could map to incorrect locations. In simulated reads, since the original locations were known, we evaluated the rate of misplacement. In 25-bp single-end reads, 2.3% and 3.5% of the 1- and 2-mismatch hits, respectively, were mapped incorrectly (Fig. 2C). This rate was reduced to 0.6% and 0.8% when using reads that were 50 bp in length. A similar survey using simulated 1-mismatch paired-end reads with insert sizes of 100 bp and 10 kb had an incorrect placement of 0.4% and 0.06%, respectively; and 2-mismatch reads of these two insert sizes had a misplacement rate of 0.3% and 0.06%, respectively (Fig. 2D).
We calculated the frequency of incorrect alleles in the simulated data to detect misidentified SNPs from real ones. More than 95% of the erroneous alleles appear only once in both single-end (35-bp read length) and paired-end (200-bp insert size) reads. Given these findings, we set a filtering threshold that removed all low-frequency alleles that had fewer than four reads as support. We used this strategy to call SNPs in the Asian genome, and only about 0.036% of all the incorrect alleles remained. In a simulation of random DNA fragmentation, using 36× sequencing, only ∼0.008% of real heterozygous alleles were removed by this frequency filter. Next, to distinguish high-frequency errors from heterozygous SNPs, we used binomial distribution (P = 0.0001) to detect a frequency discrepancy of these two alleles at any site and found that 87.3% of the remaining incorrect alleles were also removed. In all, 99.93% of the erroneous alleles caused by misplaced reads were filtered out using these frequency thresholds.
An additional error source in our SNP identification process is the presence of incorrectly aligned reads that contain indels. This error source is related to our read alignment method. Because there are nearly five to 10 times more SNPs than small insertions or deletions (Dawson et al. 2001), we first carried out an ungapped alignment. For those reads that could not be mapped by this method, we then allowed up to a 3-bp insertion or deletion to assign a best hit. Because of this “ungapped prior to gapped alignment” strategy, some reads that truly contain an indel may have been erroneously aligned during the ungapped mapping stage. To evaluate the potential impact of this on our SNP detection, we simulated 10,000 small indels and found that 0.6% of the indel-containing reads did have a best hit during the ungapped mapping stage. By using the same frequency filter as we used above (requiring at least four reads for support), only three (0.03%) of the incorrect SNP alleles generated by read misplacement remain.
Recalibration of Illumina GA quality scores
Errors accumulate during the sequencing process, and the later cycles near the 3′-end of reads have a much higher error rate than do earlier cycles. The raw quality scores of Illumina GA sequencing are calculated from the signal intensities. These quality scores do not accurately represent the true error rate. To correct for this, we evaluated the deviation in the Asian genome sequencing data and designed a method to recalibrate the standard quality scores using the observed mismatch rate from the alignment of mapped reads on the reference genome. To avoid as many true SNPs as possible in our recalibration, we excluded all the mismatches between the sequenced reads and the reference genome that are currently present in dbSNP, and thus are known SNP sites.
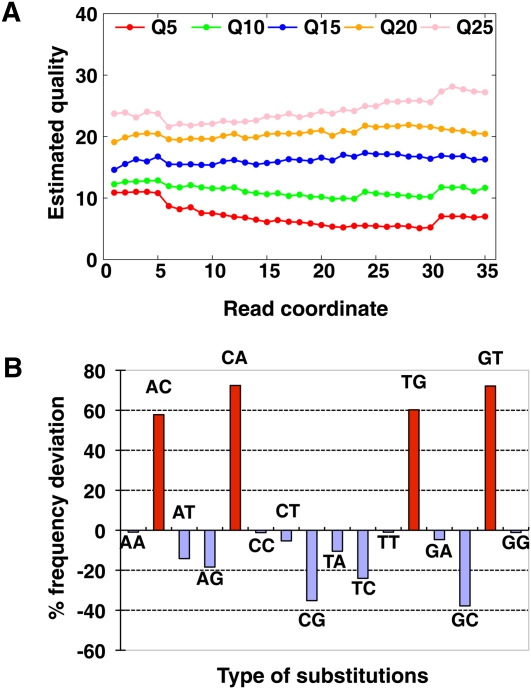
The Illumina GA pipeline can recalibrate the quality scores by separating sequencing cycles into several bins. The calibrated quality scores still had an obvious deviation from the real mismatch rate at each sequencing cycle, and the deviation fluctuated over the cycles (Fig. 3A). Here, we recalibrated the scores cycle-by-cycle again by loading the alignment together with either the raw sequencing scores or the recalibrated scores such as by the Illumina GA pipeline.
Figure 3.
Inaccuracy of sequencing quality score and biased substitution errors. (A) Estimated quality of Illumina GA pipeline recalibrated quality values 5, 10, 15, 20, and 25 along sequencing cycles. We extracted bases with each quality value from raw reads of the Asian genome sequencing, then estimated the real quality by the mismatch rate in the alignment as [−10log10(mismatch rate)]. (B) Deviation of quality score to estimated mismatch rate of each substitution combination. The percentage of deviation was calculated by [(Error rate by alignment mismatch rate) − (Error rate according to quality value)]/(Error rate according to quality value). The substitution of A on read to C on reference was represented as AC in the figure.
In addition to increased sequencing errors in later cycles having an impact on the calculated quality scores, the method of nucleotide detection can also affect the quality score. Illumina GA technology uses two lasers to excite the dye attached to each of the four nucleotides. The four intensity signals are not independent because the frequency emission of these four dyes overlaps. A and C use the same laser, while G and T use another laser, so the sequencing errors of A ↔ C or G ↔ T substitution are more frequent than the other types of substitutions. We found that the quality scores of A ↔ C and G ↔ T substitution were ∼58%–72% overestimated than the observed substitution rate from alignment, while the quality scores of C ↔ G substitution were ∼36% underestimated (Fig. 3B). For example, among the bases with quality 10 (or equal to error rate 0.1), the observed substitution rates between the reads and the reference sequence of A → C, C → A, G → T, and T → G are 4.62%, 5.27%, 5.29%, and 4.62%, respectively, while the rate of the other types of substitutions is ∼1.62% to 2.48%. Thus, we also calibrated the quality score by separating each type of substitution.
Penalty for duplicate reads
PCR was used to add adapter and amplify the library for sequencing. There can be an increase in the number of duplicate clones in a library if the starting amount of DNA is small, as is the case when obtaining DNA from a gel slice to get uniform fragment length or if there are too many PCR cycles. The presence of duplicate clones will significantly influence the randomness of the sequencing process. Some of the genomic region will have an unexpectedly high depth. This can also result in large frequency differences between the two alleles of a heterozygous site. In particular, DNA damage or amplification errors from early PCR cycles could then be present in multiple reads; such repeated identical errors would be hard to distinguish from real SNPs. Thus, we set a penalty to the reads that have an identical mapping location on the reference genome. If the library and sequencing process is random, then the location distribution on the reference is expected to exhibit a Poisson distribution.
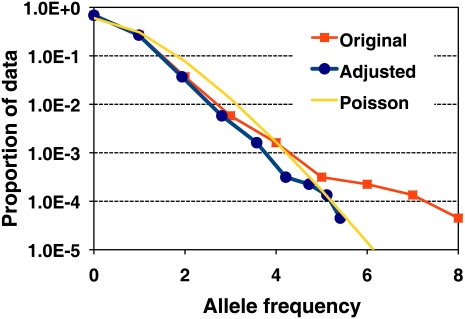
In the 36× Asian genome sequencing using read lengths up to 35 bp, 0.39% of the chromosomal positions were covered by six or more read mapping start points; however, in theory, the percentage should be ∼0.07%. We therefore used an empirical exponential adjustment to reduce the contribution of reads with an identical genomic start point. Using the Illumina 1M BeadChip on the same DNA sample, we examined the homozygous alleles that showed different allele types in the reads alignment and found that the frequency of these incorrect alleles fit the Poisson distribution after this adjustment (Fig. 4).
Figure 4.
Effective allele frequency of incorrect alleles before and after adjusting reads with identical mapping location. We calculated the frequency of the alleles in the Asian genome sequencing reads that are different from the genotyping results. The frequency contributed by the n-th reads with the same mapping location was multiplied by θn, where 0 ≤ θ ≤ 1. The original, adjusted frequency and Poisson distribution are shown.
Evaluation in human genome deep resequencing
We tested our SNP detection method, which incorporated all of the above corrections, in the Asian genome data with 36× deep sequencing of a male individual. According to the sequencing quality scores, the estimated error rate of the detected SNPs is lower than 1%. PCR amplification and validation of a few dozens of randomly selected SNPs cannot reflect the accuracy rate precisely. So, we did the validation by comparing the inferred consensus sequence to the genotyping result using the same DNA sample on an Illumina 1M BeadChip. We assumed that all the genotyped alleles were correct and separated all the conflicting sites into false-negative (FN) and false-positive (FP) categories. FN is considered a call of a heterozygous site where one allele is missing in the GA sequencing consensus calling, and FP is considered a call of an incorrect allele. Although the FN and FP rates are defined over the genotyping sites, they are effective overall indicators of SNP calling accuracy for the whole genome.
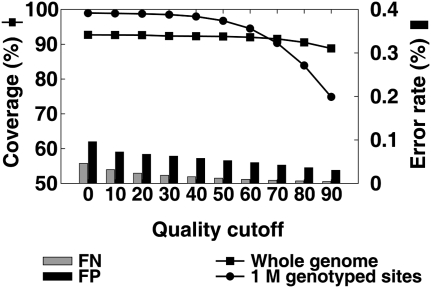
Figure 5 shows the percent of the reference genome that is covered by the assembled nonrepetitive consensus sequence, the percent of the Illumina 1M BeadChip genotyped alleles covered by the consensus sequence, and the error rate of the consensus sequence under different quality filtering assuming that all genotyping results were correct. Without using a quality filter, the coverage of the whole genome is lower than the coverage of the genotyping sites. This is because the genotyping sites are biased to a unique portion of the genome. By increasing the quality cutoff from Q0 to Q40, the coverage of the genotyping sites decreased only slightly: from 98.98% to 97.02%; but the coverage decreased very rapidly when a higher cutoff was used. This can be explained by the use of a low prior probability setting of the SNP sites, which resulted in relatively lower quality scores than at other sites. The rate of FP and FN showed a continuous decrease with an increase in the quality filtering threshold (from 0.067% by Q0 to 0.059% by Q20 for FP, from 0.115% by Q0 to 0.083% by Q20 for FN). Based on these data, we set a quality cutoff at Q20 to obtain the best trade-off between coverage and rate of undercall and overcall.
Figure 5.
Coverage of whole genome, coverage of the genotyped sites, and error rate as a function of quality filter. The consensus sequence of the Asian genome was calculated from the 36× sequencing reads. The error rate was estimated by assuming the genotyping result on the same sample was right. A false-positive (FP) is to call an incorrect allele, while a false-negative (FN) is the heterozygous site with one allele missing.
Five additional filter steps were used to remove unreliable portions of the consensus sequence: (1) We required at least two reads for haploid chromosome X/Y and four reads for diploid autosomes. (2) The overall depth, including randomly placed repetitive hits, had to be less than 100. (3) The approximate copy number of flanking sequences had to be less than two. (This was done in order to avoid misreading SNPs as heterozygotes caused by the alignment of similar reads from repeat units or by copy number variations [CNVs].) (4) There had to be at least one paired-end read. (5) The SNPs had to be at least 5 bp away from each other.
For haploid chromosome X
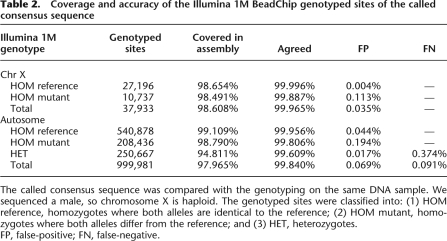
We sequenced a male genome, which has only one X chromosome, so consensus calling for the X chromosome is the same as for a haploid genome. There are four different genotypes and only one allele at each site. Among the 37,933 Illumina 1M BeadChip loci on the X chromosome, 98.61% were well covered in the assembled consensus sequence with a 99.97% agreement (Table 2). The consensus sequence covered 88.07% of the X chromosome of the reference genome. The remaining unassembled chromosomal regions are highly repetitive and thus had very few unique read mappings. The Y chromosome is mainly composed of repeat sequences and was therefore poorly assembled; thus, these results are not included.
Table 2.
Coverage and accuracy of the Illumina 1M BeadChip genotyped sites of the called consensus sequence
The called consensus sequence was compared with the genotyping on the same DNA sample. We sequenced a male, so chromosome X is haploid. The genotyped sites were classified into: (1) HOM reference, homozygotes where both alleles are identical to the reference; (2) HOM mutant, homozygotes where both alleles differ from the reference; and (3) HET, heterozygotes.
FP, false-positive; FN, false-negative.
For diploid autosomes
To evaluate the accuracy of consensus calling and SNP detection in a diploid genome, we compared the assembled consensus sequence of all autosomes of the Asian genome to the genotyping results and to the NCBI reference. The inferred consensus sequence covered 92.25% of the whole autosomal reference and 97.97% of the Illumina 1M BeadChip loci. In a comparison of the array-based genotyped alleles and GA sequencing-called alleles, ∼99.84% of the overlapping loci were in agreement (Table 2). Of the genotyped homozygous loci that were identical to the reference, 0.044% were called as heterozygote by our GA sequencing consensus; while in the genotyped homozygous loci that are different from the reference, 0.104% were called as heterozygote, and 0.090% were called as homozygote but of the incorrect allele type. Among the 250,667 genotyped heterozygous loci, there was a 0.017% FP rate and 0.374% FN rate in the GA sequencing consensus. Overall, the 999,981 genotyped sites, the FP and FN rates were 0.069% and 0.091%, respectively. As has been shown in the analysis of the Asian genome data, a subset of the possible falsely called SNPs, which are inconsistent with the array-based genotyping, was PCR-amplified and sequenced again using traditional Sanger sequencing technology to assess the accuracy of the GA sequencing versus genotyping methods for SNP identification (Wang et al. 2008). Among the 57 examined loci, 49 (86.0% loci) showed consistent allele types with GA sequencing consensus rather than the array genotyping.
In all, the consensus and SNP calling by this method showed good consistency with the array-based genotyping results; and for the possible falsely called loci, the GA sequencing consensus appeared more often to be correct.
MAQ is another tool developed for short reads mapping and consensus assembly (Li et al. 2008a). We ran MAQ on the same read data set and also compared the inferred consensus sequence to the genotyping results. On the haploid X chromosome, SOAPsnp has an obviously higher coverage of the Illumina 1M BeadChip loci than MAQ (98.61% vs. 95.93%). The FP rate of SOAPsnp (0.04%) is also lower than MAQ (0.18%). On the diploid autosomes, SOAPsnp had a slightly higher coverage (97.97% vs. 97.92%), but both a lower FP rate (0.07% vs. 0.15%) and FN rate (0.09% vs. 0.17%) than MAQ.
Using dbSNP prior probability for low-depth sequencing
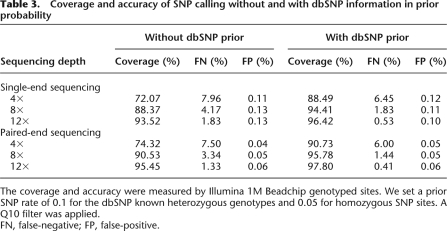
At a low sequencing depth, the real heterozygous alleles will be difficult to distinguish from sequencing errors. In our method, we used the estimated SNP rate based on its prior probability, so the FP rate was very low. But using this in the called consensus of low-depth sequencing will mean that some of the heterozygous alleles are likely to fail to pass the quality filter or will pass, but with the alternative allele missing. We therefore used a Q10 quality filter instead of Q20, which was used for high-depth sequencing regions, and found that the percentage of genotyping sites covered by the unfiltered consensus sequence was improved from 32.4% by Q20 to 72.1% for 4× single-end reads (Table 3). At the same time, the FN rate of heterozygous alleles increased significantly from 1.19% using Q20 to 7.96%, meaning that 13.49% of the heterozygous sites in the consensus sequence with a quality filter between Q10 and Q20 were incorrectly called as homozygous. Our results indicate that the quality filter used in regions of low-depth sequencing can have a large impact; therefore, it is important to make a decision based on a trade-off between coverage and FN rate of heterozygotes.
Table 3.
Coverage and accuracy of SNP calling without and with dbSNP information in prior probability
The coverage and accuracy were measured by Illumina 1M Beadchip genotyped sites. We set a prior SNP rate of 0.1 for the dbSNP known heterozygous genotypes and 0.05 for homozygous SNP sites. A Q10 filter was applied.
FN, false-negative; FP, false-positive.
According to a comparison of the identified SNPs in the Asian genome with those present in dbSNP, ∼90% of the SNPs in a newly sequenced human individual genome are already in dbSNP, thus using dbSNP information for determining prior probability is useful for capturing more SNPs. We therefore set a prior SNP rate of 0.1 for the dbSNP known heterozygous genotypes and 0.05 for homozygous SNPs to improve our SNP calling in regions of low-depth sequencing. Assuming that a genotype is G/T at a dbSNP site, then the prior probability of GG and TT is estimated to be 0.454; GT is 0.0909; AT, CT, AG, CG is 9.1 × 10−5; AA, CC is 4.55 × 10−7; and AC is 9.11 × 10−8. Consensus calling using these new prior probabilities showed that the coverage of the genotyping sites was improved from 88.37% to 94.41% with single-end and 90.53% to 95.78% with paired-end sequencing of 8× reads (Table 3). Correspondingly, the FN rate of heterozygotes was reduced from 4.17% to 1.83% and 3.34% to 1.44% with single-end and paired-end sequencing, respectively. Thus, using dbSNP sites in prior probability for low-depth sequencing provides a solid improvement in the coverage of SNP sites and improves the accuracy of heterozygous site detection.
Computational complexity
The software is implemented in standard C++ language. SOAP alignment results of raw short reads onto the corresponding reference genome were loaded into SOAPsnp, and it output the inferred consensus sequence. To facilitate SNP calling for a multi-sample population data set, there is also an option to have allele likelihoods for each genomic locus output in a flat tabular format. Analysis time is proportional to the total amount of sequencing data: For example, our analysis of the 36× Asian genome sequencing data consumed ∼200 CPU hours with a RAM usage of <2 GB. The computing process can also be parallelized by splitting the input data into chromosomes or genomic fragments.
Discussion
Ultrahigh throughput and the characteristics of short read length make next-generation sequencing technologies particularly suitable for large-scale resequencing, such as for use in human personal genome sequencing or in the sequencing of a cohort of individuals to capture rare mutations and to build a detailed genetic variation map of a population. These sequencing technologies could also be used for genome-wide association studies. Although the cost of sequencing is still not low enough to perform whole-genome sequencing on thousands of individuals in one study, the target region capture method does provide a current way to focus on specific regions of interest, such as coding, regulatory, or non-repeat regions (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007; Porreca et al. 2007). SNP detection is a key step in all such studies.
In this study, we described a method for consensus calling and SNP detection of massively parallel sequencing-by-synthesis Illumina GA technology. This method took into account the error patterns inherent in this sequencing technology and integrated all this information into a single quality score to measure the accuracy of each nucleotide position in the consensus sequence. The quality of this method has been evaluated using the Asian genome sequencing data and shows a higher accuracy than that in previous studies using traditional Sanger sequencing technology.
The typical error pattern seen in Illumina GA sequencing differs from that of other sequencing-by-synthesis methods and thus requires the availability of a SNP detection method that properly takes this into account. For example, the typical sequencing errors in reads that are generated by the Roche 454 Genome Sequencer 20 are a miscalculation of homopolymer runs, which display as insertions or deletions rather than substitutions. So the quality score of 454 reads represents the probability that the base should be called according to the observed signal intensity, and a method taking this aspect into account for calibrating quality scores and SNPs for Roche 454 sequencing technology has been developed (Brockman et al. 2008), while for Illumina GA sequencing technology, common errors are substitutions resulting from cross-talk between signals.
SNP detection accuracy is related to the sequencing error rate and read length. In the Asian genome sequence, the error rate over all the mappable reads was ∼1.4%, and read length averaged 35 bp. We have recently been able to reduce this error rate to 0.5%∼0.8% in reads of 35 bp and to 0.5%∼1.5% for 50∼75-bp read length in a typical run, and thus expect to be able obtain very accurate SNP calling from low-depth (such as 4∼10×) paired-end sequencing in the near future with continuous improvement in data quality.
The method described in this study was developed primarily to handle the consensus assembly and SNP detection of one haploid or diploid genome with a known reference sequence. This program, however, also provides the option to output the likelihood of each allele type at each genomic location in a Genome Likelihood Format (GLF), which was proposed as the standard format for use in the 1000 Genomes Project (http://www.1000genomes.org). Thus, for multi-individual data sets, the likelihood information of each individual can be integrated and used to build a statistic frame to infer the genotype for each allele.
We also showed that using dbSNP genotypes for prior probability calculation substantially helps in distinguishing real heterozygotes from errors in regions of low-depth sequencing. The use of additional information for prior probability under the general Bayesian probability framework could likely aid in further improving accuracy of posterior probability calculation. For example, we could use different polymorphism rates for different portions of the genome, such as a lower polymorphism rate for gene regions; we could also use HapMap allele frequencies or haplotype block information for SNP calling of an individual belonging to a specific population. Furthermore, we could use joint probability for genotype calculation of multiple individuals from Mendelian segregation.
For assessment of our SNP detection method, we also estimated the impact of potential errors by ungapped alignment of reads containing small indels, but we have not yet built a model to call the indels, and thus calling for indels is not yet part of our SOAP package. With paired-end sequencing and increasing read lengths, it is feasible to accurately identify small indels from GA sequencing. We have also found that with very deep paired-end sequencing, we can detect structural variations including insertions, deletions, inversions, and rearrangements. Ultimately, de novo assembly of each sequenced genome would facilitate creating a complete picture of all kinds of genetic variations between any two individuals.
Methods
Data sets
Sequence data
NCBI build 36.1 was used as the reference of the human genome in this study. The chromosome sequences were downloaded from the UCSC database (http://genome.ucsc.edu/). We used version 128 of dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) data in the model of applying known SNPs in prior probability. The Asian genome data were sampled from an anonymous male Han Chinese and sequenced by the Illumina Genome Analyzer. These data are available in the EBI/NCBI Short Read Archive (accession no. ERA000005), and in the YH database (http://yh.genomics.org.cn).
The Asian genome genotype validation
An array-based technology, the Illumina 1M BeadChip, was used to genotype the Asian DNA sample for validation of SNP calling. Two technical duplicates were performed, and 1,038,923 sites that were identified in both experiments with consistent genotypes were used for the comparison against the alleles called by the Illumina GA sequencing.
PCR amplification and Sanger sequencing were performed to further validate a subset of the SNPs that showed inconsistent genotypes between the called consensus and genotyping. In total, 57 sites were amplified successfully and sequenced using AB 3730xl. The genotypes were called manually from the trace files.
Simulated data
Both single-end reads with different lengths and paired-end reads with different span sizes were simulated from chromosome 12 of the human genome randomly. We carried out the following process to create simulated reads that were similar to that which would be generated by GA sequencing:
Use chromosome 12 of the NCBI human genome as a reference, and produce an identical copy of the chromosome sequence.
Generate SNPs in this copy sequence with an estimated rate of 0.001. Both the sites and mutated allele types were chosen at random.
Reads were generated from random locations for both forward and reverse strains of the copy. For single-end reads, read lengths of 15, 20, 25, 30, 25, 40, 45, and 50 bp were chosen. For paired-end reads, the read length was fixed at 35 bp, which is the typical read length of current Illumina GA sequencing, and span sizes of 100, 200, 500, 1000, and 10,000 bp were chosen. The span size was not fixed, but it obeyed a normal distribution with 10% standard deviation in each data set. Thirty-six-fold coverage reads were generated for each of the data sets
Quality from the real Asian genome sequencing reads was selected at random and assigned to each of the simulated reads. Then an error rate for each base on each read was calculated from the assigned quality score by 10−Q/10, and this error was introduced according to its rate.
The simulation of small indels with sizes of 1 to 3 bp used a similar process, but produced 1000 insertions or deletions in step 2 rather than SNPs.
Consensus calling and SNP detection
Read alignment
The short reads were aligned onto the reference using the SOAP program. To obtain reliable alignment hits, at most two mismatches were allowed between the read and the reference. The alignments with the least number of differences were defined as “best hits.” If there was only one single best hit for a read, then the read was taken as uniquely placed; a read with multiple equal best hits was taken as repeatedly placed. For paired-end reads, two reads belonging to a pair were aligned together with both in the correct orientation and with a proper span size on the reference. In this study, we only used those reads with unique ungapped alignment for consensus calling and SNP detection.
Basic statistic model
Under a Bayesian model, the probability of genotype Ti by observing data D from an individual at a locus can be expressed as
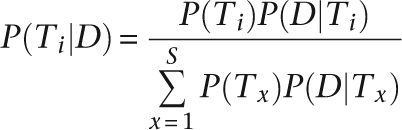 |
S is the total number of genotypes. If we define haploid genotype as Hm, then for a haploid genome, there are four kinds of genotypes: Ti = Hm ∈ {A, C, G, T}, S = 4; while for a diploid genome, Ti = HmHn ∈ {AA, CC, GG, TT, AC, AG, AT, CG, CT, GT}, S = 10. At each genomic location, prior probability P(Ti) of each genotype Ti was set according to the reference genotype and the estimated SNP rate between the sequenced individual and the reference genome. An example has been given in the Results section. The likelihood P(D|Ti) for the assumed genotype Ti was calculated from the observed allele types in the sequencing reads. We defined the likelihood of observing allele dk in a read for a possible haploid genotype H as P(dk|H). Supposing the two sets of chromosomes of a genome are independent, the likelihood P(dk|T) at a locus of a diploid genome can be calculated as
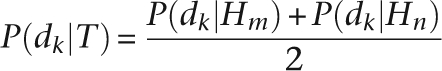 |
So, for a set of n total observed alleles at a locus, D = {d1, d2, …, dn},
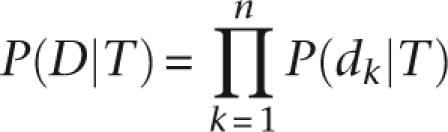 |
Thus the posterior probability can be derived from a Bayesian formula. The genotype Ti with the highest posterior probability P(Ti|D) was chosen as the consensus, and the phred-like quality score was calculated as −10 log10[1 − P(Ti|D)].
Quality calibration matrix and likelihood calculation
For each observed allele dk from a read mapped on a genomic location, there are four attributes: (1) ok, observed allele type; (2) qk, quality score; (3) ck, sequencing cycle (coordinate on read); and (4) tk, the tk-th observation of the same allele from reads with the same mapping location. All four attributes are useful for the calculation of likelihood: We first suppose sequencing errors are independent and fit attributes 1, 2, and 3 into the model. Then in the next section, we will discuss the method used to deal with potential dependent errors by considering attribute d. So the likelihood P(dk|H) now is
We built a four-dimensional matrix to store the likelihood of observing an allele dk with type ok, quality score qk, and at the ck-th cycle on the read for each assumed genotype H. By using the unique alignments, we counted the number of substitutions and estimated the mismatch rate for each combination of quality score qk, read coordinate ck, and substitution type. So P[(ok, ck)|(H, qk)] becomes a known value, which could be looked up in the matrix. Each raw sequencing quality score was in effect rescaled by each sequencing cycle and for each substitution combination.
P(qk|H) is the probability of an allele H to have an observation with quality score qk. The quality distribution of each assumed allele is unknown. Here, we assumed that the distributions from A, C, G, and T are the same; then P(qk|H) is the function of qk only, which can be written as f(qk) and be reduced in Bayesian formula.
Dealing with dependent errors
The same alleles from reads with the same mapping locations were ordered by the sequencing quality scores from low to high. An empirical treatment was used to reduce the quality of the tk-th observation:
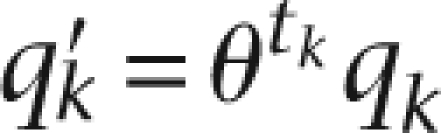 |
Here, θ is called a dependency coefficient. The adjusted quality score q′k, instead of the original qk, was used in the likelihood matrix. θ is set between 0 and 1. Specifically, θ = 0 means the completely dependent model, and θ = 1 is the completely independent model.
Sum rank test for HET
Since the quality scores of erroneous bases are lower than that for correct bases, we used the sum rank test to check the heterozygous sites of the called consensus. All observed appearances of the two alleles in the reads were ordered according to the quality score, then the sum rank of the less frequent allele was tested. The calculated P-value was integrated into the consensus quality score by subtracting −10log10(p).
Acknowledgments
This project is supported by the Chinese Academy of Science (GJHZ0701-6; KSCX2-YWN-023); the National Natural Science Foundation of China (30725008; 90403130; 90608010; 30221004; 90612019; 30392130); the Chinese 973 program (2007CB815701; 2007CB815703; 2007CB815705); the Chinese 863 program (2006AA02Z334; 2006AA10A121; 2006AA02Z177); the Chinese Municipal Science and Technology Commission (D07030200740000); the Danish Platform for Integrative Biology; the Ole Rømer grant from the Danish Natural Science Research Council; a pig bioinformatics grant from the Danish Research Council and the Solexa project (272-07-0196); and the Lundbeck Foundation Centre of Applied Medical Genomics for Personalized Disease Prediction, Prevention and Care (LUCAMP). Laurie Goodman edited the manuscript.
Footnotes
[SOAPsnp is freely available from http://soap.genomics.org.cn under GPL license. The raw sequence data used in this report have been deposited in the EBI/NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. ERA000005, and the SNP set has been deposited in dbSNP (release 130). These data are also available at http://yh.genomics.org.cn.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.088013.108.
References
- Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Altshuler D., Pollara V.J., Cowles C.R., Van Etten W.J., Baldwin J., Linton L., Lander E.S. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Bentley D., Balasubramanian S., Swerdlow H., Smith G., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R., et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman W., Alvarez P., Young S., Garber M., Giannoukos G., Lee W.L., Russ C., Lander E.S., Nusbaum C., Jaffe D.B. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., McLellan M.D., Ding L., Wendl M.C., Kasai Y., Wilson R.K., Mardis E.R. PolyScan: An automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F., Lander E., Rogers J., Waterston R. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Dawson E., Chen Y., Hunt S., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P., et al. A SNP resource for human chromosome 22: Extracting dense clusters of SNPs from the genomic sequence. Genome Res. 2001;11:170–178. doi: 10.1101/gr.156901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Hodges E., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Jimenez-Sanchez G., Childs B., Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008a;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008b;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Marth G.T., Korf I., Yandell M.D., Yeh R.T., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- Ning Z., Cox A.J., Mullikin J.C. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou D.T., Steinberg K.M., Middle C., Cutler D.J., Albert T.J., Zwick M.E. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Porreca G.J., Zhang K., Li J.B., Xie B., Austin D., Vassallo S.L., LeProust E.M., Peck B.J., Emig C.J., Dahl F., et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Shendure J., Mitra R.D., Varma C., Church G.M. Advanced sequencing technologies: Methods and goals. Nat. Rev. Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Sloan J.S., Robertson P.D., Scheet P., Nickerson D.A. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat. Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang W.Y., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: Theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Li R., Li Y., Tian G., Goodman L., Fan W., Zhang J., Li J., Zhang J., et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckx S., Del-Favero J., Rademakers R., Claes L., Cruts M., De Jonghe P., Van Broeckhoven C., De Rijk P. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.J., Makhijani V., Roth G.T., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wheeler D.A., Yakub I., Wei S., Sood R., Rowe W., Liu P.P., Gibbs R.A., Buetow K.H. SNPdetector: A software tool for sensitive and accurate SNP detection. PLoS Comput. Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Boerwinkle E. Neighboring-nucleotide effects on single nucleotide polymorphisms: A study of 2.6 million polymorphisms across the human genome. Genome Res. 2002;12:1679–1686. doi: 10.1101/gr.287302. [DOI] [PMC free article] [PubMed] [Google Scholar]