Abstract
Here we describe the use of reverse-phase liquid chromatography mass spectrometry (RP-LC-MS) to simultaneously characterize variants and post-translationally modified isoforms for each histone. The analysis of intact proteins significantly reduces the time of sample preparation and simplifies data interpretation. LC-MS analysis and peptide mass mapping have previously been applied to identify histone proteins and to characterize their post-translational modifications. However, these studies provided limited characterization of both linker histones and core histones. The current LC-MS analysis allows for the simultaneous observation of all histone PTMs and variants (both replacement and bulk histones) without further enrichment, which will be valuable in comparative studies. Protein identities were verified by the analysis of histone H2A species using RPLC fractionation, AU-PAGE separation and nano-LC-MS/MS.
Keywords: Histone, RPLC-MS, post-translational modification, AU-PAGE, nano-LC-MS/MS
INTRODUCTION
Histones are an essential building block of eukaryotic chromatin and regulate higher order chromatin structure [1]. They influence DNA transcription, replication, repair, and recombination. As a result a significant effort has been devoted to tackling the bewildering structural and functional complexity of histone and their post-translational modifications. Their characterization is complex due to the existence of sequence variants and their high degree of post-translational modifications (PTMs). Alterations in histone PTMs have been documented in many types of cancer and may in fact play an important role in tumorogenesis [2]. The National Human Genome Research Institute (NHGRI) maintains a histone database (http://research.nhgri.nih.gov/histones/) that list 21 variants of human H2A, 14 of H2B, and at least three variants of H3 [3], in addition to proteins similar to core histones or histone-like.
Traditionally, immunoassay methodologies such as immunoprecipitation, Western-Blotting, and immunofluorescence have dominated the biologists’ toolbox for the characterization of histones and their PTMs [4-6]. However, these approaches have disadvantages that limit their application. Immunoassays rely on highly specific antibodies that are obtainable commercially for known modification sites. These antibodies may be adversely affected by the presence of multiple PTMs. For example, H3 S10 phosphorylation impacts the specificity of antibodies that target modifications at K9 [7]. Thus, special antibodies have been developed that target both methylation of K9 in addition to S10. In principle, the characterization of these two amino acids (considering acetylation, and 1−3 methylations at K9 with/without S10 phosphorylation) would require 10 antibodies of which only 4 are currently available commercially. Histone characterization is further complicated due to the existence of sequence variants [8-12]. These variants often differ by only a few amino acids, which may not be in the antigenic epitope. Thus, detection of specific variants using antibodies is challenging. More direct approaches have been employed including micro-sequencing and sequencing by mass spectrometry [9-13]. However, sequencing based methodologies are usually tedious and time-consuming. Proteins or peptides larger than 25 amino acids (AAs) in sequence are required to be digested to smaller fragments (5−25 AAs) either enzymatically or chemically for unambiguous analysis.
As the importance of chromatin structure and function in human diseases is increasingly appreciated in this epigenomic era, the need for more rigorous methods that are capable of profiling histones present in clinically relevant materials has become apparent. Such profiles of histones can be used as an assay to determine changes in chromatin that are a function of disease or treatment. Several investigators have dedicated their research efforts to the development of LC-MS methods for the analysis of histones [14-16]. These studies have primarily focused on the characterization of core histones. For example, Zhang and Tang have applied micorbore C4 RP LC-MS to analyze the core histones from the chicken blood [17]. Bonenfant et al. applied C18 RP LC-MS to the study of histones derived from Jurkat cells [16]. Hydrophilic-interaction liquid chromatography (HILIC) coupling with off-line mass spectrometry has been successful in separating and characterizing different histone isoforms [18,19]. However, the high salt required to elute the histones adds an additional desalting step prior to MS analysis. Top down proteomics has also been successfully applied to the characterization of histone forms by Kelleher and coworkers [20-22]. However, their approach required pre-purified histone fractions (such as H2A, H2B and H3) with large sample requirements. Thus a rapid characterization in a single run would be impractical as is necessitated in the analysis of clinical samples.
In this paper we describe a LC-MS based method for profiling histones with improved chromatographic resolution capable of profiling histone variants and their post-translationally modified isoforms. Subsequent validation of differentially expressed proteins was performed by RPLC, AU-PAGE and nano-LC-MS/MS.
EXPERIMENTAL
Sample Preparation
Bovine calf thymus was purchased from Worthington Biochemical Corp (Lakewood, NJ, USA). B-cells from patients with Chronic Lymphocytic Leukemia (CLL) and normal healthy volunteers were procured as reported previously [15]. Raji cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured using a standard growth media [23]. Kasumi-1 cells were grown as in the literature [24]. HeLa cells were monolayer cultured in DMEM containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA).
Histone Preparation
Histones were purified using a standard acid extraction procedure [14,25] as well as Fast Performance Liquid Chromatography (FPLC) with a hydroxyapatite column [26]. Histone H2A from a patient’s CLL cells was fractionated by reverse-phase HPLC (HP1100, Agilent, CA, USA) by use of a Discovery Bio wide pore C18 column (1.0 mm × 150 mm, 5 μm, 300 Å, Supleco Inc, Bellefonte, PA, USA) with a UV-Vis detector. The gradient separation was performed at a flow rate of 50 μL/min with mobile phases A (0.1% TFA in water) and B (0.1% TFA in acetonitrile), in which B increased from 30% to 45% in 2 min, to 60% in 20 min and was maintained at 60% for 4 min. Between each run, the column was washed at 100% B for 2 min and equilibrated at 30% B for 30 min. The H2A fraction was combined and dried in a speed-vac prior to AU-PAGE separation.
Liquid Chromatography Mass Spectrometry (LC-MS)
Histone extracts were separated by reverse-phase HPLC (Waters 2690, Waters, Milford, MA, USA) and detected by ESI-TOF mass spectrometer. HPLC separation was carried out using a flow rate of 50 μL/min on a 1.0 mm × 150 mm C18 column (Discovery Bio wide pore C18 column, 5 μm, 300 Å, Supleco Inc, Bellefonte, PA, USA). The gradient was composed of mobile phase A (0.1% TFA in water) and mobile phase B (0.1% TFA in acetonitrile), where B linearly increased from 30% to 45% in 2 min, 60% in 20 min and was held at 60% for 4 min. Between each run, the column was washed at 95% B for 2 min and equilibrated at 30% B for 30 min. A Micromass LCT (Micromass, Wythenshawe, UK) mass spectrometer with an orthogonal electrospray source (Z-spray) was coupled to the outlet of the HPLC. Histones were infused into the electrospray source at the flow rate 50 μL/min without splitting. ESI was performed at the optimal conditions of 3 kV capillary voltage, 100 °C source temperature and 50 V cone voltage. Data were acquired in continuum mode at the rate of 1 spectrum sec−1. All spectra were obtained in the positive ion mode. NaI was used for external mass calibration over the m/z range 500−2500.
Acetic acid-Urea Polyacrylamide Gel Electrophoresis (AU-PAGE)
Fractionated H2A was further separated by acetic acid urea-polyacrylamide gel electrophoresis (AU-PAGE) [27]. The resolving gel consisting of 15% acrylamide (acrylamide:bisacrylamide = 29:1), 6 M Urea and 5% acetic acid. The gel was run for one hour following addition of a scavenger solution composed of 8 M Urea, 5% acetic acid and 0.6 M β- mercaptoethanol. After casting the stacking gel composed of 7.5% acrylamide (acrylamide:bisacrylamide = 29:1) in water, ∼5 μg of histone H2A was loaded onto each lane. The gels were run at 300 V (constant voltage) overnight (18 hours) at 4 °C with a running buffer of 5% acetic acid under reversed current mode. The gels were fixed in 50% ethanol/10% acetic acid at room temperature for 4 hours, then washed twice (30 minutes each) with 50% methanol/5% acetic acid, stained with Coomassie Brilliant Blue for 30 minutes and then destained in 30% methanol/10% acetic acid until the background was clear.
In-gel Tryptic Digestion
Each of the H2A gel bands separated by AU-PAGE were excised into 1 mm × 1 mm pieces and digested with trypsin. The gel pieces were washed twice (one hour each) in a freshly made solution (50% methanol/5% acetic acid) followed by two cycles of dehydration with acetonitrile and rehydration with 100 mM ammonium bicarbonate (5 minute each). The gel pieces were dried in a speed-vac for 2−3 minutes prior to addition of 600 ng trypsin (20 ng/μL in 50 mM NH4HCO3) to each gel band. The mixture was then hydrated on ice for 10 minutes and another 20 μL of 50 mM ammonium bicarbonate was added. The digestion was carried out at 37 °C for 1.5 hours. Finally, the digested peptides were extracted with 50% ACN/5% formic acid three times. The extract was dried completely in the speed-vac and redissolved in 10 μL of HPLC water.
Nano-LC-MS/MS
The in-gel tryptic digests of each H2A band were subject to nano-LC-MS/MS analysis that was performed on a Shimadzu capillary LC instrument (Columbia, MD, USA) coupled with a Thermo Finnigan LCQ DECA XP+ ion trap (San Jose, CA, USA). Peptide separation was conducted on a self packed reverse-phase C18 nano-column (5 cm, 300 Å, 5 μm, I.D. 75 μm) using a 120 min gradient of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). Mobile phase B increased linearly from 5% to 60% in 80 min, was held at 60% for 5 min, held at 90% for 5 min and finally returned to 5%. The column was washed for half an hour between each run at 5% B. One μL of the digest was injected onto the column. Electrospray voltage was maintained at 1.1 kV and capillary temperature was set at 200 °C. The mass spectrometric detection range was 200−2000 Da. Three data-dependent MS/MS scans using dynamic exclusion were carried out between each full MS scan. The generated product ions were analyzed by the Mascot (Matrix Science Inc., Boston, MA, USA) and an in-house search algorithm, MassMatrix. The search parameters include variable modifications: acetylation of lysine; mono-, di- and tri-methylation of lysine; mono- and di-methylation of arginine; phosphorylation of serine and threonine; and oxidation of methionine. Each of the MS/MS spectra matched with those in the database was manually validated.
RESULTS AND DISCUSSION
Reverse-Phase Liquid Chromatography Mass Spectrometry
LC-MS has the ability to measure the molecular weight for many if not all of the most abundant histone isoforms during a single experiment. From the molecular weights the possible identities for each variant and its modified isoforms may be inferred. The species present in the LC-MS profile were matched with sequences from the databases that corresponded in mass to a specific variant and its potential modified isoforms (acetylation, methylation and phosphorylation). It has been established that mass alone is not the best tool for protein identification.
For the most part many of the isoforms have unique masses. Situations in which isobaric species could be present are noted and designated as a mixture of all possible species. Thus, species assignments are tentatively based upon mass used as temporary identities for subsequent profiling. Full characterization of the interesting species can then be carried out by peptide mass fingerprinting and/or tandem MS. The LC-MS technique has excellent reproducibility and is well suited as a technology for profiling the distributions of histones in clinical specimens, especially allowing one to discern changes induced by the disease or other external stimuli. For instance, large difference in relative abundance were observed for histone H3 isoforms in fractions 8−9 and 11−12 between the normal B-cells and the leukemia cells including CLL and Kasumi-1 (see Figure 2). Thus, the approach has immediate clinical value as an assay to monitor changes in histones due to epigenetic therapies that target histone modifying enzymes.”
Figure 2.
Comparison of LC-MS profiles of histones from bovine thymus tissue and different cell types. The profiles with fractions labeled with  corresponded to the histones that were acid extracted from the different sources. Fractions labeled with
corresponded to the histones that were acid extracted from the different sources. Fractions labeled with  were only observed in the histone mixture prepared using HAP chromatography. The detailed annotations for each fraction are shown in Figure 3-6.
were only observed in the histone mixture prepared using HAP chromatography. The detailed annotations for each fraction are shown in Figure 3-6.
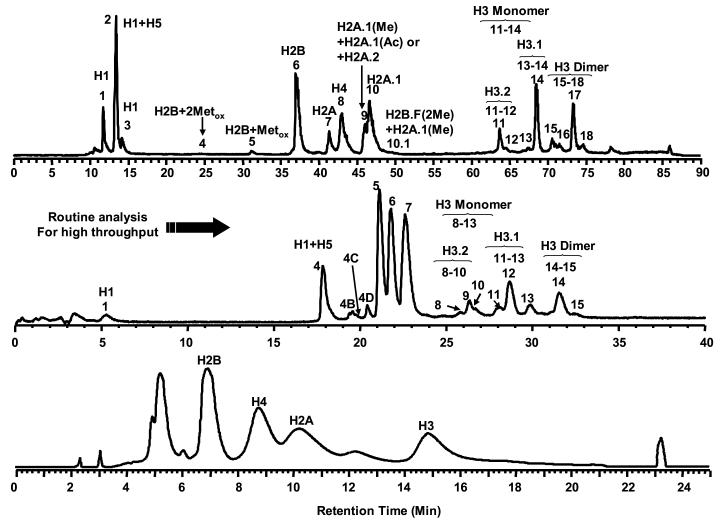
The separation of highly basic histones under reverse-phase conditions presents a challenge to ESI-LC-MS. These lysine and arginine rich proteins require the use of end-capped columns to achieve an effective separation. In addition, the use of ion-paring agents such as TFA and HFBA are necessary but produce adducts that complicate the mass spectra and decrease the limit of detection [17,28,29]. Zhang et al. reported an approach that used a deactivated C18 column with low TFA concentration in the mobile phase to achieve a reasonable separation of the core histones [14,15]. This early work revealed differences in the distribution of H2A isoforms that appeared to be unique to patients with chronic lymphocytic leukemia (CLL). Since that report, further optimization of the chromatography to achieve higher resolution has been performed. Specifically, the use of a 1.0 mm × 150 mm discovery C18 column has resulted in a 4.7-fold improvement in peak capacity over the deactivated LC C18-DB column used in earlier reports. Peak capacity was further enhanced by a factor of 20 by performing the chromatography at low flow rates (e.g. 10 μL/min). The use of the discovery C18 column also allowed for separations at TFA concentrations ≤ 0.05% thus reducing the presence of TFA adducts (data not shown). Consequently, histone variants and modification isoforms that were previously obscured were resolved and detected by ESI-MS (Figure 1).
Figure 1.
Improved HPLC separation of histones derived from bovine thymus tissue. Upper: LC-MS, 10 μL/min, 1.0 mm × 150 mm Discovery C18 column (5 μm, 300 Å). Middle: LC-MS, 50 μL/min, 1.0 mm × 150 mm Discovery C18 column (5 μm, 300 Å). Bottom: LC-MS, 1 mL/min, 4.6 mm × 250 mm C18-DB column (5 μm, 120 Å).
The reproducibility of the developed method was assessed by continuous assay of bovine histones (as standard) that are most readily available and have been well studied by our group. During a 48-hour batch sample run, the retention time of each peak varied within 5% RSD. H2A variants were selected to determine reproducibility in relative abundances as these variants are suspected markers for CLL patients. No significant deviation (less than 3% RSD) of the relative abundance of H2A variants was observed. The optimized LC-MS assay was used to compare histone abundance and modifications from samples obtained from bovine thymus, normal human B cells, primary human CLL tumor cells and several tumor cell lines (Kasumi-1, Raji and HeLa) [30-32].
Effect of Acid Extraction on Histone
Since acid extraction may lead to loss of labile side-chain modification such as phosphorylation [33], a comparison between acid extracted and hydroxyapaptite (HAP) separated histone from HeLa cells was conducted. Samples were purified by both methods and then analyzed by LC-MS. The total ion chromatograms (TICs) are shown in Figure 2 along with bovine thymus histones, which were used as a standard for all LC-MS experiments. The TICs and mass spectral profiles for each sample were nearly identical with notable exceptions. The RPLC fractions labeled with dashed arrow were only observed in the histones purified using HAP separation. Both the acid extracted and HAP purified samples contained linker and core histones. A greater number of H1 variants were observed in the HAP, whereas the three most abundant variants (H1.5, H1.2 and H1.4) were observed in the acid extract. This difference is rationalized by the fact that HAP chromatography fractionates the weakly DNA associated linker histones from the strongly bound core histones resulting in an improved dynamic range of detection. The two approaches yielded similar histone H1 modification patterns. In particular, the distribution of putative phosphorylated H1 variants was nearly identical in the acid extracted or HAP isolated histones (Supplementary Figure S-1). Recent improvements in the isolation of cellular nuclei and combined HAP chromatography have enhanced the speed and reliability of this method. However, not all laboratories may be equipped for the automated chromatography required for HAP analysis, leaving acid extraction as a primary protocol.
Core histone profiles
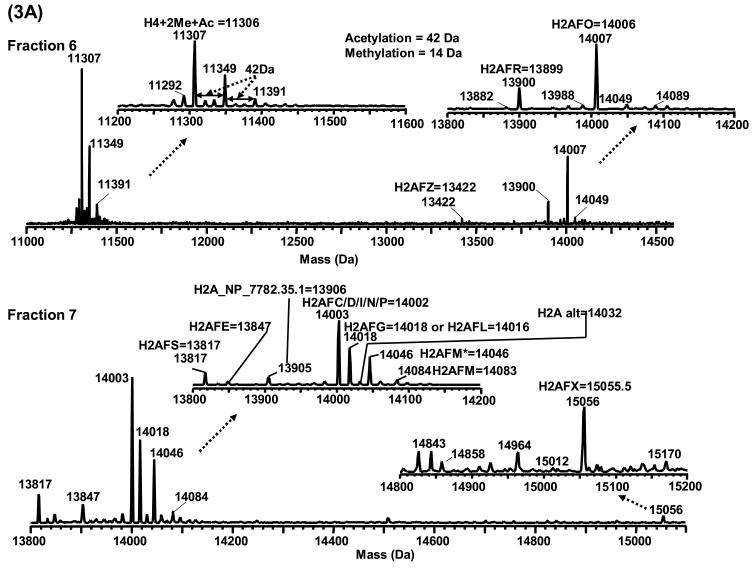
The LC-MS profiles showed several species that corresponded in mass to H2A and H2B variants and/or their post-translationally modified isoforms. The majority of H2A eluted in fractions 6 and 7, and all H2A variants except H2AFZ were shifted in mass by 42 Da, presumably due to N-terminal acetylation. Despite the similarity in the LC-MS profiles of histones derived from different sources, histones purified from HeLa cells (Figure 2) were used as an example for the following discussion. The LC-MS profiles along with proposed assignments are shown in Figures 3 and 4 and listed in Table 1. The most prominent species in fraction 7 were assigned as H2AFC/D/I/N/P (14,002 Da) followed by H2AFL (14,016 Da) and/or H2AFG (14,018 Da) and H2AFM* (14,046 Da). Species corresponding in mass to H2A family members E, S, M, alt, X and H2A_NP_7782.35.1 were also present but at much lower relative abundance. The species present in fraction 6 correlated in mass with the variants H2AFO (14,006 Da) and H2AFR (13,899 Da). The exchange family member Z (13,420 Da) was occasionally observed in fraction 6 in HeLa cells, normal human B cells and primary tumor cells from a patient with chronic lymphocytic leukemia. Family member X (H2AFX) and Z (H2AFZ) are two interesting H2A subtypes whose sequences deviate significantly from the homologous bulk H2A variants [34,35]. They are considered minor variants because of their low abundance. However, the limit of detection (LOD) of the current method was sufficient to detect these minor variants. Compared to H2A, H2B has fewer reported sequence variants. Fraction 5 from the LC-MS profile contained species that corresponds in mass to H2BFA (13,775 Da), H2BFQ (13,789 Da) and/or H2BFN (13,791 Da), H2BFT (13,759 Da) and/or H2BFJ (13,761 Da), and H2BFB (13,805 Da). Unlike H2A, no species corresponding to N-terminally acetylated H2B were observed.
Figure 3.
Mass spectra of 3A) H4 in fraction 6 and H2A in fraction 7; 3B) H3 in fractions 8−13. Fraction # is the same as shown in Figure 2. The histones presented in this figure were derived from HeLa cells.
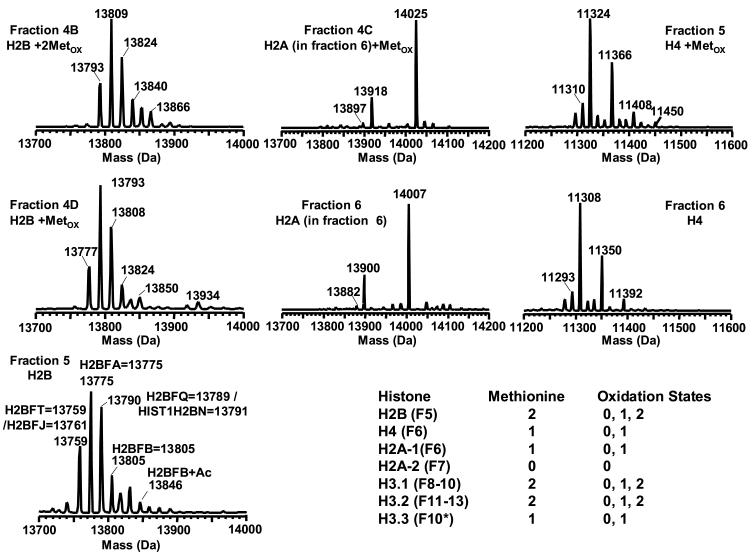
Figure 4.
Methionine oxidized isoforms of H2B in fractions 4B, 4D and 5; H2A in fractions 4C and 6; and H4 in fractions 5 and 6. Fraction # is the same as shown in Figure 2. The included table indicates the number of methionine residues contained in each histone species eluted in different fractions. “Fn” in the table refers to the fraction number. The histones presented in this figure were derived from HeLa cells.
Table 1.
The H2A and H2B variants characterized by LC-MS*
| Fractions | |||
|---|---|---|---|
| 5 (H2B) | 6 (H2A) | 7 (H2A) | |
| 1 | H2BFT, 13759 / H2BFJ, 13761 | H2AFO, 14006 | H2AFI/C/D/N/P, 14002 |
| 2 | H2BFA, 13775 | H2AFR 13899 | H2AFG, 14018 / H2AFL, 14016 |
| 3 | H2BFQ, 13789 / HIST1H2BN, 13791 | H2AFV.1, 13420 / H2AFZ, 13422 | H2AFM*, 14046 |
| 4 | H2BFB, 13805 | H2AFM, 14083 | |
| 5 | H2AFS, 13817 | ||
| 6 | H2AFE, 13847 | ||
| 7 | H2A_NP7782.35.1, 13906 | ||
| 8 | H2A alt, 14032 | ||
| 9 | H2AFX, 15055.5 | ||
For the LC-ESI-TOF MS, the detected molecular weight of each protein deviated from the theoretical molecular weight by 1−2 Da.
Multiple PTMs have been recorded for histone H2A and H2B. H2A and H2B have been reported to possess acetylation (H2A: K5, K9 and H2B: K5, K12, K15, K20), phosphorylation (H2A: S1 and H2B: S10), monomethylation (H2A: K15 and H2B: K5, K43) and ubiquitination (H2A: K119 and H2B: K120/K123) [1,14,16,36-40]. Thus the unambiguous assignment of the species in these profiles is complicated by possible presence of these modifications along with the variants. While the masses correspond to the expected values for the H2A/H2B variants, it is likely that isoforms of modified variants could exist and further complicate the mass spectral interpretation. For example, H2AFM* (14,046 Da) and monoacetylated H2AFC/D/I/N/P (14,002 + 42 = 14,044 Da) (if there is any) are isobaric. Likewise, monophosphorylated H2AFC/D/I/N/P (14,002 + 80 = 14,082 Da) is isobaric with H2AFM (14,083 Da). For histone H2B, the species at 13,791 Da could not be exclusively assigned to either H2BFQ (13,789 Da) or H2BFN (13,791 Da). The same situation occurs with H2BFT (13,759 Da) and H2BFJ (13,761 Da), and also the monoacetylated H2BFA (13,775 + 42 = 13,817 Da) and unmodified H2BFF (13,819 Da). In addition, the mass shift between adjacent peaks in H2B profile is around 15 Da. LC-MS at the current mass resolving power (<10,000) was not capable of differentiating lysine methylation (Δmass = 14 Da) and methionine oxidation (Δmass = 16 Da). Thus, as previously stated these specific assignments are tentative. Further characterization of these assignments is warranted if they prove clinically relevant. Two species at 14,049 Da and 14,089 Da in fraction 6 corresponded uniquely in mass to monoacetylated and monophosphorylated H2AFO (Figure 3A) and the species at 13,846 Da in fraction 5 corresponded uniquely with monoacetylated H2BFB (Figure 4).
H3 produced the most complicated mass spectra of all the core histones due to the presence of multiple variants and its high degree of post-translational modifications, in particular acetylation and methylation [20,41]. Three human variants of H3 have been reported including H3.1, H3.2, and H3.3. H3.1 and H3.2 differ by one amino acid residue (H3.1 C96 vs H3.2 S96) and are deposited onto DNA during S-phase [42]. H3.3 differs from H3.1 by five substitutions (A31S, S87A, V89I, M90G and C96S) [43,44]. These amino acid substitutions introduce changes in hydrophobicity of the H3 variants resulting in different retention times when separated by reverse-phase chromatography. The substitution of S for C96 results in decreased hydrophobicity of H3.2 (H3.1 hydrophobicity = 0.680 and H3.2 hydrophobicity = 0.359) (http://psyche.uthct.edu/shaun/SBlack/aagrease.html). The calculated “sequence specific retention” (http://hs2.proteome.ca/ssrcalc-cgi/SSRCalc3.pl) for these two variants yielded a relative hydrophobicity (RH) of H3.2 (RH = 55.54) that was slightly smaller than that of H3.1 (RH = 55.62). This calculated relative order of elution is supported by the LC-MS profile in that the species corresponding in mass with H3.2 eluted in fractions 8−10, whereas the H3.1 isoforms eluted later in fractions 11−13 (Figure 2). Interestingly, in fraction 10 from the HeLa cells a bimodal distribution of modified H3 was observed. One possible explanation for the difference in the distributions could be the elution of the minor variant, H3.3. The observed masses of the species at the low mass end of the distribution correlate well with modified isoforms of H3.3. This supposition is consistent with prior observations by Lindner et al. [45]. Overall, the LC-MS profile of H3 shows a complex pattern of PTMs. The 14 Da mass shifts between adjacent peaks could be correlated to different numbers of methylations, either multiple monomethylations or the combination of mono-, di-, and/or trimethylations. The isobaric acetylation and trimethylation were indistinguishable at the current mass resolving power and chromatographic resolution. Thus, it is impossible to unambiguously assign a unique isoform to each peak in the H3 distributions.
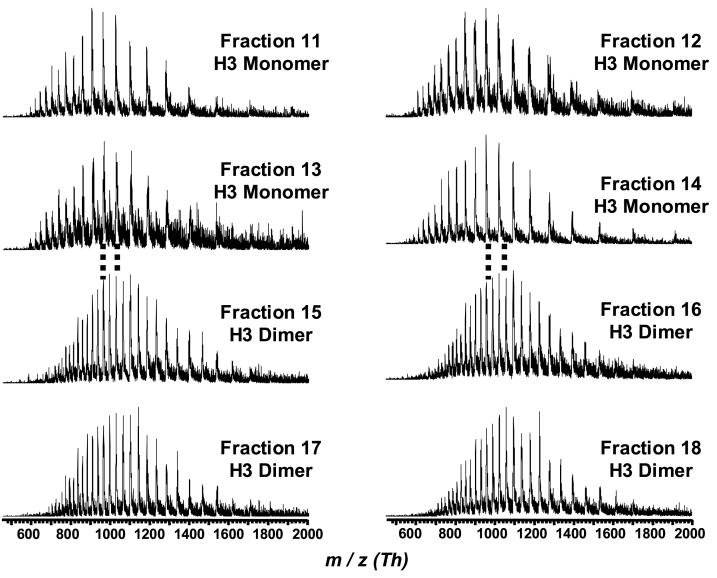
H3 dimers were also observed in bovine thymus histones in fractions 14 and 15 (Figure 1 and 5). These dimers have been attributed to the formation of an intermolecular disulfide bridge that lies on the dyad axis of the nucleosome [46]. No H3 dimers were observed by PAGE approaches that employ reducing conditions. In this regard, the LC-MS technique has an advantage over PAGE. Interestingly, no H3 dimers were observed with any of the human cells.
Figure 5.
Mass spectra (m/z) of H3 dimers and monomers present in bovine thymus tissue. The LC-MS conditions and peak annotations were the same as shown in upper chromatogram of Figure 1.
Histone H4 is unique with respect to the other core histones. There are no reported sequence variants for H4. Thus, its mass spectrum is representative of its post-translational modifications and not complicated by the presence of multiple variants. The distribution of species located in LC-MS fraction 6 corresponded in mass to post-translationally modified histone H4 isoforms (Figure 3A). H4 has been shown to be predominantly dimethylated at K20 and acetylated at K16, K12, K8 and K5 in human cells [47-50]. All the masses observed for H4 isoforms were 42 Da higher than their theoretical masses, consistent with N-terminal acetylation of H4 [51]. The most abundant species observed at 11,306 Da corresponded in mass with the dimethylation (DiMe) and N-terminal acetylation (N-Ac) of H4. The next most abundant peaks, 11,348 Da and 11,390 Da, corresponded to additional acetylation upon H4. Other minor species were observed at 11,278, 11,292, 11,320 and 11,334 Da that corresponded in mass to the H4 isoforms with N-Ac, N-Ac + Me (monomethylation), N-Ac + Ac (monoacetylation) and N-Ac + Ac + Me, respectively. Due to isobaric nature of three methyl groups and one acetyl group (both add 42 Da), the species at 11,320 Da could also be due to N-Ac + TriMe and / or N-Ac + DiMe + Me and / or NAc + 3Me, and the species at 11,334 Da could be due to N-Ac + TriMe + Me and / or N-Ac + DiMe + 2Me and / or N-Ac + 2DiMe and / or N-Ac + 4Me. Previous reports that used peptide mass mapping and tandem mass spectrometry favor the assignments of 11,306 Da as N-Ac + K20DiMe, 11,348/9 Da as N-Ac + K16Ac + K20DiMe and 11390/1 Da as N-Ac + K16Ac + K12Ac + K20DiMe [47,48].
Methionine Oxidation
In addition to the extensively studied methylation and acetylation of core histones, methionine oxidation is another probable modification. It has been well documented that oxidation of a single methionine changes the physical properties of peptides and proteins [52-56]. In particular, the oxidation of methionine to methionine sulfoxide decreases the hydrophobicity of the protein and consequently its retention in reverse-phase chromatography [56]. The degree of oxidation may be used to determine the number of methionines and thus indirectly assist in assignment of protein identification. Oxidized forms of histones were observed in both acid extracted and HAP purified samples.
In the HAP purified histones from HeLa cells, the methionine oxidized forms that were observed included: 1) monooxidized H4 in fraction 5, 2) monooxidized H2B variants in fraction 4D, 3) dioxized H2B variants in fraction 4B, and 4) monooxidized H2A in fraction 4C (Figure 4). The number of methionine residues oxidized was determined by mass and change in retention/hydrophobicity as tabulated in Figure 4. The decreased retention for the oxidized histone variant was consistent with other reports [56]. Since all H2B variants and isoforms contain two methionines, there should be three oxidation states (0, 1 and 2) corresponding to the methionine oxidation. This assertion was supported by the data that showed the presence of species corresponding in mass with 1) dioxidized H2B (Δmass = 32 Da) in fraction 4B, 2) monooxidized H2B (Δmass = 16 Da) in fraction 4D, and 3) native H2B in fraction 5. Similar trends were observed for H2A and H4 that each contains a single methionine residue. Oxidized H2A (containing 1 Met) appeared at fraction 4C while its native form was present in fraction 6. For H4, fraction 5 and 6 corresponded to the oxidized and native forms, respectively. No oxidized species were observed for variants that possessed no methionine residues (H2AFC/D/I/N/P, H2AFL or H2AFG, H2AFM*, H2AFM, H2AFE, H2AFS, H2A alt, H2AFX and H2A_NP_7782.35.1).
The bulk H3.1 and H3.2 contained two methionine residues and should be present in three different oxidation states (non-, mono-, and dioxidized), whereas replacement H3.3 that contains only one methionine and there should be present in two states (non- and monooxidized). Although species resulting from methionine oxidation (Δmass = 16 Da) and lysine methylation (Δmass = 14 Da) are indistinguishable under the current mass resolving power and mass accuracy, the change in RPLC retention caused by the former could help differentiate these two modifications. The data did show shift in mass of 16 Da between fractions 8 and 9 where the most abundant peaks were centered on 15,344 and 15,327 Da, respectively. A similar trend was observed for H3 in fractions 11 and 12 with the most abundant peaks centered on 15,360 and 15,343 Da. The distribution of species within each pair of fractions was nearly identical leading to the conclusion that these species were likely to be due to isoforms that differ by a methionine oxidation altering their retention. Unlike the other core histones, it was not possible to see all the possible oxidation states for H3. There have been no reports regarding histone methionine oxidation and its functional correlation.
It is possible that this modification was introduced during sample preparation. However, the extent of oxidation in acid extraction was comparable to that of HAP. Furthermore methionine oxidation is involved in important biological processes and should be characterized in greater detail. One of the benefits of the profiling approach is that such species can be readily observed and flagged for further analysis using targeted experiments in order to assess the source/role of these species. However, such studies were beyond the scope of this report. Due to their relatively low abundance, we did not assess the extent and variability of the oxidized histones. Semiquantitation based upon the relative abundance of these species would have resulted in large relative standard deviations. Therefore, we carried out a purely qualitative analysis.”
Linker histone profiles
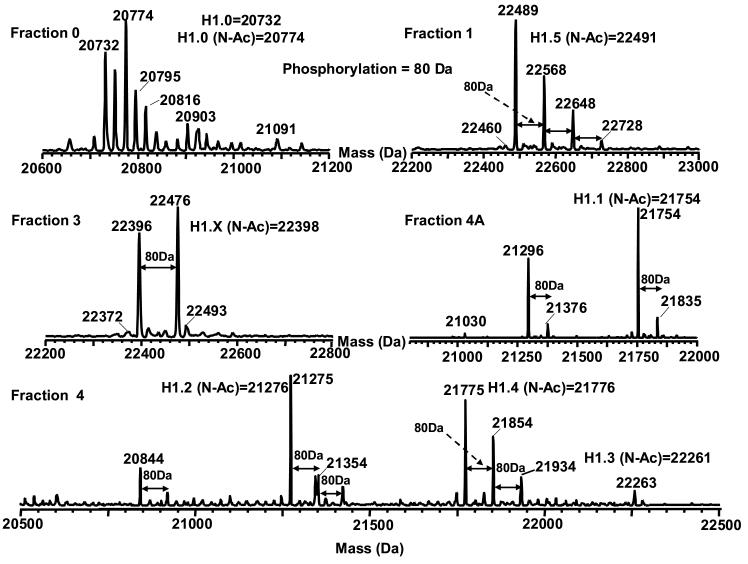
The linker histone H1 (including subtypes H5 and H10 [57]) have been correlated with chromatin condensation and transcription repression in eukaryotes [58]. It is known that its function with regard to the organization of chromatin structure is mediated by phosphorylation [59,60]. At least seven human H1 variants (five somatic and two germinal) have been identified to date and their sequences are available in the Histone Database (http://research.nhgri.nih.gov/histones/). Representative mass spectra obtained from both acid extracted and HAP purified histones from HeLa cells is shown in Figure 6. The LC-MS of the acid extracted histones indicated the presence of several species that correlated in mass with H1.5, H1.2 and H1.4. H1.5 showed several higher mass peaks consistent with multiple phosphorylations (at multiple residues). In contrast, H1.2 and H1.4 exhibited phosphorylation to a lesser extent (at one or two residues). The masses for all the H1 variants were 42 Da higher than their theoretical masses. H1 variants have been reported to be N-terminally acetylated, which would precisely account for this mass difference [61].
Figure 6.
Mass spectra of histone H1 and H1-like proteins. The histones presented in this figure were derived from HeLa cells and the fraction # is the same as shown in Figure 2. N-Ac refers to N-terminal acetylation.
We compared the acid extracted results with those from HAP chromatography. The results from the two purification strategies were nearly identical with the exception that some lower abundant linker histones were observed with HAP. These species included H1.X, H1.0, H1.1 and H1.3. In addition, mono-phosphorylation was observed on H1.X and H1.1 but not on H1.0 and H1.3. These results are consistent with those reported previously by Hunt and coworkers who used immobilized metal ion affinity chromatography (IMAC) combined with tandem mass spectrometry to characterize histone phosphorylation [61]. The LC-MS profiles were able to detect the presence of phosphorylation without enrichment by IMAC. However, location of the modification could not be deduced from the profile.
Characterization of Differential Expressed Histone Form as Determined by LC-MS Profiling
While profiling can rapidly identify potential histone forms of interest, subsequent experiments are needed to properly characterize the species in question. Because of the immediate application of the current LC-MS method to the analysis of clinically important specimens, we present an example of the workflow for post-profiling characterization of histone forms. The following example set out to characterize the H2A forms present in RPLC fraction 7 derived from the B-cells of a CLL patient. This fraction has been shown by LC-MS profiling to contain H2A forms that are differentially expressed in CLL patients as compared with normal individuals [62]. The pipeline for characterization initiated with RPLC fractionation followed by AU-PAGE separation and nano-LC-MS/MS detection.
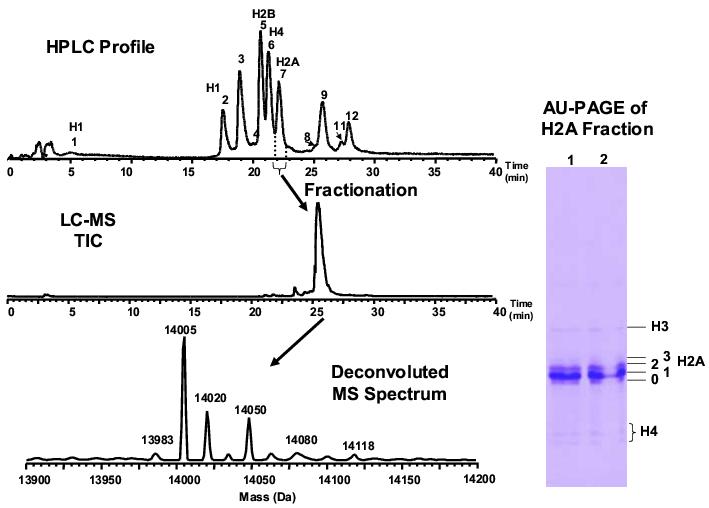
AU-PAGE is prominent in the separation of histone subtypes according to the mass and charge difference. However, due to the highly similar electrophoretic behavior of H2A and H2B, they overlap when separated by AU-PAGE. To avoid this complication, the species were first fractionated by RP-HPLC. The fractionated H2A as characterized by LC-MS (Figure 7) contained > 95% purity with small abundance of H4 and H3. The H2A fraction was then separated by AU-PAGE into four distinct bands (Figure 7). Each gel band was excised, in-gel tryptic digested and analyzed by nano-LC-MS/MS that was used to validate the histone identities determined solely by LC-MS profiling. The high homology of H2A variants presents a great challenge to this characterization. The fractionation of H2A followed by the AU-PAGE separation was crucial to detect the low abundant peptides that were unique to each H2A form. AU-PAGE serves as an additional fractionation for the different variants and their isoforms. Because some isoforms contain homologous sequences, their further fractionation allows assignment of these sequences (and their PTMS) to specific variants. This analysis would not be possible by direct LC-MS/MS. The sequence variation within the human H2A variants are listed in Table 2 [63]. Specific peptide sequences were used to determine the presence of a unique form. For example, the presence of peptides containing T6 indicated the unique presence of variant H2AFX (H2A family member X).
Figure 7.
Left) LC-MS profile of isolated H2A in fraction 7 (see Figure 2). The purity of H2A in the fraction is up to 95% due to the small amount of adjacent H3 and H4 species. RPLC conditions: Discovery C18 column 5 μm, 300 Å 1.0 mm × 150 mm; flow rate 50 μL/min. The conditions were optimized using easily available standards of bovine histones. Right) AU-PAGE of H2A fractionated by RPLC. The 0−3 labels represent different bands of H2A. The protein amount in band 3 was too little to be detected by MS. The concomitant H3 and H4 were well resolved from H2A. The histone H2A variants presented in this figure were derived from CLL cells.
Table 2.
Sequence variation in human H2A variants
| gi number | H2A variant% | #AA | MW#(Da) | 6*Q/T** | 10 A/V | 16 S/T | 37−38 | 40 A/S | 51 M/L | 87 V/I | 99 K/R/G | 123/4 H | 125 | 126 A/T | 127 | 128 | 129 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gi|121959 | H2AFC/D/I/N/P | 129 | 14002 | Q | A | T | GN | A | L | I | K | H | K | A | K | G | K |
| gi|4504251 | H2AFO | 129 | 14006 | Q | A | S | GN | A | M | I | K | H | K | A | K | G | K |
| gi|55930954 | H2AFL | 129 | 14016 | Q | A | S | GN | A | L | I | R | H | K | A | K | G | K |
| gi|121978 | H2AFG | 129 | 14018 | Q | A | T | GN | S | L | I | K | H | K | A | K | G | K |
| gi|19557656 | H2AFM* | 129 | 14046 | Q | A | T | GN | S | L | I | R | H | K | A | K | G | K |
| gi|121986 | H2AFM | 129 | 14083 | Q | A | T | AH | S | L | I | R | H | K | A | K | G | K |
| gi|12654707 | H2A alt | 129 | 14032 | Q | A | S | GN | S | L | I | R | H | K | A | K | G | K |
| gi|10800144 | H2AFE | 127 | 13847 | Q | A | T | GN | A | L | I | K | H | K | T | K | Δ | Δ |
| gi|49659927 | H2AFR | 128 | 13899 | Q | A | S | GN | A | M | I | K | Δ*** | K | A | K | S | K |
| gi|28195394 | H2A_NP_7782.35.1 | 129 | 13906 | Q | A | S | GN | A | L | V | G | H124K**** | P | G | K | N | K |
| gi|18105045 | H2AFS | 127 | 13817 | Q | A | T | GN | A | L | I | K | H | K | A | K | Δ | Δ |
| gi|4504253 | H2AFX | 142 | 15055.5 | T | A | S | GN | A | L | I | G | X | |||||
| gi|29553970 | H2AFJ.2 | 128 | 13930 | Q | V | S | GN | A | L | I | K | ΔΔQ | K | T | K | S | K |
| gi|8922758 | H2AFJ.1 | 150 | 16020 | Q | V | S | GN | A | L | I | K | X | |||||
| Less Homology | 14 | 38 | 127 | ||||||||||||||
| A/T | T/S | A/V | |||||||||||||||
| gi|4504255 | H2AFZ | 127 | 13422 | T | T | A | |||||||||||
| gi|6912616 | H2AFV.1 | 127 | 13420 | A | S | V |
Residue site
Possible amino acid residue at a specific site
Mutation site
Residue substitution
The sequences of H2A variants were retrieved from NHGRI Histone Database.
All the H2A MWs (except H2AFZ) were calculated with acetylation at N-terminal serine.
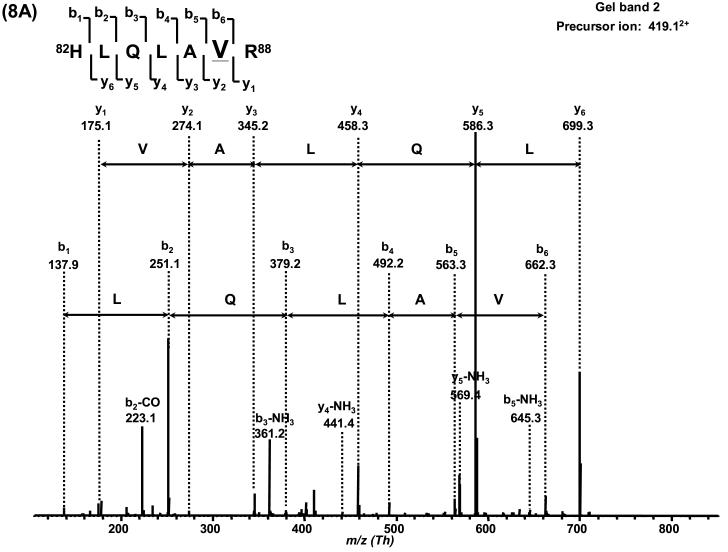
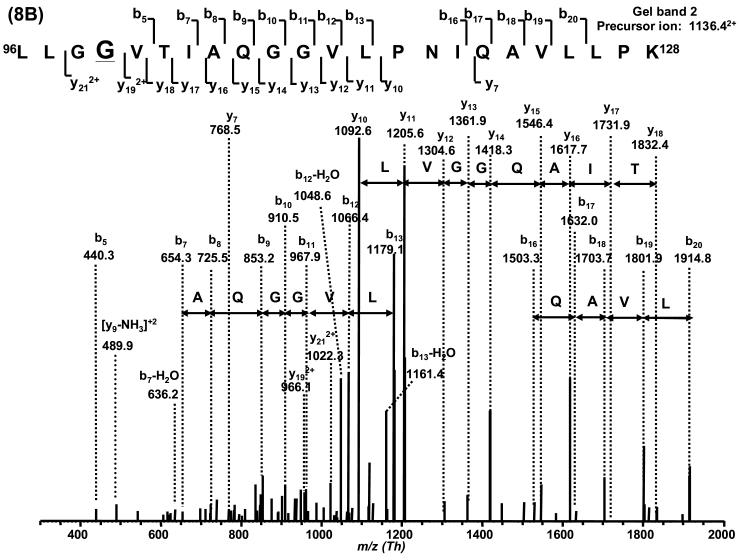
Based on the LC-MS mass spectrum of this H2A fraction, H2AFC/D/I/N/P (14,002 Da) is the major variant, and H2AFL (14,016 Da) or H2AFG (14, 018 Da) and H2AFM* (14,046 Da) are the second most abundant variants. Other H2A variants are present at lower abundance. With the combination of sequence information obtained by nano-LC-MS/MS, H2AFC/D/I/N/P was determined to be present in gel band 1. H2A_NP_7782.35.1 (13,906 Da) was confined to gel band 2 due to its unique V87 in 82HLQLAVR88 (419.122+Th) and G99 in 96LLGGVTIAQGGVLPNIQAVLLPK118 (1,136.392+Th). H2AFX was thus inferred to be present in band 1 due to the presence of a peptide with residue G99 (G99 is common to both H2AFX and H2A_NP_7782.35.1). The S40 containing variants including H2AFM* (14,046 Da) and H2A alt. (14,032 Da) could only possibly be present in gel band 1 as no peptide containing S40 was observed in gel band 0 and 2. The only possible sequence that contained G37N38, A40 (36KGNYAER42, 419.452+Th), I87 (82HLQLAIR88, 426.382+Th) and R99 (89NDEELNKLLGR99, 650.972+ Th) in gel band 0 was H2AFL. If there were any H2AFG (14,018 Da), it should be confined to gel band 1 as indicated by the presence of S40 in 36KGNYSER42 (427.452+ Th). The variants H2AFS (13,817 Da) and H2AFE (13,847 Da) may be present in any of the bands since no specific sequence tag was observed. No unique sequence for H2AFM (14,083 Da, A37H38) were observed. The average sequence coverage of H2A was ∼50%.
Post-translational modifications of H2A were also observed in gel band 2. The presence of the monomethylated peptide 82HLQLAVRMe88 (426.342+ Th) inferred that H2A_NP_7782.35.1 (13,906 Da) was monomethylated at R88. Monoacetylation was also observed on two common sequences 89NDEELNKAcLLGK99 (658.722+ Th) and 89NDEELNKLLGR99 (672.672+ Th) at K95. This modification could occur on H2AFC/D/I/N/P, H2AFL, H2AFE and/or H2AFS due to the sequence homology for fragment 89−99. The possibility that the peak at 14,016/8 Da was the methylated isoform of H2AFC/D/I/N/P was excluded by double SILAC labeling experiments (data not shown). Based on the results above, the peak at 14,046 Da may be a mixture of variant H2AFM* and monoacetylated H2AFC/D/I/N/P. Table 3 summarizes these observations and Table 4 lists all the peptides detected by nano-LC-MS/MS. Figure 8 shows two representative MS/MS spectra of the sequence specific peptides and all other tandem mass spectra are shown in the supplementary material.
Table 3.
Summary of the results obtained by LC-MS, AU-PAGE and nano-LC-MS/MS--H2A variants observed in the CLL patient.
| Relative Abundance | MW | Gel band 0 | Gel band 1 | Gel band 2 |
|---|---|---|---|---|
| H2AFC/I/N/P | 14002 | -- | X | Y (+Ac) |
| H2AFL | 14016 | X | -- | Y (+Ac) |
| H2AFG | 14018 | -- | Y | -- |
| H2AFM* | 14046 | -- | X | -- |
| H2AFE | 13847 | Y | Y | Y or Y (+Ac) |
| H2AFS | 13817 | Y | Y | Y or Y (+Ac) |
| H2A_28195394 | 13906 | -- | -- | X |
| H2AFX** | 15056 | -- | X | -- |
| H2A alt** | 14032 | -- | X | -- |
| H2AFM | 14083 | -- | -- | -- |
| AA | Band 0 | Band 1 | Band 2 |
|---|---|---|---|
| 37−38 GN | GN | GN | GN |
| 40 A/S | A | A | |
| 87V/I | I | I | |
| 99 K/R /G | K, R | ||
| 123/4 H | H | ||
| 125 K | K |
X (Present with 100% confidence)
Y (Possibly present)
“--” (Not present)
H2AFX and H2A_alt are in the similar low abundance.
Table 4.
Peptides identified by nano-LC-MS/MS in each gel band of histone H2A**
| Gel band | Observed (m/z) | Mr, Da (expt) | Mr, Da (calc) | Delta | Miss* | Peptide*** |
|---|---|---|---|---|---|---|
| H2A_0 | 419.442+ | 836.87 | 836.41 | 0.46 | 1 | 36KGNYAER42 |
| 426.242+ | 850.47 | 849.52 | 0.96 | 0 | 82HLQLAIR88 | |
| 861.31+ | 860.30 | 860.39 | −0.08 | 0 | 89NDEELNK95 | |
| 431.322+ | 860.62 | 860.39 | 0.24 | 0 | 89NDEELNK95 | |
| 944.50+ | 943.49 | 943.52 | −0.03 | 0 | 21AGLQFPVGR29 | |
| 473.072+ | 944.13 | 943.52 | 0.61 | 0 | 21AGLQFPVGR29 | |
| 637.222+ | 1272.43 | 1271.67 | 0.76 | 1 | 89NDEELNKLLGK99 | |
| 651.122+ | 1300.23 | 1299.70 | 0.53 | 1 | 89NDEELNKLLGR99 | |
| 966.352+ | 1930.69 | 1930.16 | 0.53 | 0 | 100VTIAQGGVLPNIQAVLLPK118 | |
| H2A_1 | 430.33+ | 429.32 | 429.30 | 0.03 | 0 | 96LLGK99 |
| 498.49+ | 497.48 | 497.33 | 0.15 | 0 | 78IIPR81 | |
| 355.882+ | 709.75 | 708.32 | 1.43 | 0 | 37GNYAER42 | |
| 419.592+ | 837.17 | 836.41 | 0.76 | 1 | 36KGNYAER42 | |
| 426.242+ | 850.47 | 849.52 | 0.96 | 0 | 82HLQLAIR88 | |
| 431.622+ | 861.23 | 860.39 | 0.85 | 0 | 89NDEELNK95 | |
| 473.262+ | 944.51 | 943.52 | 0.99 | 0 | 21AGLQFPVGR29 | |
| 637.002+ | 1271.99 | 1271.67 | 0.32 | 1 | 89NDEELNKLLGK99 | |
| 651.122+ | 1300.22 | 1299.70 | 0.52 | 1 | 89NDEELNKLLGR99 | |
| 847.232+ | 1692.44 | 1691.90 | 0.55 | 1 | 82HLQLAIRNDEELNK95 | |
| 966.322+ | 1930.63 | 1930.16 | 0.47 | 0 | 100VTIAQGGVLPNIQAVLLPK118 | |
| 1931.94+ | 1930.93 | 1930.16 | 0.77 | 0 | 100VTIAQGGVLPNIQAVLLPK118 | |
| 1136.592+ | 2271.17 | 2270.37 | 0.79 | 0 | 96LLGGVTIAQGGVLPNIQAVLLPK118 | |
| 434.212+ | 866.41 | 865.44 | 0.97 | 1 | 119KTESHHK125 | |
| 427.452+ | 852.89 | 852.41 | 0.48 | 1 | 36KGNYSER42 | |
| H2A_2 | 498.46+ | 497.45 | 497.33 | 0.12 | 0 | 78IIPR81 |
| 355.572+ | 709.12 | 708.32 | 0.80 | 0 | 37GNYAER42 | |
| 419.802+ | 837.58 | 836.41 | 1.17 | 1 | 36KGNYAER42 | |
| 850.45+ | 849.44 | 849.52 | −0.08 | 0 | 82HLQLAIR88 | |
| 426.382+ | 850.75 | 849.52 | 1.24 | 0 | 82HLQLAIR88 | |
| 426.262+ | 850.51 | 849.52 | 0.99 | 0 | 82HLQLAVR(Me)88 | |
| 861.38+ | 860.37 | 860.39 | −0.01 | 0 | 89NDEELNK95 | |
| 431.602+ | 861.18 | 860.39 | 0.80 | 0 | 89NDEELNK95 | |
| 944.51+ | 943.50 | 943.52 | −0.02 | 0 | 21AGLQFPVGR29 | |
| 472.962+ | 943.90 | 943.52 | 0.38 | 0 | 21AGLQFPVGR29 | |
| 637.242+ | 1272.47 | 1271.67 | 0.80 | 1 | 89NDEELNKLLGK99 | |
| 650.972+ | 1299.92 | 1299.70 | 0.22 | 1 | 89NDEELNKLLGR99 | |
| 658.722+ | 1315.43 | 1313.68 | 1.75 | 1 | 89NDEELNK(Ac)LLGK99 | |
| 672.672+ | 1343.32 | 1341.71 | 1.61 | 1 | 89NDEELNK(Ac)LLGR99 | |
| 1930.982+ | 1929.97 | 1930.16 | −0.19 | 0 | 100VTIAQGGVLPNIQAVLLPK118 | |
| 966.552+ | 1931.08 | 1930.16 | 0.92 | 0 | 100VTIAQGGVLPNIQAVLLPK118 | |
| 1136.392+ | 2270.77 | 2270.37 | 0.39 | 0 | 96LLGGVTIAQGGVLPNIQAVLLPK118 | |
| 419.122+ | 836.23 | 835.50 | 0.73 | 0 | 82HLQLAVR88 |
Miss: the # of miscleavage sites;
The protein abundance in band 3 was too low to be detected by MS.
Any peptide hit with Mascot Score < 18 was not considered as a match.
Figure 8.
Verification of H2A variants by MS/MS. The histone H2A variants presented in this figure were derived from CLL cells.
The current study was also compared to the other recent efforts using LC-MS to identify histones proteins [64]. In the work of Naldi et al., 8 histone forms were baseline separated and their mass analysis correlated the species to post-translationally modified isoforms. However, our results suggest that histone H2A and H2B are present as both modified forms and sequence variants. The presence of variants was verified by the analysis of the H2A in fraction 7 by use of RPLC fractionation, AU-PAGE separation and nano-LC-MS/MS. The liquid chromatographic separation of histones in our study was further improved by optimization of gradients and lowering the flow rate (e.g. 10 μL/min, Figure 1 Upper) such that sequence variants were readily resolved. In addition, lower concentration of TFA (≤ 0.05%) and the combination of TFA and HFBA additives were tested to achieve comparable separation. While the overall chromatograms were similar to that in Figure 1. However, 0.1% TFA proved to be have slightly better resolution than the other conditions tested.
Histone Nomenclature
There are 21 sequences for human H2A, 14 for H2B, 12 for H3 and 3 for H4 cataloged in the NHGRI histone database. The current nomenclature for the histone variants is quite confusing and varies across sources. H3 variants are usually referred to as H3.1, H3.2 and H3.3. However, in the database, there are H3, H3/o, two H3 family member 3A, etc. By performing sequence comparison and mass calculation, H3 in the database corresponds with H3.1 (15,273 Da), H3/o with H3.2 (15,257 Da) and H3F3A_ gi|4504279 to H3.3 (15,197 Da). From this point of view, a more consistent and clear annotation of the identity of histones in the literature and the database is necessary. Among all the core histones, the nomenclature for H4 is the most interesting. Almost all of references assert that H4 is invariant. However, the database contains H4 (11,237 Da), H4 family member L (H4FL, 11,262 Da) [65], H4 family member J (H4FJ, 11,236 Da) [66] and HIST1H4F [67]. HIST1H4F is identical to H4FL, H4 and H4FL differ in sequence by an Ala-to-Pro substitution at position 76, and H4FL and H4FJ differ by an Asn-to-Asp substitution at position 25. The latter two variants differ in mass by 1 Da, which is not resolvable under the current mass spectrometry conditions. Thus the questions arise: Do these variants exist? Is their existence obscured by the isobaric species in the mass spectrum? With N-terminal acetylation, the theoretical masses of these three possible H4 variants are 11,279, 11,304, and 11,278 Da. As species corresponding in mass to 11,278 Da and 11,279 Da are isobaric (considering the isotopic distributions), the variant 11,304 Da would overlap their modified isoform with N-Ac + DiMe (11,306 Da). Thus methods that provide definitive identification for these species are necessary. Finally, the NHGRI histone database list more variants than what were observed in the LC-MS profiles presented herein. It is possible that not all the documented histone variants are observable under the current experimental conditions due to their low abundance. The gi numbers of all the human histones mentioned in this paper were indicated in Table 2 for H2A, and in supplementary Table 5-1 for linker histones and Table 5-2 for other core histones.
CONCLUSION
LC-MS appears to be an effective research and clinical tool for profiling histones. Seven linker histone H1 variants with multiple phosphorylations were observed without further enrichment. Thirteen of the possible H2A sequence variants were observed including the minor species H2AFZ and H2AFX. Six H2B variants were observed along with their acetylated isoforms. Three H3 variants were observed with multiple modifications. Histone H4 was also observed with the characteristic isoform distribution consistent with predominant dimethylation of K20 and acetylation of lysine residues on the N-terminal domain. All the methionine-oxidized species were observed at shorter retention times as separated by reverse-phase chromatography. The protein identification and confidence level was confirmed by RPLC fractionation, AU-PAGE and nano- LC-MS/MS on H2A species in RPLC fraction 7.
The current LC-MS analysis of intact proteins significantly reduces the time required for sample preparation and simplifies data interpretation. It minimizes the possibility of introducing putative modifications during sample preparation. The complete characterization of all the histones in a single analysis allows for comparisons between normal and disease specimens.
Supplementary Material
ACKNOWLEDGMENTS
Acknowledgements are given to the funding support of the Ohio State University and the Camille and Henry Dreyfus Foundation. The authors are recipients of a V Foundation/American Association for Cancer Research Translational Cancer Research Grant and are also supported by National Institutes of Health R21 CA110496. JCB is supported by the Leukemia & Lymphoma Society and the D. Warren Brown foundation. RF is supported by National Institute of Health grants CA56542, CA67007, GM63556, and the Milo Gladstein Bloom’s Syndrome Foundation. The authors wish to thank Richard Sessler for technical assistance and Nanette Kleinholz at Campus Chemical Instrumentation Center for assistance with the LC-MS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Henry KW, Berger SL. Nat. Struct. Biol. 2002;9:565. doi: 10.1038/nsb0802-565. [DOI] [PubMed] [Google Scholar]
- 2.Freitas MA, Sklenar AR, Parthun MR. J. Cell. Biochem. 2004;92:691. doi: 10.1002/jcb.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan S, Sink DW, Trout KL, Makalowska I, Taylor PM, Baxevanis AD, Landsman D. Nucleic Acids Res. 2002;30:341. doi: 10.1093/nar/30.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. J. Biol. Chem. 2002;277:29496. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- 5.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Mol. Cell. 2000;5:917. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 6.Davie JK, Edmondson DG, Coco CB, Dent SY. J. Biol. Chem. 2003;278:50158. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- 7.Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. J. Cell. Sci. 2005;118:2247. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- 8.Millard CB, M. A., Meier HL, A. R., Broomfield CA. Biochim Biophys Acta. 1994;1224:389. doi: 10.1016/0167-4889(94)90273-9. [DOI] [PubMed] [Google Scholar]
- 9.Kuster B, Mann M. Curr. Opin. Struct. Biol. 1998;8:393. doi: 10.1016/s0959-440x(98)80075-4. [DOI] [PubMed] [Google Scholar]
- 10.Burlingame AL, Zhang X, Chalkley RJ. Methods. 2005;36:383. doi: 10.1016/j.ymeth.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Adams GW, Mayer P, Medzihradszky KF, Burlingame AL. Methods Enzymol. 1995;258:90. doi: 10.1016/0076-6879(95)58039-5. [DOI] [PubMed] [Google Scholar]
- 12.Galasinski SC, Louie DF, Gloor KK, Resing KA, Ahn NG. J. Biol. Chem. 2002;277:2579. doi: 10.1074/jbc.M107894200. [DOI] [PubMed] [Google Scholar]
- 13.Millard CB, Meier HL, Broomfield CA. Biochim. Biophys. Acta. 1994;1224:389. doi: 10.1016/0167-4889(94)90273-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Chromosoma. 2003;112:77. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Freitas MA, Wickham J, Parthun MR, Klisovic MI, Marcucci G, Byrd JC. J. Am. Soc. Mass Spectrom. 2004;15:77. doi: 10.1016/j.jasms.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Mol. Cell. Proteomics. 2005;5:541. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K, Tang H. J. Chromatogr. B. 2003;783:173. doi: 10.1016/s1570-0232(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 18.Lindner H, Sarg B, Helliger W. J. Chromatogr. A. 1997;782:55. doi: 10.1016/s0021-9673(97)00468-8. [DOI] [PubMed] [Google Scholar]
- 19.Lindner H, Sarg B, Meraner C, Helliger W. J. Chromatogr. A. 1996;743:137. doi: 10.1016/0021-9673(96)00131-8. [DOI] [PubMed] [Google Scholar]
- 20.Thomas CE, Kelleher NL, Mizzen CA. J. Proteome Res. 2006;5:240. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 21.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. J. Proteome Res. 2006;5:233. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 22.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. J. Proteome. Res. 2006;5:248. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 23.Roychowdhury S, Baiocchi RA, Vourganti S, Bhatt D, Blaser BW, Freud AG, Chou J, Chen CS, Xiao JJ, Parthun M, Chan KK, Eisenbeis CF, Ferketich AK, Grever MR, Caligiuri MA. J Natl Cancer Inst. 2004;96:1447. doi: 10.1093/jnci/djh271. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Shen T, Huynh L, Klisovic MI, Rush LJ, Ford JL, Yu J, Becknell B, Li Y, Liu C, Vukosavljevic T, Whitman SP, Chang KS, Byrd JC, Perrotti D, Plass C, Marcucci G. Cancer Res. 2005;65:1277. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 25.Sures I, Gallwitz D. Biochemistry. 1980;19:943. doi: 10.1021/bi00546a019. [DOI] [PubMed] [Google Scholar]
- 26.Simon RH, Felsenfeld G. Nucleic Acids Res. 1979;6:689. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren C, Ghoshal K, Zhang L, Parthun MR, Jacob ST, Freitas MA. J Am Soc Mass Spectrom. 2005;16:1641. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas O, Farenc C, Calas M, Vial HJ, Bressolle F. Clin. Chem. 2005;51:593. doi: 10.1373/clinchem.2004.042580. [DOI] [PubMed] [Google Scholar]
- 29.Argirov OK, Lin B, Ortwerth BJ. J. Biol. Chem. 2004;279:6487. doi: 10.1074/jbc.M309090200. [DOI] [PubMed] [Google Scholar]
- 30.Karpova MB, Schoumans J, Ernberg I, Henter JI, Nordenskjold M, Fadeel B. Leukemia. 2005;19:159. doi: 10.1038/sj.leu.2403534. [DOI] [PubMed] [Google Scholar]
- 31.Drexler HG, Minowada J. Leuk. Lymphoma. 1998;31:305. doi: 10.3109/10428199809059223. [DOI] [PubMed] [Google Scholar]
- 32.Canavan TP, Doshi NR. Am. Fam. Physician. 2000;61:1369. [PubMed] [Google Scholar]
- 33.Fujitaki JM, Fung G, Oh EY, Smith RA. Biochemistry. 1981;20:3658. doi: 10.1021/bi00515a055. [DOI] [PubMed] [Google Scholar]
- 34.Ausio J, Abbott DW, Wang X, Moore SC. Biochem. Cell. Biol. 2001;79:693. [PubMed] [Google Scholar]
- 35.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Curr. Opin. Genet. Dev. 2002;12:162. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 36.Goll MG, Bestor TH. Genes Dev. 2002;16:1739. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 37.Turner BM. Cell. 2002;111:285. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 38.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Cell. 2005;120:25. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 39.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Mol. Cell. 2005;20:601. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Cao R, Tsukada YI, Zhang Y. Mol. Cell. 2005;20:845. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. J. Biol. Chem. 2006;281:9287. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 42.Franklin SG, Zweidler A. Nature. 1977;266:273. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 43.Pusarla RH, Bhargava P. Febs J. 2005;272:5149. doi: 10.1111/j.1742-4658.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- 44.Ahmad K, Henikoff S. Proc. Natl. Acad. Sci. USA. 2002;99(Suppl 4):16477. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindner H, Helliger W, Puschendorf B. J. Chromatogr. 1988;450:309. doi: 10.1016/s0021-9673(01)83585-8. [DOI] [PubMed] [Google Scholar]
- 46.Camerini-Otero RD, Felsenfeld G. Proc Natl Acad Sci U S A. 1977;74:5519. doi: 10.1073/pnas.74.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. J Am Soc Mass Spectrom. 2005;16:1641. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K, Williams KE, Huang L, Yau P, Siino JS, Bradbury EM, Jones PR, Minch MJ, Burlingame AL. Mol. Cell. Proteomics. 2002;1:500. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 49.Sarg B, Helliger W, Talasz H, Koutzamani E, Lindner HH. J. Biol. Chem. 2004;279:53458. doi: 10.1074/jbc.M409099200. [DOI] [PubMed] [Google Scholar]
- 50.Talasz H, Lindner HH, Sarg B, Helliger W. J. Biol. Chem. 2005;280:38814. doi: 10.1074/jbc.M505563200. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Science. 2004;306:279. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 52.Shechter Y. J. Biol. Chem. 1986;261:66. [PubMed] [Google Scholar]
- 53.Brot N, Weissbach H. Arch. Biochem. Biophys. 1983;223:271. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 54.Brot N, Weissbach H. Biofactors. 1991;3:91. [PubMed] [Google Scholar]
- 55.Brot N, Weissbach L, Werth J, Weissbach H. Proc. Natl. Acad. Sci. USA. 1981;78:2155. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anantharamaiah GM, Hughes TA, Iqbal M, Gawish A, Neame PJ, Medley MF, Segrest JP. J. Lipid Res. 1988;29:309. [PubMed] [Google Scholar]
- 57.Linder C, Thoma F. Mol. Cell. Biol. 1994;14:2822. doi: 10.1128/mcb.14.4.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zlatanova J, Van Holde K. J. Cell. Sci. 1992;103(Pt 4):889. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]
- 59.Mizzen CA, Dou Y, Liu Y, Cook RG, Gorovsky MA, Allis CD. J. Biol. Chem. 1999;274:14533. doi: 10.1074/jbc.274.21.14533. [DOI] [PubMed] [Google Scholar]
- 60.Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA. Mol. Cell. 1999;4:641. doi: 10.1016/s1097-2765(00)80215-4. [DOI] [PubMed] [Google Scholar]
- 61.Garcia BA, Busby SA, Barber CM, Shabanowitz J, Allis CD, Hunt DF. J. Proteome Res. 2004;3:1219. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Freitas MA, Wickham J, Parthun MR, Klisovic MI, Marcucci G, Byrd JC. J Am Soc Mass Spectrom. 2004;15:77. doi: 10.1016/j.jasms.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Mol Cell Proteomics. 2005 doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Naldi M, Andrisano V, Fiori J, Calonghi N, Pagnotta E, Parolin C, Pieraccini G, Masotti L. J. Chromatogr. A. 2006 doi: 10.1016/j.chroma.2006.06.100. In print. [DOI] [PubMed] [Google Scholar]
- 65.Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Genomics. 1997;40:314. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 66.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Proc. Natl. Acad. Sci. USA. 2002;99:16899. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halleck A, Ebert L, Mkoundinya M, Schick M, Eisenstein S, Neubert P, Kstrang K, Schatten R, Shen B, Henze S, Mar W, Korn B, Zuo D, Hu Y, LaBaer J. RZPD Deutsches Ressourcenzentrum fuer Genomforschung GmbH, Im Neuenheimer Feld 580. 2004. D-69120 Heidelberg, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.