Abstract
Motion during magnetic resonance imaging (MRI) scans routinely results in undesirable image artifact or blurring. Since high-resolution, three-dimensional (3D) imaging of the mouse requires long scan times for satisfactory signal-to-noise ratio (SNR) and image quality, motion-related artifacts are likely over much of the body and limit applications of mouse MRI. In this investigation, we explored the use of self-gated imaging methods and image coregistration for improving image quality in the presence of motion. Self-gated signal results from a modified 3D gradient-echo sequence showed detection of periodic respiratory and cardiac motion in the adult mouse—with excellent comparison to traditional measurements, sensitivity to respiration-induced tissue changes in the brain, and even detection of embryonic cardiac motion in utero. Serial image coregistration with rapidly-acquired, low-SNR volumes further enabled detection and correction of bulk changes in embryo location during in utero imaging sessions and subsequent reconstruction of high-quality images. These methods, in combination, are shown to expand the range of applications for 3D mouse MRI, enabling late-stage embryonic heart imaging and introducing the possibility of longitudinal developmental studies from embryonic stages through adulthood.
Keywords: respiratory motion, cardiac motion, embryo, embryo heart, embryonic imaging, in utero imaging, brain, spinal cord
Biomedical research frequently focuses on mouse models of human disease and fetal development to investigate the influences of particular molecular and genetic factors. The use of magnetic resonance imaging (MRI) to study these models continues to increase (1,2), providing detailed morphological measurements for phenotyping (3,4) or evaluation of time course data (5–7). In combination with new magnetic labeling methods, MRI also provides a unique approach for in vivo tracking of transplanted or endogenous cell populations (8–11). However, mouse imaging requires high-resolution images (100 µm or better) and are routinely limited by signal-to-noise ratio (SNR)—in spite of small-volume radiofrequency coils and high magnetic field strengths. Consequently, it is not uncommon for high-resolution in vivo mouse images to require long acquisition times (1–3 h) in order to achieve satisfactory SNR and isotropic, three-dimensional (3D) image resolution. In light of the rapid rates of physiological motion in mice, such long scan times result in a high likelihood of motion-related compromises in image quality over most of the body.
Imaging methods designed to accommodate subject motion are generally of two types, designed to deal either with periodic motions or with more random displacements. Periodic motions, including heart beats and respiration events, are characterized by a “resting” tissue geometry that is reestablished following each motion event. Images free of motion artifact can be generated by selecting—either prospectively or retrospectively—only data in a particular phase of the motion. In the case of the heart, it is common to use a series of such images at several phases of the heart cycle to produce a movie loop of the whole cardiac cycle. However, many types of motion are not periodic, these include gastrointestinal peristalsis, uterine contractions, shifts of the head or torso, embryonic movement in utero, and even cardiac motion in some models of arrhythmia. Many clinical applications avoid artifact from these nonperiodic motions by acquiring images more quickly than the motion, an option not generally feasible for 3D mouse imaging. Alternatively, if the location of the imaging subject can be determined over time, image data can be corrected to provide consistent alignment for the image reconstruction process. For long 3D MRI scans in the mouse, both types of motion can be important. In this investigation, we sought to determine whether simple MR-based detection of both types of motion—through the combined use of self-gating and image coregistration—would enable improved 3D mouse MRI and thus broaden the potential for MR studies of mouse development and disease models.
Periodic motions from the diaphragm and heart can be detected by a variety of methods. Frequently, external devices such as ECGs and respiratory billows or pillows are used to provide a surrogate marker of tissue motion. However, the manipulation of such devices—particularly ECG leads—can be cumbersome at the small size of the mouse and the acquired signal may not accurately reflect tissue motion in the imaging volume. Furthermore, some types of motion are not amenable to detection with such external devices. For this reason many techniques that provide an MR-based signal for detection, correction, and/or gating of motion have been investigated. Navigator echoes representative of tissue motion can be acquired along with image data during acquisition by adding extra gradients and/or radiofrequency pulses (12–17). More recently, “self-gated” methods have been proposed that seek to achieve the same aim but avoid the addition of any gradient or RF events in the pulse sequence (18–21). In principle, many types of physiological motion could be detected from these gating signals and the image data could then be prospectively (22) or retrospectively (23,24) manipulated to generate artifact-free image reconstructions.
Demonstration of self-gated acquisitions have been presented for two-dimensional (2D) imaging in the mouse as well as in the human. Mouse investigations have demonstrated variations of 2D bright-blood cardiac imaging with both radial and Cartesian sampling schemes (25,26). In the human, investigation of 3D methods have been recently explored (27). As many important mouse imaging applications also require 3D image acquisitions, a thorough consideration of the potential for self-gated mouse imaging in 3D scans is warranted. In this work, we therefore explore the use of self-gated methods particularly for 3D mouse imaging applications with long scan times. Notably, since one of the most exciting benefits of self-gated imaging is expected to be detection of motion for situations in which the use of peripheral monitoring devices is difficult or impossible, we sought to determine if a self-gated signal would be sufficiently sensitive to enable cardiac imaging of the mouse embryo in utero.
Only a limited number of studies have reported in utero embryonic MRI in the mouse (28–32), but the potential benefits are well-recognized (33). Three-dimensional scans with maternal respiratory gating and manganese enhancement can provide excellent anatomical visualization in the nervous system (32). In general, however, image quality during long high-resolution scans can be adversely affected by nonperiodic, bulk rotations and translations of the embryo. Artifact due to such motion is not unique to mouse embryo imaging; studies of dynamic contrast enhancement and functional MRI (fMRI), for instance, require an exact orientation of serially-acquired images for proper analysis of intensities over a time series. To ensure proper alignment, images can be registered together during postprocessing to eliminate or limit the effects of motion in these studies (34–36). In this investigation, we considered whether a similar approach with rapid serial acquisition of 3D volumes, each with relatively low SNR, and registration postprocessing would enable correction for embryonic motion during in utero MRI scans.
In order to provide an assessment of the potential of self-gating and retrospective image registration to accommodate motion during longer 3D image acquisitions in mice, we experimented with several different 3D imaging applications. Roughly in order of increasing motion/gating complexity, they include:
thoracic/lumbar spinal cord imaging;
adult and neonate brain imaging;
bright and dark blood adult cardiac imaging; and
in utero embryonic cardiac imaging.
Collectively, these 3D imaging applications demonstrate imaging in the presence of a single type of periodic motion (respiratory), two periodic motions in combination (respiratory and cardiac), different types of image contrasts (bright vs. dark blood), and motion not readily detectable by external devices (in utero cardiac imaging). The latter, in particular, enables novel imaging applications and studies with regard to normal and altered development.
MATERIALS AND METHODS
Imaging Methods
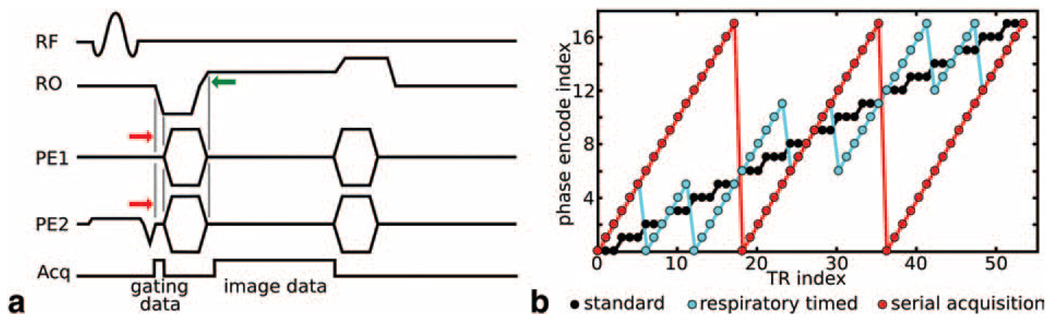
For this study we implemented a simple modification to a standard 3D gradient-echo sequence (Fig. 1a). It is similar to ones implemented previously for 2D imaging (20,24), in which signal is acquired during or after the slice refocusing gradient but prior to the readout dephase and phase encoding gradients. In this case, a standard 3D gradient-echo sequence was modified to include a small time delay between the start of the readout dephase gradient pulse and the two phase-encode pulses. This permits signal acquisition during the initial part of the readout dephasing and provides low-resolution spatial information in the readout dimension. This additional information can be acquired with no time cost if the delay on the phase-encode gradient lobes is less than the gradient rise time. In our implementation, a delay of 200–250 µs was used, permitting acquisition of 15 to 25 data points with different encoding for gating purposes. Using the readout gradient for acquisition of spatial information—as opposed to the slice-/slab-select refocusing gradient—is advantageous for 3D imaging applications since the slab-select gradient is either very low in amplitude or absent altogether.
FIG. 1. 3D image acquisition sequence.
a: All images were acquired using a modified 3D gradient echo sequence. The phase encoding gradients on both axes (PE1 and PE2) were shifted to a slightly later time (red arrows) to permit acquisition of gating data during the initial portion of the readout (RO) dephasing gradient. A shift equal to the gradient rise time (200 µs on our system) avoids time cost by overlapping the falling edge of the phase encode gradient with the rising edge of the readout gradient (green arrow). b: The phase encode acquisition order during the scan was prescribed in one of three ways according to the application. The standard ordering (black) used repeated acquisition of each phase encode until all averages/repeats were collected and then proceeded to the next phase encode. Respiratory-timed encoding (cyan) was designed for retrospective respiratory gating and acquired phase encode averages/repeats every ~400–600 ms (about one-half a respiratory period) to ensure at least one acquisition for each phase encode with minimal respiratory effects. This was achieved by acquiring N consecutive phase encodes together and then repeating the set for averages/repeats (b: shows N = 6 with three repeats, typical imaging scans in this work used N ≈ 8 with three repeats). For in utero imaging, phase encodes were incremented linearly (red) until a full image was acquired, and then the entire image was repeated (serial acquisition). In (a), RF = radiofrequency, Acq = acquisition, RO = readout gradient, PE1 = first phase-encode gradient, PE2 = second phase-encode gradient.
All imaging sequences were implemented on a 7T magnet (Magnex Scientific Ltd., Yarnton, UK) equipped with a Bruker Biospec Avance II console (Bruker BioSpin MRI, Ettlingen, Germany) and actively-shielded gradients (BGA9-S; Bruker BioSpin MRI). Adult cardiac and spine images and all in utero images were acquired using a spine surface coil for receive and a volume resonator (72-mm inner diameter quadrature resonator; Bruker BioSpin MRI) for transmit. Adult and neonate brain images were acquired using a quadrature, transmit/receive Litzcage coil (25-mm inner diameter; Doty Scientific, Columbia, SC, USA). Imaging parameters, including repetition time (TR), echo time (TE), matrix size, number of image repeats/averages (NR), and total acquisition time (TA), are provided in the figure legends with the image data. Dark-blood contrast in heart images—both adult and embryonic—was produced by prescribing saturation slices (10–15-mm-thick, 3.0–3.6-kHz sinc pulse) covering incoming blood vessels as described elsewhere (37,38).
Phase-Encode Acquisition Order
In this work, all gating and imaging data were acquired continuously and reconstruction was performed with retrospective gating. Consequently, it was necessary to sample each phase encode at least twice in the aim of acquiring each k-space line during a quiescent phase of the respiratory cycle. It should be noted that this oversampling does not necessarily entail an unreasonable time cost, as averaging is usually necessary in 3D mouse MRI to obtain satisfactory SNR. When respiratory motion was expected, the order of phase encodes was modified such that the period between oversampled acquisitions was about one-half the expected respiratory period. This phase encode acquisition scheme is shown schematically in Fig. 1b and was used for adult spine, adult brain, and neonate brain imaging. For adult cardiac imaging, the standard order (depicted as black in Fig. 1b) was used, with phase encodes repeated consecutively to ensure an acquisition of each k-space line at every phase of the cardiac cycle.
For the in utero embryonic image acquisitions, the potential for physical displacement of the embryo over the acquisition due to uterine, gastrointestinal, or embryonic motion suggested an alternative encoding strategy. In these scans, an entire 3D image was collected as rapidly as possible by minimizing the field-of-view, maintaining a short TR, and acquiring one full image prior to collecting repeats/averages (Fig. 1b, red). This permitted acquisition of single 3D images in 2 to 3 min, a series of which could be processed as described below to form one or more higher-quality images for the session.
Analysis and Display of Gating Signal
Self-gating data were postprocessed to produce scalar data representative of periodic physiological motion at each TR of the image acquisition. These data, referred to here as physiological traces, were used for gated reconstructions and permitted comparison with traditional measurements from external devices. Respiratory traces were produced from the first five points of the self-gate signal (i.e., primarily unencoded signal) by averaging them together and then computing the square-difference from the local median (determined over 512 TR periods). For gated reconstructions, a threshold—typically corresponding to about one quartile—was selected to determine which acquisitions were affected by respiratory motion. Affected lines (25–30% of acquired data) were then excluded from the reconstruction. In the rare event that all acquisitions of a particular k-space line were above threshold (occurring in less than 1% of lines), the line nearest the threshold was selected for inclusion in the reconstruction. Cardiac traces were computed by comparison to reference data selected from the scan representative of a full cardiac period. Reference data was represented by a 2D data array, which included 10 to 12 consecutive TR periods in one dimension and three to five data points near k ≈ 0.3/mm in the other dimensions. Note that we use the convention:
where γ is the gyromagnetic ratio, and G is the time-dependent gradient waveform (integrated over time t).
For each point on the cardiac trace, a 2D-correlation was calculated between the local and reference data. Cardiac phase were then assigned based on the peaks of the resulting periodic trace.
In the adult cardiac mouse imaging experiments, we compared the cardiac phase determined from the self-gating signal with that determined from ECG measurements. As the phase from the self-gating signal is assigned relatively, without exact reference to systole or diastole, a constant phase shift between the ECG and self-gating data is likely. We, therefore, calculated the coherence of the two signals, a metric that has also been used in time-series analysis of fMRI data (39,40). The coherence indicates relationships between two time series and is defined here as:
, where f̄xy is the average cross-spectrum of time series x and y, and f̄xx and f̄yy represent the average power spectra of time series x and y.
A coherence value of 0 indicates the two time series are independent, while a coherence value of 1 indicates they are exactly related through a linear amplitude and phase transformation. In our assessment, a coherence of unity indicates that the ECG and self-gating phase assignments are equivalent for the purpose of generating an image reconstruction. To calculate coherence values, each phase time series was split into segments of 1024 points. The cross-spectra and power spectra were computed for each segment and then averaged together for a minimum of 80 segments. We then evaluated the coherence at the frequency of the heart cycle indicated by the ECG time series.
In order to eliminate the need for respiratory pillows and ECG electrodes for some imaging applications, it was advantageous to plot self-gated physiological traces in real time on the image console. This permits monitoring of mouse physiology throughout the imaging experiment and adaptation of isoflurane anesthesia levels as required. To demonstrate the feasibility of such a “wireless” monitoring method, we implemented a simple scrolling display of respiratory traces (using the Python programming language, release 2.3) and ran it on the console during image acquisitions.
Embryonic Image Registration and Reconstruction
For reconstruction of in utero image acquisitions, each of the serially-acquired embryo images were registered to eliminate shifts in the embryo position over the course of the scan. For this purpose, a six-parameter registration was performed using software produced by the Montreal Neurological Institute (MNI_AutoReg; http://www.bic.mni.mcgill.ca/software/mni_autoreg) (41,42). A coarse, manually drawn mask covering the embryo heart, inflow and outflow tracts, liver dome, and spine were used for each registration. Image-space translation and rotation parameters from the six-parameter transforms were used to compute equivalent k-space transformations. Specifically, translations were transformed to linear phase ramps that were multiplied with k-space data. Rotations were applied without alteration. The resulting transformed k-space lines were averaged together after discarding lines affected by maternal respiration and sorting based on the embryonic cardiac cycle. Images for each phase of the cardiac cycle were then reconstructed from the averaged k-space data.
Animal Handling
All animal studies described in this report were approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. We used 6- to 10-week-old ICR mice for spinal cord, brain, and adult cardiac imaging. In utero embryonic imaging was performed in ICR females at embryonic day 16 (E16) to E18, where E0.5 is defined as noon of the day after overnight mating. Animals were prepared for imaging in an induction chamber with 4% to 5% isoflurane and then transferred to the imaging holder and coil assembly where they were maintained under anesthesia with 1.0% to 1.5% isoflurane. For some imaging studies, mice were monitored with a respiratory pillow and ECG electrodes connected to a Biopac monitoring system (Biopac Systems, Inc., Goleta, CA, USA). This provided external data for direct comparison to MRI-derived physiological measures. Alternatively, where comparisons were not required, we also used the physiological monitoring display software described above, which generated physiological traces from the self-gating signal without the need for peripheral hardware.
RESULTS
Self-Gated Detection of Periodic Physiological Motion
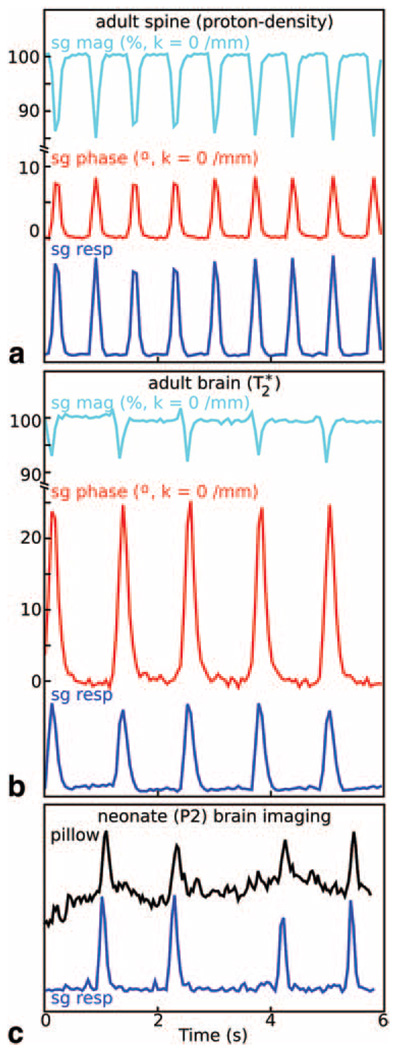
In all applications, the self-gating signal showed high sensitivity to respiratory events (Fig. 2), which were detectable independent of the excitation volume or location in the body. Interestingly, this was true even in the immobilized head during T2*-image acquisitions (Fig. 2b). For imaging of neonatal mouse pups, the self-gated detection of respiratory motion was observed to be superior to a respiratory pillow, which had limited sensitivity to the pups’ breathing motion (Fig. 2c). In all other applications, the self-gating respiratory signal was at least as sensitive as the respiratory pillow. Hence, the respiratory pillow could be eliminated from the imaging setup to provide faster and simpler animal preparation.
FIG. 2. Respiratory motion characterization with the self-gating signal.
All applications investigated showed a strong sensitivity to respiratory events near k ≈ 0/mm in the self-gating data, which was consequently used to compute self-gated respiratory traces (sg resp). a: Raw and processed self-gating data acquired during an in vivo spine image. In T2*-weighted adult brain images, respiratory events were apparent even in the absence of motion and the self-gated signal (b) showed a more prominent phase shift (sg phase) with less significant loss in magnitude (sg mag) relative to protondensity-weighted spine images (a). Notably, detection and monitoring of respiration during neonate imaging was significantly improved by using the sg resp trace as opposed to a traditional respiratory pillow (c: showing data from a 2-day-old neonate). All unmarked vertical axes have arbitrary units. The phase (shown in degrees) and magnitude (shown as a percentage of the median) in the adult spine (a) and adult brain (b) examples are scaled identically.
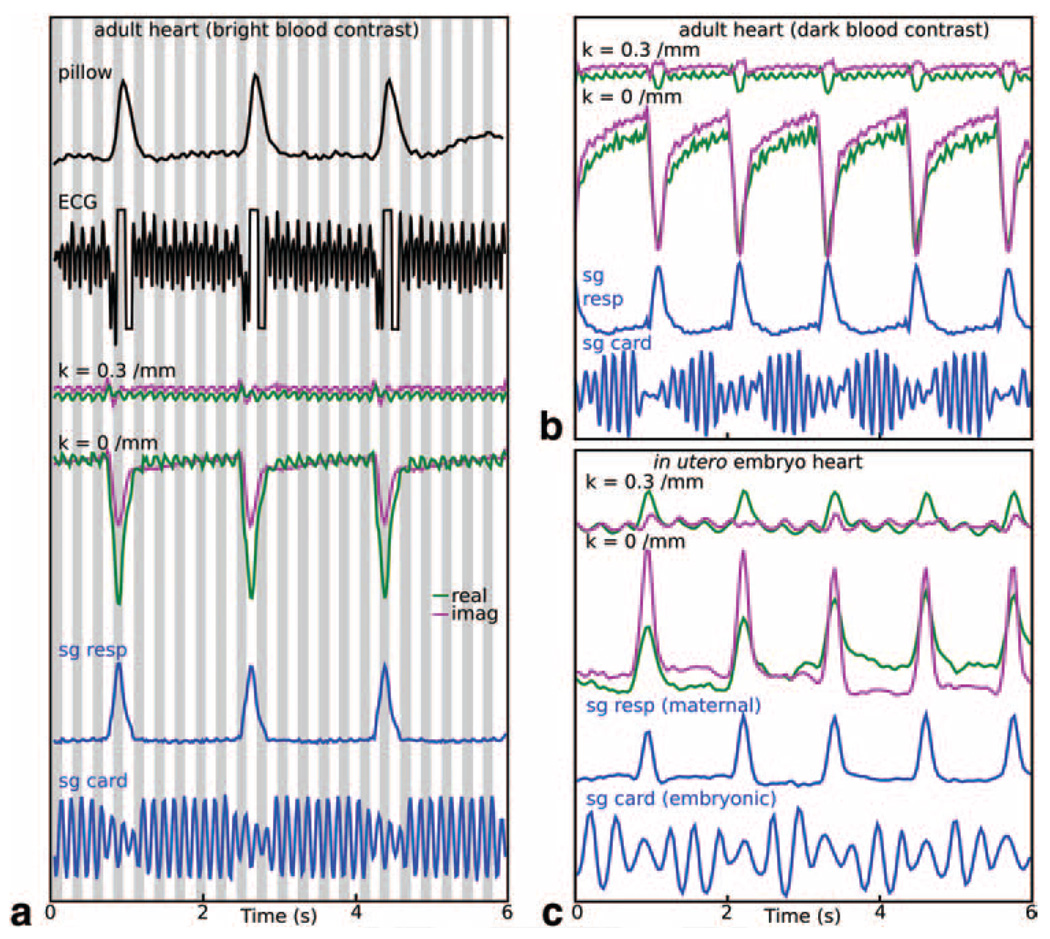
Cardiac motion produced a smaller change in the gating signal than respiratory motion, but still provided sufficient sensitivity for gating in 3D cardiac studies (Fig. 3). In the adult mouse, cardiac data acquired with bright-blood contrast resulted in a modulation of the gating signal that was apparent over most of the readout ramp (Fig. 3a). However, the self-gating signal from k ≈ 0.3/mm provided the best detection of the cardiac cycle through respiratory events. Similar characteristics were apparent from images prepared with dark-blood contrast (Fig. 3b), except in this case self-gating data from k ≈ 0.3/mm was necessary to generate a satisfactory cardiac waveform due to an overall decrease in heart motion sensitivity. In both bright- and dark-blood cases, assignment of cardiac phase based on the self-gating signal compared well with ECG-based assignments (with coherence values C2 = 0.98 and 0.90, respectively). The small periodic motion of the embryonic heart also produced a signal change sufficiently large for computation of an embryonic cardiac waveform (Fig. 3c), although the encoded portion of the gating signal and careful placement of the image slab proved critical for this application.
FIG. 3. Cardiac motion characterization with the self-gating signal.
Physiological traces representative of cardiac motion were most reliably produced by using self-gating data near k ≈ 0.3/mm and computed cardiac traces (sg card) compared well with ECG measures (a: where alternating gray and white bars highlight the ECG derived cardiac cycles). Respiratory traces (sg resp) during the same scans also compared will with respiratory pillow measurements. Bright-blood contrast preparation provided a more prominent cardiac signature in the gating signal than dark-blood contrast (b), although both permitted generation of a satisfactory cardiac trace. During in utero embryonic heart imaging (with dark-blood contrast), a periodic signal representative of the embryonic heart phase could be detected in the k ≈ 0.3/mm self-gating signal, which was used to generate an embryonic cardiac trace for cine image reconstructions of the embryo heart. All unmarked vertical axes have arbitrary units. Vertical scaling of raw self-gating signal at k = 0/mm and k = 0.3/mm are identical within each panel.
Respiratory-Gated Imaging in the Thoracic and Lumbar Spinal Cord
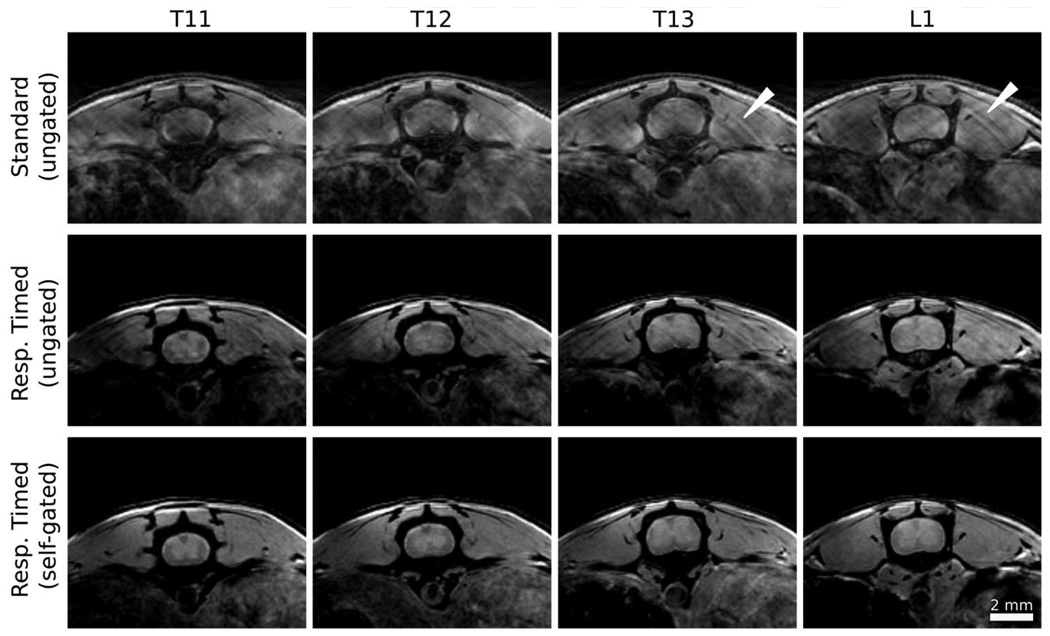
Images were acquired in the thoracic-lumbar region of the spinal cord with three different acquisition/reconstruction strategies (Fig. 4). Standard 3D image acquisitions (without gating) were always observed to have significant artifact over the length of the cord. Adaptation of the phase encoding order according to the timing of the respiratory events (as in Fig. 1b, cyan) substantially improved image quality, even in the absence of gating. Further improvement was observed by using the gating signal to discard lines acquired during respiratory events. This resulted in images with little or no respiratory artifact throughout the thoracic and lumbar regions of the spinal cord and permitted visualization of gray and white matter using a proton-density-weighted imaging protocol.
FIG. 4. Self-gated 3D imaging of the thoracic and lumbar spinal cord.
Ungated images acquired with the standard phase encoding order (top row) show obvious motion artifact (white arrows) that obscures the spinal cord and surrounding tissue. Adjustment of the phase encode order to the respiratory-timed scheme (Fig. 1b, cyan) provides significant improvement even without gating (second row). Exclusion of data acquired during respiratory events detected by the self-gating signal further improve image quality, and better distinguishes gray and white matter in the cord. Image parameters: TE/TR = 3.4/80 ms, flip angle = 15°, matrix size = 256 × 256 × 135, NR = 3, TA = 2 h 18 min, and resolution = 70 × 70 × 280 µm3.
Respiratory-Gated T2*-Weighted Imaging in the Adult Brain
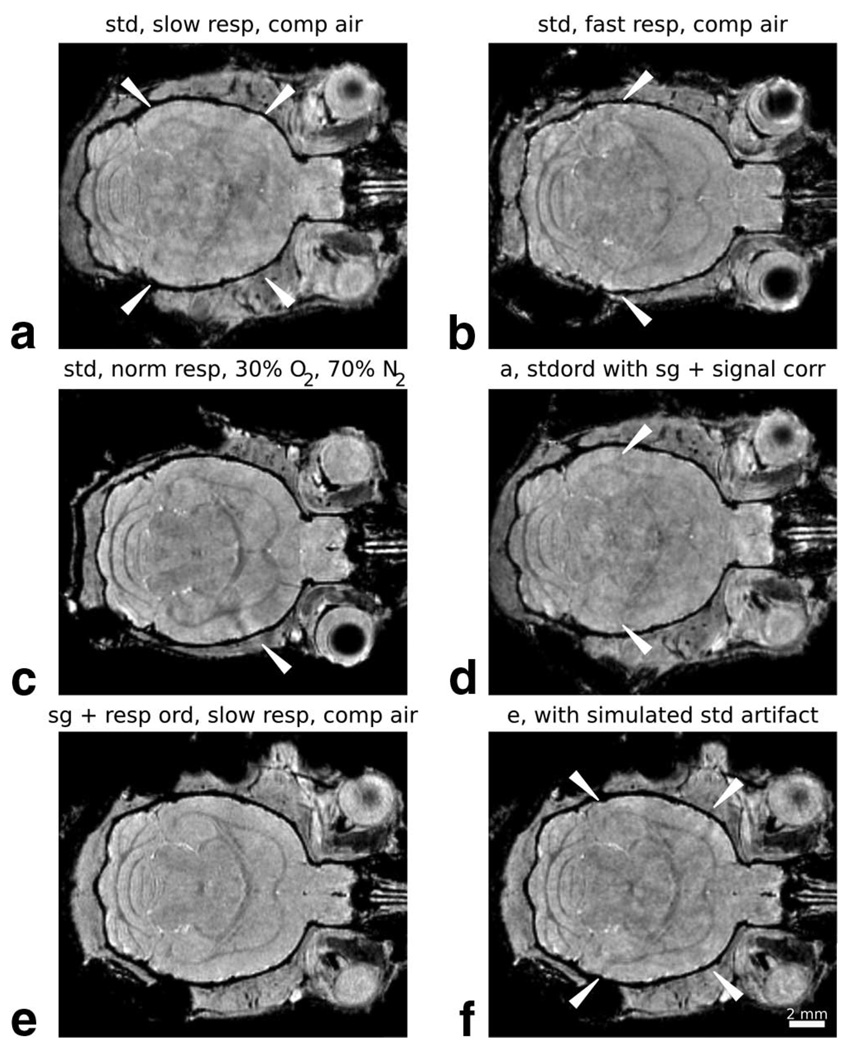
Standard T2*-weighted, 3D gradient-echo images of the adult brain in our laboratory were frequently observed to have artifact, as in Fig. 5a–c, where a mottled intensity pattern was evident. These artifacts persisted in spite of measures to immobilize the head. Furthermore, the artifact level was observed to depend both on the respiration rate (Fig. 5b) and the type of inhalation gas (Fig. 5c), but could not be routinely eliminated by manipulation of these parameters. Strong respiratory signatures were observed in self-gating data from these acquisitions, largely due to an apparent signal phase shift (Fig. 2b). The artifact level could be reduced, but not eliminated, by using the self-gating signal for gating the standard acquisition and for correcting image data phase and magnitude (Fig. 5d). Images acquired with self-gating signal and the respiratory-timed phase-encoding order routinely resulted in a homogeneous tissue signal, free of obvious artifact (Fig. 5e). Furthermore, modification of the gated image (Fig. 5e) by periodically applying signal changes suggested by Fig. 2b (~23° phase shift and 10% magnitude change) resulted in a mottled artifact similar to that seen in the standard, ungated acquisitions. This suggests that the artifact may indeed the result from periodic respiration-induced tissue signal change, and not directly from tissue motion.
FIG. 5. Self-gated T2*-weighted imaging in the adult brain.
Artifacts were routinely observed in 3D images of the adult brain with T2*-weighting. a–c: Examples are provided, with white arrows delineating the region of most obvious artifact. Slow respiration rates (a: 1.9 ± 0.4 s respiratory period) produced more intense artifact than fast ones (b: 1.0 ± 0.1 s). Increasing the oxygen content of the administered gas further improved image quality (c: 1.3 ± 0.1 s; see also Ref. 36). d: Artifact in images acquired with a standard phase encode order could also be reduced by using the self-gating signal to select for data least altered by respiratory events and to correct the magnitude/phase of image data. Images with minimal artifact (e: 2.3 ± 0.3 s) were routinely achieved by using self-gating in combination with the respiratory-timed encoding scheme of Fig. 1b. The clean image data of (e) was also used to simulate standard image acquisitions by applying a 23° phase shift and 10% magnitude change to image data periodically (distributed through k-space with 1.6 ± 0.2 s period). This results in the artifact shown in (f), reminiscent of the mottled artifacts evident in (a–c). Image parameters: TE/TR = 8/80 ms, flip angle = 15°, matrix size = 256 × 256 × 135, NR = 3, TA = 2 h 18 min, and isotropic resolution = 100 µm. Abbreviations: std = standard phase encode order with no gating, resp = respiration rate, comp air = compressed air, norm = normal, stdord = standard phase encode order, sg = self-gating, signal corr = phase and magnitude sg-based image data correction, resp order = respiratory-timed encoding order.
3D Bright- and Dark-Blood Cardiac Imaging
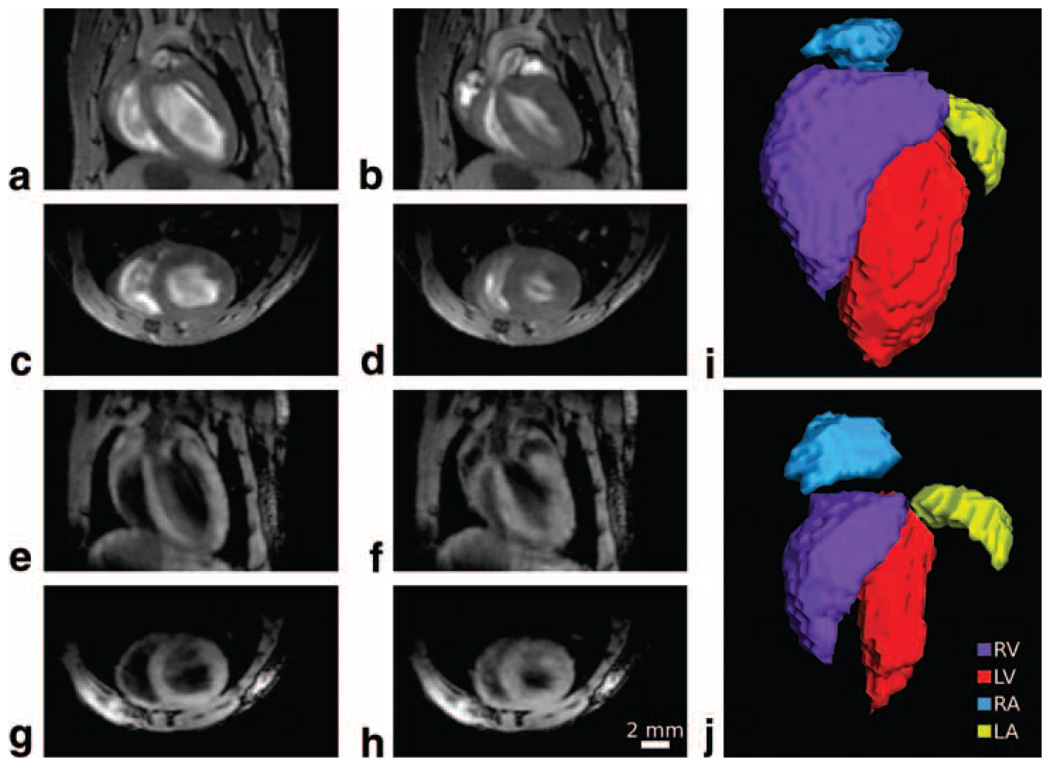
Three-dimensional cine images of the adult heart were produced with both bright- and dark-blood contrast. Respiratory-gated reconstructions of the data were performed at each of 10 different cardiac phases to produce a 3D cine reconstruction of the heart cycle. Comparison of retrospective reconstructions with traditional ECG measurements and with self-gated cardiac phase measurements showed no apparent differences. Individual slices from the frames at systole and diastole are shown in Fig. 6 with both contrast mechanisms. A 3D view of the bright-blood image is also provided to emphasize the 3D character of the data as well as the change in chamber shape at each extreme of the cardiac cycle.
FIG. 6. 3D cardiac imaging with bright- and dark-blood contrast.
Orthogonal slices from 3D images reconstructed using the self-gating signal are shown at both diastole (a,c,e,g) and systole (b,d,f,h) and with bright-blood (a–d) and dark-blood (e–h) image contrast. 3D segmentations of the chambers from the bright-blood image show changes in the chamber shape from diastole (i) to systole (j). Bright-blood image parameters: TE/TR = 2.1/20 ms, flip angle = 25°, matrix size = 148 × 170 × 70, NR = 40, TA = 2 h 39 min, isotropic resolution = 175 µm, reconstructed with 10 3D image frames per cardiac cycle. Dark-blood image parameters: TE/TR = 2.6/20 ms, flip angle = 25°, matrix size = 160 × 148 × 70, NR = 40, TA = 2 h 18 min, isotropic resolution = 175 µm, reconstructed with 10 3D image frames per cardiac cycle.
In Utero Embryonic Cardiac Imaging
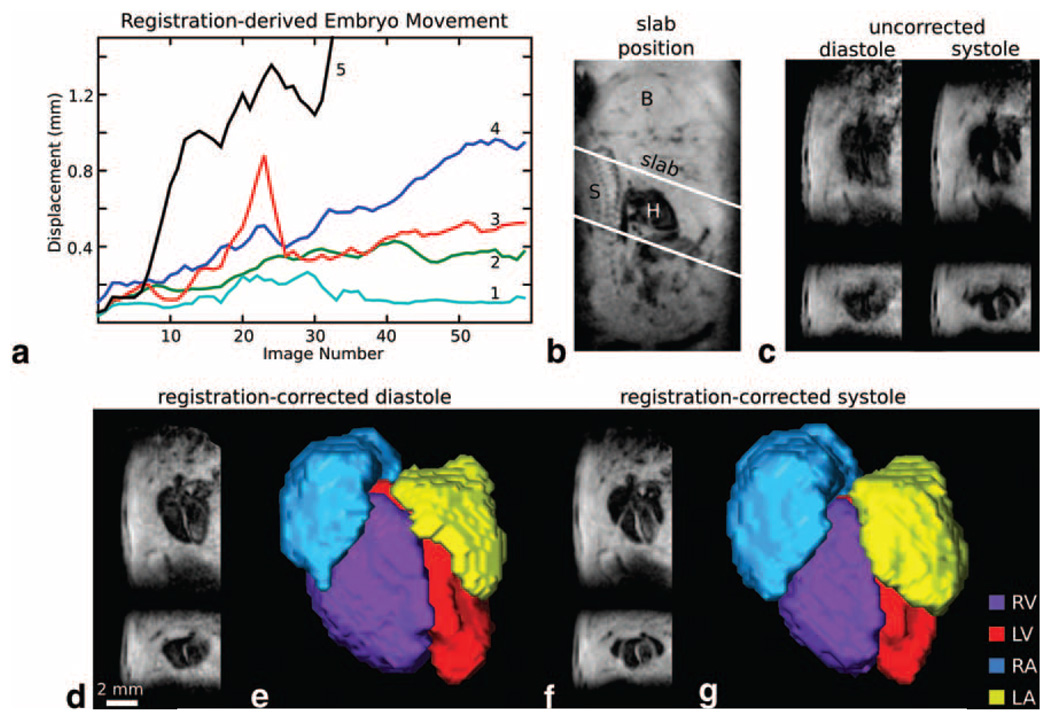
In addition to detection of cardiac motion via a self-gating signal, 3D imaging of the embryonic heart in utero generally required detection and correction of bulk embryo motion to generate images with satisfactory quality. Six-parameter registration of repeated images provided a transformation to map the position of the embryo heart in individual images to the initial imaging orientation. Extraction of the translations and rotations from these image registrations provided an estimation of embryo motion over the course of the imaging session. Comparison of embryonic displacement data across several imaging sessions showed a range of embryo displacement patterns (Fig. 7a). For the in utero cardiac imaging protocol, displacements of as much as ~2 mm could be accommodated, with larger displacements likely to result in migration of the embryonic heart outside of the imaging field-of-view. The majority of in utero imaging cases we attempted (13/15) had displacements within this range. Larger fields-of-view, for imaging of the whole embryo or the embryo head for instance, were able to accommodate larger displacements but at the expense of slower image acquisition and poor detection of embryonic heart signal.
FIG. 7. In utero cardiac imaging in the E17 embryo.
Embryonic displacement during the cardiac scan was followed by image registration of serially-acquired 3D images. a: Five examples of distance displacements from the first image. Occasionally, image sessions show little embryo displacement over the session (curve 1), but most frequently several voxels displacement are observed over the coarse of the scan session (curves 2–4). In some instances very large or rapid displacements were observed (curve 5) that moved the embryo heart outside of the initial imaging slab (prescribed as in (b)). Images without correction for embryo displacement generally show blur artifact as in (c), where long- and short-axis images from the same 3D data set are shown at both diastole and systole. d,f: Correction of image displacement with registration provides improved image quality. e,g: Segmentations of the chambers emphasize the changes in 3D shape over the cardiac cycle. Image parameters: TE/TR = 6.0/40 ms, flip angle = 16°, matrix size = 192 × 78 × 48, NR = 64, TA = 2 h 40 min, isotropic resolution = 130 µm, reconstructed with six 3D image frames per cardiac cycle. In (b), B = brain, S = spine, and H = heart.
To generate a 3D cardiac cine image, image data was binned according to the phase of the embryonic cardiac cycle as determined by the self-gating data. Dark-blood contrast with T2*-weighted imaging parameters were used for embryo cardiac imaging because it was not possible to achieve satisfactory bright-blood contrast consistently in 3D slabs. Reconstruction of images was performed at six different phases of the heart cycle. Long-axis and short-axis slices at systole and diastole are provided in Fig. 7, along with 3D renderings of the heart chambers. In comparison with the adult heart (Fig. 6), the relative shape and size of the embryonic heart chambers was observed to be markedly different, with larger atrial chambers and more symmetric ventricles. Additionally, the change in chamber size from systole to diastole was much less remarkable in the embryonic heart.
DISCUSSION
The ability to view anatomical detail and tissue morphology in three-dimensions—in vivo and noninvasively—is one of the chief advantages of mouse MRI in biomedical research. In the mouse, this type of imaging frequently requires long scan times and multiple image averages in order to ensure adequate SNR and image quality. In this work, we have explored the use of self-gating data and image coregistration to permit reconstruction of images in the presence of both periodic physiological motion and nonperiodic rigid-body motion. This method was demonstrated in a number of applications, notably including in utero cardiac imaging. Further development of these and similar methods promise to extend the range of applications addressed by 3D MRI methods.
The self-gating signal provides a sensitive and universal means of detecting motion in the imaging volume, with little or no time cost in 3D Cartesian sequences. It can be sensitive to a variety of MR signal-related changes, including tissue motion, inflow, and intrinsic tissue signal changes. Consequently, while the elimination of peripheral monitoring devices is one very attractive feature of self-gating, the acquisition of gating data is expected to be beneficial even when traditional physiological measurements are readily available. It is interesting, for example, that we observed a prominent respiratory signal throughout the body, even for cases in which tissue motion was limited or absent. This observation indicates that the self-gating signal may be sensitive to tissue or blood oxygenation levels linked to the respiratory cycle. This signal may therefore be helpful during acquisition of fMRI data sets in order to monitor and perhaps even correct data over the course of the experiment, thereby reducing to some extent the background variability of mouse fMRI data (43–45).
The sensitivity of the self-gated data to changes in the image volume is a function of several parameters. As the motion of the embryonic heart seemed to be near the limit of what could be reliably detected in this study, it is worth considering several of these parameters. First, the adult cardiac imaging data presented here suggests the signal is strongly modulated by fully-relaxed signal moving into or out of the excitation volume. Hence, the type of image contrast plays an important role. Likewise, the sensitivity profile of the coil also has a profound effect; an inhomogenous receive sensitivity profile is expected to be advantageous because it increases the tissue-geometry-dependence of low-order k-space data. We expect that smaller coils would lead to higher motion sensitivity for the same reason, and that use of a multiple coil array would provide an improved gating signal over a single coil. Additionally, the direction of movement relative to the gradient directions is also important. Improvements in the data acquired here might be obtained by modifying the sequence to include self-gating signal from multiple gradient directions. A modification of the phase-encode gradient shape to permit acquisition on the dephasing gradient ramp in each of these directions has been suggested (46), and could be used to incorporate data from those directions as well. With a coil array and low-level encoding gradients in each direction, one could even generate a low-resolution image to use for gating. Last, it should be noted that the size of objects undergoing motion-related changes certainly affects the gating signal. Relatively large objects have significant information in low-order k-space that can be used for gating. Smaller structures in a volume require higher-order k-space data—at inherently lower SNR—before detection of changes is likely to be feasible. In this work, signal at k ≈ 0.3/mm was used for detection of cardiac motion. We expect that this k-space position is most sensitive to objects of at least a few millimeters in size, roughly corresponding to the dimensions of an adult heart chamber or an embryonic heart. Future improvements that provide a clearer depiction of the embryonic heart cycle will have to take each of these factors into account.
In order to accommodate both periodic and nonperiodic motions in embryonic imaging in utero, the registration-based image analysis proved necessary to detect and correct for embryonic displacements. Further development in this area should enable routine in utero imaging studies. We found that a six-parameter, rigid-body registration was satisfactory for correcting displacements of isolated organs such as the heart. A more sophisticated registration would likely be required if the entire embryo were of interest, owing to the potential for changes in embryo body position (such as motion of the head with respect to the thorax). However, such analyses are likely to require higher SNR in individual image frames for reliable performance. We expect that significant improvement in image quality could be achieved with a embryo-dedicated coil configuration. An increase in SNR might further permit an improved temporal resolution of the registration-based embryo tracking. In this work, a low-SNR image was acquired every 2 to 3 min, whereas an image every few tens of seconds would be necessary to meticulously track embryo displacements from gastrointestinal and other events. While it is not likely that a full-resolution 3D image could be achieved in such a short time frame, a more sophisticated partially- or undersampled image acquisition scheme could also increase temporal resolution. Provided a sufficiently high level of reliability from the registration analysis, it would be beneficial to adapt the prescription of the imaging volume during the scan to follow the location and orientation of the embryo in real time. This would avoid the loss of some data sets in the current scheme, where the heart or other structure of interest may drift beyond the initial field-of-view. In this regard, measures to constrict embryonic motion are also worth investigating (and have been experimented with previously in our laboratory (32)). These areas of future development should provide improved in utero embryo images, beyond the encouraging cardiac results presented here.
The ability to image the embryo in utero opens some exciting opportunities for research with mouse MRI. Given that engineered mouse mutations can frequently result in phenotypes lethal in early postnatal stages, in utero imaging should permit functional and morphological characterization of a much broader array of mutant mice than is currently possible. Further, longitudinal imaging and analysis of developmental processes will allow quantitative characterization of the dramatic changes in size and morphology that occur over early development, from late embryonic stages, and through adulthood. Although there are several challenges in pursuing such developmental studies in vivo, the results here indicate the potential for excellent image quality and even for functional cardiac imaging of the embryo in utero.
CONCLUSIONS
Methods of retrospective self-gating and image registration can be applied to acquire 3D images with high resolution in the presence of physiological motion. These MR-based motion detection methods also provide flexibility and sensitivity appropriate for a wide range of applications, including imaging in the spinal cord, brain, heart, and embryo in utero. These and similar MR methods for 3D imaging in the presence of motion will extend the range of mouse MRI applications to include all regions of the body and important dynamic processes such as cardiac and respiratory motion as well as in utero and postnatal development of normal and mutant mice.
ACKNOWLEDGMENTS
We thank Dr. Glyn Johnson, Ph.D., New York University School of Medicine, for feedback on the manuscript.
Grant sponsor: New York State Department of Health, New York State Spinal Cord Injury Trust Fund; Grant numbers: C020926, C022053; Grant sponsor: National Institutes of Health (NIH); Grant number: R01 HL078665.
REFERENCES
- 1.Nieman BJ, Bock NA, Bishop J, Chen XJ, Sled JG, Rossant J, Henkelman RM. Magnetic resonance imaging for detection and analysis of mouse phenotypes. NMR Biomed. 2005;18:447–468. doi: 10.1002/nbm.981. [DOI] [PubMed] [Google Scholar]
- 2.Turnbull DH, Mori S. MRI in mouse developmental biology. NMR Biomed. 2007;20:265–274. doi: 10.1002/nbm.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieman BJ, Lerch JP, Bock NA, Chen XJ, Sled JG, Henkelman RM. Mouse behavioral mutants have neuroimaging abnormalities. Hum Brain Mapp. 2007;28:567–575. doi: 10.1002/hbm.20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerch JP, Carroll JB, Dorr A, Spring S, Evans AC, Hayden MR, Sled JG, Henkelman RM. Cortical thickness measured from MRI in the YAC128 mouse model of Huntington’s disease. Neuroimage. 2008;41:243–251. doi: 10.1016/j.neuroimage.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Lau JC, Lerch JP, Sled JG, Henkelman RM, Evans AC, Bedell BJ. Longitudinal neuroanatomical changes determined by deformation-based morphometry in a mouse model of Alzheimer’s disease. Neuroimage. 2008;42:19–27. doi: 10.1016/j.neuroimage.2008.04.252. [DOI] [PubMed] [Google Scholar]
- 6.Schneider JE, Stork LA, Bell JT, Hove MT, Isbrandt D, Clarke K, Watkins H, Lygate CA, Neubauer S. Cardiac structure and function during ageing in energetically compromised guanidinoacetate N-methyltransferase (GAMT)-knockout mice—a one year longitudinal MRI study. J Cardiovasc Magn Reson. 2008;10:19. doi: 10.1186/1532-429X-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falangola MF, Dyakin VV, Lee SP, Bogart A, Babb JS, Duff K, Nixon R, Helpern JA. Quantitative MRI reveals aging-associated T2 changes in mouse models of Alzheimer’s disease. NMR Biomed. 2007;20:343–351. doi: 10.1002/nbm.1163. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro EM, Gonzalez-Perez O, Garcia-Verdugo JM, Alvarez-Buylla A, Koretsky AP. Magnetic resonance imaging of the migration of neuronal precursors generated in the adult rodent brain. Neuroimage. 2006;32:1150–1157. doi: 10.1016/j.neuroimage.2006.04.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotkin JR, Cahill KS, Tharin SA, Shapiro EM. Cellular magnetic resonance imaging: nanometer and micrometer size particles for noninvasive cell localization. Neurotherapeutics. 2007;4:428–433. doi: 10.1016/j.nurt.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulte JWM, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–489. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 11.Heyn C, Ronald JA, Mackenzie LT, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55:23–29. doi: 10.1002/mrm.20747. [DOI] [PubMed] [Google Scholar]
- 12.Sachs TS, Meyer CH, Hu BS, Kohli J, Nishimura DG, Macovski A. Real-time motion detection in spiral MRI using navigators. Magn Reson Med. 1994;32:639–645. doi: 10.1002/mrm.1910320513. [DOI] [PubMed] [Google Scholar]
- 13.Anderson AW, Gore JC. Analysis and correction of motion artifacts in diffusion weighted imaging. Magn Reson Med. 1994;32:379–387. doi: 10.1002/mrm.1910320313. [DOI] [PubMed] [Google Scholar]
- 14.Firmin D, Keegan J. Navigator echoes in cardiac magnetic resonance. J Cardiovasc Magn Reson. 2001;3:183–193. doi: 10.1081/jcmr-100107467. [DOI] [PubMed] [Google Scholar]
- 15.Welch EB, Manduca A, Grimm RC, Ward HA, Jack CR., Jr Spherical navigator echoes for full 3D rigid body motion measurement in MRI. Magn Reson Med. 2002;47:32–41. doi: 10.1002/mrm.10012. [DOI] [PubMed] [Google Scholar]
- 16.Fu ZW, Wang Y, Grimm RC, Rossman PJ, Felmlee JP, Riederer SJ, Ehman RL. Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magn Reson Med. 1995;34:746–753. doi: 10.1002/mrm.1910340514. [DOI] [PubMed] [Google Scholar]
- 17.Kim WS, Mun CW, Kim DJ, Cho ZH. Extraction of cardiac and respiratory motion cycles by use of projection data and its applications to NMR imaging. Magn Reson Med. 1990;13:25–37. doi: 10.1002/mrm.1910130105. [DOI] [PubMed] [Google Scholar]
- 18.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med. 1999;42:963–969. doi: 10.1002/(sici)1522-2594(199911)42:5<963::aid-mrm17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self-gated cardiac cine MRI. Magn Reson Med. 2004;51:93–102. doi: 10.1002/mrm.10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brau AC, Brittain JH. Generalized self-navigated motion detection technique: preliminary investigation in abdominal imaging. Magn Reson Med. 2006;55:263–270. doi: 10.1002/mrm.20785. [DOI] [PubMed] [Google Scholar]
- 21.Glover GH, Lai S. Self-navigated spiral fMRI: interleaved versus single-shot. Magn Reson Med. 1998;39:68–78. doi: 10.1002/mrm.1910390305. [DOI] [PubMed] [Google Scholar]
- 22.Wiesmann F, Ruff J, Hiller KH, Rommel E, Haase A, Neubauer S. Developmental changes of cardiac function and mass assessed with MRI in neonatal, juvenile and adult mice. Am J Physiol Heart Circ Physiol. 2000;278:H652–H657. doi: 10.1152/ajpheart.2000.278.2.H652. [DOI] [PubMed] [Google Scholar]
- 23.Bishop J, Feintuch A, Bock NA, Nieman B, Dazai J, Davidson L, Henkelman RM. Retrospective gating for mouse cardiac MRI. Magn Reson Med. 2006;55:472–477. doi: 10.1002/mrm.20794. [DOI] [PubMed] [Google Scholar]
- 24.Heijman E, de Graaf W, Niessen P, Nauerth A, van Eys G, de Graaf L, Nicolay K, Strijkers GJ. Comparison between prospective and retrospective triggering for mouse cardiac MRI. NMR Biomed. 2007;20:439–447. doi: 10.1002/nbm.1110. [DOI] [PubMed] [Google Scholar]
- 25.Hiba B, Richard N, Janier M, Croisille P. Cardiac and respiratory double self-gated cine MRI in the mouse at 7T. Magn Reson Med. 2006;55:506–513. doi: 10.1002/mrm.20815. [DOI] [PubMed] [Google Scholar]
- 26.Hiba B, Richard N, Thibault H, Janier M. Cardiac and respiratory self-gated cine MRI in the mouse: comparison between radial and rectilinear techniques at 7T. Magn Reson Med. 2007;58:745–753. doi: 10.1002/mrm.21355. [DOI] [PubMed] [Google Scholar]
- 27.Uribe S, Muthurangu V, Boubertakh R, Schaeffter T, Razavi R, Hill DLG, Hansen MS. Whole-heart cine MRI using real-time respiratory self-gating. Magn Reson Med. 2007;57:606–613. doi: 10.1002/mrm.21156. [DOI] [PubMed] [Google Scholar]
- 28.Hogers B, Gross D, Lehmann V, Zick K, de Groot HJM, Gittenberger-de Groot AC, Poelmann RE. Magnetic resonance microscopy of mouse embryos in utero. Anat Rec. 260:373–377. doi: 10.1002/1097-0185(20001201)260:4<373::AID-AR60>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.Chapon C, Franconi F, Roux J, Marescaux L, Le Jeune JJ, Lemaire L. In utero time-course assessment of mouse embryo development using high resolution magnetic resonance imaging. Anat Embryol. 2002;206:131–137. doi: 10.1007/s00429-002-0281-6. [DOI] [PubMed] [Google Scholar]
- 30.Ahrens ET, Srinivas M, Capuano S, Simhan HN, Schatten GP. Magnetic resonance imaging of embryonic and fetal development in model systems. Methods Mol Med. 2006;124:87–101. doi: 10.1385/1-59745-010-3:87. [DOI] [PubMed] [Google Scholar]
- 31.Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benajamin LE, Neeman M. MRI detection of transcriptional regulation of gene expression in transgenic mice. Nat Med. 2007;13:498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- 32.Deans AE, Wadghiri YZ, Berrios-Otero CA, Turnbull DH. Mn enhancement and respiratory gating for in utero MRI of the embryonic mouse central nervous system. Magn Reson Med. 2008;59:1320–1328. doi: 10.1002/mrm.21609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari VA, Steven A. Imaging the embryonic heart: How low can we go? How fast can we get? J Mol Cell Cardiol. 2002;35:141–143. doi: 10.1016/s0022-2828(02)00306-1. [DOI] [PubMed] [Google Scholar]
- 34.Maas LC, Frederick BD, Renshaw PF. Decoupled automated rotational and translational registration for functional MRI time series data: the DART registration algorithm. Magn Reson Med. 1997;37:131–139. doi: 10.1002/mrm.1910370119. [DOI] [PubMed] [Google Scholar]
- 35.Kim B, Boes JL, Bland PH, Chenevert TL, Meyer CR. Motion correction in fMRI via registration of individual slices into an anatomical volume. Magn Reson Med. 1999;41:964–972. doi: 10.1002/(sici)1522-2594(199905)41:5<964::aid-mrm16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 36.Hayton P, Brady M, Tarassenko L, Moore N. Analysis of dynamic MR breast images using a model of contrast enhancement. Med Image Anal. 1997;1:207–224. doi: 10.1016/s1361-8415(97)85011-6. [DOI] [PubMed] [Google Scholar]
- 37.Felmlee J, Ehman R. Spatial presaturation: a method for suppressing flow artifacts and improving depiction of vascular anatomy in MR imaging. Radiology. 1987;164:559–564. doi: 10.1148/radiology.164.2.3602402. [DOI] [PubMed] [Google Scholar]
- 38.Nayak KS, Rivas PA, Pauly JM, Scott GC, Kerr AB, Hu BS, Nishimura DG. J Magn Reson Imaging. 2001;13:807–812. doi: 10.1002/jmri.1112. [DOI] [PubMed] [Google Scholar]
- 39.Muller K, Lohmann G, Bosch V, von Cramon DY. On multivariate spectral analysis of fMRI time series. Neuroimage. 2001;14:347–356. doi: 10.1006/nimg.2001.0804. [DOI] [PubMed] [Google Scholar]
- 40.Curtis CE, Sun FT, Miller LM, D’Esposito M. Coherence between fMRI time-series distinguishes two spatial working memory networks. Neuroimage. 2005;26:177–183. doi: 10.1016/j.neuroimage.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 41.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- 42.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;33:190–208. [Google Scholar]
- 43.Himmelreich U, Weber R, Ramos-Cabrer P, Wegener S, Kandal K, Shapiro EM, Koretsky AP, Hoehn M. Improved stem cell MR detectability in animal models by modification of the inhalation gas. Mol Imaging. 2005;4:104–109. doi: 10.1162/15353500200504196. [DOI] [PubMed] [Google Scholar]
- 44.Hu XP, Kim SG. Reduction of signal fluctuation in functional MRI using navigator echoes. Magn Reson Med. 1994;31:495–503. doi: 10.1002/mrm.1910310505. [DOI] [PubMed] [Google Scholar]
- 45.Le TH, Hu XP. Retrospective estimation and correction of physiological artifacts in fMRI by direct extraction of physiological activity from MR. Magn Reson Med. 1996;35:290–298. doi: 10.1002/mrm.1910350305. [DOI] [PubMed] [Google Scholar]
- 46.Lustig M, Cunningham CH, Daniyalzade E, Pauly JM. Butterfly: a self navigating Cartesian trajectory; Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007. (Abstract 865) [Google Scholar]