Abstract
Recent studies indicate that two clusters of single nucleotide polymorphisms in the neuronal sortilin-related receptor gene (SORL1) are causally associated with late-onset Alzheimer's disease (AD). At the cellular level, SORL1 is thought to be involved in intracellular trafficking of amyloid precursor protein. When this gene is suppressed, toxic amyloid β production is increased, and high levels of amyloid β are associated with a higher AD risk. Extending the cellular findings, gene expression studies show that SORL1 is differentially expressed in AD patients compared with controls. Furthermore, several genetic studies have identified allelic and haplotypic SORL1 variants associated with late-onset AD, and these variants confer small to modest risk of AD. Taken together, the evidence for SORL1 as a causative gene is compelling. However, putative variants have not yet been identified. Further research is necessary to determine its utility as a diagnostic marker of AD or as a target for new therapeutic approaches.
Introduction
The genetics of Alzheimer's disease (AD) have been explained by four susceptibility genes: amyloid precursor protein (APP), presenilin 1 (PSEN1), presenilin 2 (PSEN2), and apolipoprotein E (APOE) [1•,2•]. Recent reports have implicated the neuronal sortilin-related receptor gene (SORL1, also known as SORLA and LR11) as a susceptibility gene for late-onset AD [3,4•,5,6,7••,8•,9]. It is located on chromosome 11q23.2-q24.2 and encodes a 250-kD membrane protein expressed in neurons of the central and peripheral nervous system [10]. It is known to be involved in intracellular trafficking between the membrane and intracellular organelles, interacting with APP in endosomes and the trans-Golgi network (TGN) in both in vitro and in vivo experiments [11]. The current data suggest that underexpression of SORL1 leads to overexpression of amyloid β (Aβ), which has been associated with a higher risk of developing AD [10,12]. In this article, we briefly discuss the molecular mechanism underlying AD to explain how SORL1 may be involved in the disease process, review issues related to the genetics of common disease, and evaluate and summarize the relation between SOLR1 and AD.
Molecular Mechanisms Underlying AD
The understanding of the molecular mechanisms underlying AD began in the early 1980s with the isolation of Aβ [13,14] and the identification of the amyloid precursor protein [15–18]. Subsequently, linkage analysis studies revealed that mutations in APP can cause either early-onset AD [19] or the Dutch-type hereditary cerebrovascular amyloidosis [20]. Furthermore, missense mutations within or adjacent to the Aβ domain of APP [20] can initiate abnormalities in APP processing and the accumulation of Aβ peptide, leading to AD [21–23]. In 1995, additional mutations in PSEN1 and PSEN2 were identified [24–26]. Mutations in PSEN1 and PSEN2 modify APP processing by producing excess Aβ42, which is toxic to neurons [27–30]. In 1999, the β-site APP cleaving enzyme (BACE)—a transmembrane protease that governs the first enzymatic step in APP processing—was isolated [31,32].
The amyloid pathway involves two enzymatic steps. In the first β-cleavage step, BACE cleaves APP near the N terminus of the Aβ peptide. In the second β-cleavage step, the membrane-bound C-terminal APP fragment is cleaved by β-secretase, a complex composed of transmembrane proteins PSEN 1 and 2, nicastrin, APH1, TMP21, and PEN2 [33]. This mechanism is sufficient to explain the Aβ accumulation observed in early-onset AD. However, these molecular defects do not exist in late-onset AD, and thus cannot explain the accumulation of Aβ40 and Aβ42 in this form of the disease. However, one can posit that defects in genes involved in the trafficking mechanism for proteins can potentially cause excess accumulation of proteins such as Aβ40 and Aβ42, which in turn facilitate neuronal degeneration.
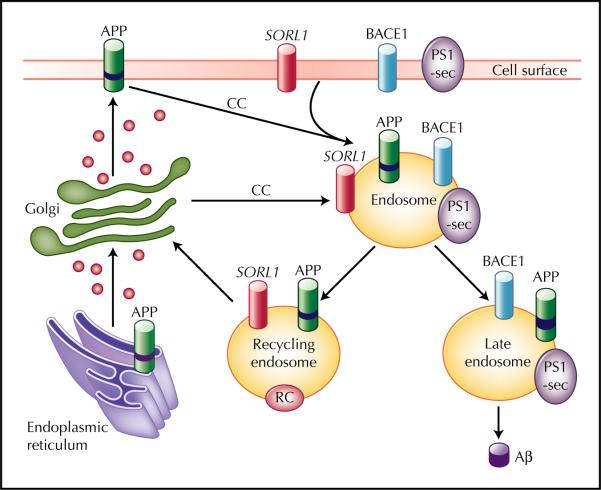
During the past two decades, the TGN and the endosome were identified as the key organelles organizing the complex movement of the transmembrane proteins via secretory and endocytic pathways (Fig. 1). Important coat complexes initiating the transport of APP and BACE through this sorting itinerary are the clathrin coat and the retromer [34,35,36•]. Clathrin coats are involved in the endocytic pathway connecting the cell surface to the endosome and the pathway connecting the TGN to the endosome [37]. The retromer is involved in the trafficking from the endosome to the TGN. Through the retromer complex, SORL1 directly binds APP and differentially regulates its sorting into endocytic or recycling pathways. In the absence of SORL1, APP is released into late endosomal pathways, where it is subjected to both β-secretase cleavage and γ-secretase cleavage, which eventually leads to Aβ production. Therefore, it is hypothesized that genetic variants in SORL might influence Aβ processing.
Figure 1.
Transmembrane sorting of sortilin-related receptor gene (SORL1) through the clathrin coats (CC) and the retromer complex (RC). Aβ—amyloid β; APP—amyloid precursor protein; BACE—β-site APP cleaving enzyme; PS—presenilin. (Adapted from Rogaeva et al. [7••].)
Challenges Facing Gene Mapping in Late-Onset AD
Following the identification of three genes for familial forms of early-onset AD and APOE for late-onset AD, progress on the identification of putative genetic variants has been limited. Unlike Mendelian or single gene disorders, for which over 1800 genes have been characterized [38], confirmed putative genes for common diseases are rare. Several reasons explain the difficulties. AD is a multifactorial disorder influenced by multiple genetic and environmental risk factors in which these factors interact to produce the phenotype (ie, gene-gene and gene-environment interactions). Additional features complicating gene identification include reduced penetrance (absence of clinical disease in individuals harboring a mutation/polymorphism), phenotypic heterogeneity (definition of being affected can be so broad that it is actually a collection of several diseases), and locus and allelic heterogeneity (disease in different families or in different individuals within a family is caused by different genetic variants). For these reasons, replication of the initial gene finding in common diseases is rarely observed.
To overcome these challenges, an array of approaches is available to uncover genetic risk factors in Mendelian and complex traits. Two complementary analytical methods, linkage analysis and association analysis, are used to detect the specific genetic variants that are involved in the disease process. Linkage analysis tests whether genetic markers cosegregate with the disease within a family. The closer the genetic marker is to the disease locus, the fewer the number of recombination events between the two loci. As a result, the disease will cosegregate with the marker. However, genetic association analysis examines whether affected individuals share the same allele more often than expected. This observed deviation can come from a deviation from the expected transmission probability of 50%, as in the family-based association method, or from deviations from expected allele frequencies obtained from unaffected controls, as in the case-control method. When the association between the disease phenotype and a marker is statistically significant, one may conclude that the marker or adjacent chromosome region may be associated with the disease. Recently, statistical methods have evolved to employ linkage and association analysis jointly to enhance power [39,40]. These advances in statistical methods, coupled with the availability of 500,000 to 1 million single nucleotide polymorphisms (SNP) in microarray chips covering the human genome, have enhanced our capability to interrogate candidate chromosomal regions [41].
Beyond the technological advances, investigators are studying endophenotypes to enhance power to detect genes underlying common diseases [42•,43]. In general, an endophenotype (ie, an intermediate phenotype) can better characterize the relation between the gene and the disease locus because such a quantitative trait is in the biological pathway toward the disease. Because endophenotypes are situated in the downstream of the biological pathway (ie, closer to the genotype than to the phenotype), endophenotypes are presumably determined by fewer genes and can serve to reduce heterogeneity, thereby reducing the complexity of genetic analysis.
Moreover, investigations of endophenotypes can identify genes with small effects because they can assess the role of genetic factors on phenotypic variations using affected and unaffected family members, providing greater statistical power than discrete disease status. For these endophenotypes to be useful, they have to be reliably assessed, moderately to highly heritable, and their relationship to the biological processes hypothesized to underlie disease must be testable. Furthermore, coherent sets of multiple endophenotypes (eg, Aβ, memory, or structural or functional brain imaging measures) can bring about an even deeper understanding of the underlying biology.
Genetic Association Studies
Rogaeva et al. [7••] first reported the allelic and haplotypic associations between AD and variants in SORL1. Subsequently, several studies supported the initial finding by showing that genetic variants in SORL1 contribute toward AD [3,4•,5,6,8•,9], but a few studies did not confirm the initial finding (Table 1). The original study included four different ethnic groups: North American and European whites, Caribbean Hispanics, African Americans, and Israeli Arabs. This investigation on more than 6000 individuals identified two different sets of haplotypes: 1) SNPs in the 5’ end of the gene (SNPs 8−10; 120,873,131 bp to 120,886,175 bp) among Caribbean Hispanics (family study), whites (case-control study), and Israeli Arabs (case-control study); and 2) SNPs in the 3’ end of the gene (SNPs 22−25; 120,962,172 bp to 120,988,611 bp) among multiple white samples (family and case-control studies) and African Americans (family study). Haplotype analysis strengthened statistical support further. However, as observed in many common diseases, these candidate SNPs conferred a modestly elevated risk of AD, ranging from an odds ratio of 1.4 to 2.2, and the allelic association was not uniform across datasets. The authors strengthened their allelic association findings with cell biology findings, which showed that suppression of SORL1 led to elevation of Aβ levels. Three subsequent studies by the same group broadly supported one or both haplotypes or some variations of the two: haplotype C-G-C at SNPs 8 to 10, or haplotype T-T-C at SNPs 23 to 25, or both. Lee et al. [5] showed that the same set of SNPs at SNPs 23 to 25 were associated with AD in whites residing in northern Manhattan. In a follow-up study, they confirmed the allelic and haplotypic associations in autopsy-confirmed cases of white ethnicity for haplotype at SNPs 8 to 10 and haplotype at SNPs 23 to 25 [4•].
Table 1.
Reported allelic associations in different studies by ethnicity
| Study / year | Haplotype 1 | Haplotype 2 | Other significant SNPs | ||||
|---|---|---|---|---|---|---|---|
|
rs668387 SNP 8*† |
rs689021 SNP 9 |
rs641120 SNP 10 |
rs3824968 SNP 23 |
rs2282649 SNP 24 |
rs1010159 SNP 25 |
||
| Significant association | |||||||
| Rogaeva et al. [7••] / 2007 | |||||||
| Whites (family dataset) | T | T†‡ | C‡ | ||||
| Caribbean Hispanics | C | G | C | ||||
| Whites (case-control datasets) | C | G | C | T | T | C | |
| Israeli Arabs | C | G | C | ||||
| African Americans | |||||||
| Lee et al. [5] / 2007 | |||||||
| Whites | C | A | T | T | T | C | rs3824966 (SNP 20) |
| Hispanics | rs12285364 (SNP 12) | ||||||
| African Americans | C‡ | G‡ | T‡ | C | C | rs12285364, rs1784933 (SNP 26) | |
| Meng et al. [6] / 2007† | |||||||
| Whites | + | + | + | ||||
| Lee et al [5] / 2007 | |||||||
| Whites | C | G | C | A | T | C | |
| Tan et al. [9] / 2007 | |||||||
| Han Chinese | A | T‡ | |||||
| Seshadri et al. [8•] / 2007§ | |||||||
| Whites | + | rs1131497 (SNP29) | |||||
| Bettens et al. [3] / 2008 | |||||||
| Whites | C | G | C | rs560573 (SNP 6), rs1614735 (SNP 27) | |||
| Weak association | |||||||
| Webster et al. [46] / 2008† | |||||||
| Whites | + | + | |||||
| Li et al. [45] / 2008† | |||||||
| Whites | T‡ | rs2070045 (SNP 19) | |||||
| No association | |||||||
| Li et al. [44] / 2008¶ | |||||||
| Whites | |||||||
SNP numbers from Rogaeva et al. [7••] are presented. Alleles are presented only when significant.
Used the nearest SNPs (indicated with a "+" sign) when different SNPs were used.
Alleles are not statistically significant in either allelic, genotypic, or haplotypic analysis.
Endophenotypes were studied.
No specific marker information for SORL1 was available from the paper.
SNP—single nucleotide polymorphism.
Six other groups examined the relation between SORL1 and AD or related traits in different populations [3,8•,9,44–46]. Three replication studies [3,8•,9] supported the initial findings, but the remaining three [44–46] showed either negative or weak results. Bettens et al. [3] directly replicated SNPs 8 to 10 and showed support for SNPs 25 to 27 in 550 Belgian patients with late-onset AD and 637 unaffected individuals. Tan et al. [9] examined 223 cases and 263 controls from a Han Chinese population to show that haplotype G-C-A at SNP 19−22−23 was associated with AD (odds ratio of 1.4; 95% CI, 1.04−1.7), but none of the haplotypes in SNP 8 to SNP 10 were associated. Using the Framingham community-based family samples, Seshadri et al. [8•] extended the existing studies of AD by examining cognitive performance in healthy elderly individuals without dementia and stroke. The authors showed that SORL1 was significantly associated with abstract reasoning ability as measured by the Similarity test (P = 3.2 × 10−6). This is a logical extension of earlier studies, because this approach is particularly powerful in detecting genetic loci that contribute to small or modest changes in cognitive functions at the preclinical stage.
The remaining three studies show little support. Webster et al. [46] used the clinical and autopsy cases and controls compiled by Reiman et al. [47] at the Translational Genomics Research Institute to examine SORL1. Based on 644 clinical and autopsy cases versus 422 controls, they observed a weak association with four SNPs (nominal P values of 0.019−0.038) that are located between SNP 8 and SNP 10 in the original paper, but they did not observe any association in the haplotype in the 3’ end of the gene (SNPs 22−25). The authors concluded that there is weak evidence for association. Interestingly, Meng et al. [6] evaluated SORL1 using the same dataset but included a somewhat larger set of patients (n = 1408). They concluded that four SNPs located between SNPs 20 and 25 in the original paper were associated with AD. The reason for the difference in the findings between the two studies is likely to be due to the fact that Meng et al. [6] included 342 additional samples.
Li et al. [44] observed no association with SORL1 in their two-stage genome-wide association study. They first examined 753 cases and 736 controls in Canadian samples, and then further examined the top 120 candidate SNPs using 418 cases and 249 controls from a United Kingdom (UK) Medical Research Council dataset. They had 48 SNPs on SORL1 but did not observe any association. However, as Rogaeva et al. [7••] reported, Li et al. [44] did observe a weak association with two SNPs in SORCS1, a gene in the sortilin pathway. Based on their report, however, it is unclear as to how many SNPs from the original paper were studied and how dense the SNP coverage was. In a separate study, Li et al. [45] examined three sets of cases and controls totaling approximately 2000 samples from either the UK or the United States. Only a weak association was observed for two SNPs: rs2070045 (SNP 19; P = 0.035) and rs2282649 (SNP 24; P = 0.022 for UK1 dataset). However, no association was observed for previously reported haplotypes when all three datasets were combined. On a closer examination, the two SNPs were weakly associated with AD in all datasets except for one UK dataset (UK2). Moreover, the associations for different SNPs for the UK2 dataset differed from those for the other two datasets (UK1 and WU), suggesting that there may be cryptic sampling heterogeneity or that simply none of the SNPs are associated with AD in these datasets. It may be of interest to examine these datasets while accounting for population stratification.
Although these allelic association studies support the association between AD and the implicated alleles and haplotypes, only one study examined the role of SORL1 in cognitive function. There has been no report of the relation between SORL1 and other neurodegenerative disorders (eg, dementia with Lewy bodies or Parkinson's disease).
Evidence for Genetic Involvement from Microarray Expression Studies
Gene expression profiling experiments measure the activity of thousands of genes at once, creating a global picture of cellular function. These studies measure the relative activity of thousands of mRNA transcripts, thereby providing a powerful tool for uncovering pathogenic genes underlying common diseases. However, as with the genetic association studies, molecular heterogeneity makes the matter of gene identification a bit more complicated, because defects in different molecular pathways can produce the same disease phenotypes. Consequently, it is difficult to identify one specific gene that underlies the phenotype of interest among many differentially expressed genes. To differentiate signals from noises, some have focused on the tissues most affected by the disease process. To this end, some researchers employ brain imaging techniques to help identify regions of physiologic dysfunction [36•]. In addition, they use unaffected regions within the same brain to reduce signal noises resulting from interindividual differences.
The first clue about which type-I transmembrane proteins might be sorted by the neuronal retromer came from studies exploring microarray data generated from human brain tissue [36•,48]. Small et al. [36•,48] demonstrated that among a list of possible retromer cargo molecules, SORL1 and BACE were the type-I transmembrane molecules whose expression levels cross-correlated most strongly with levels of neuronal VPS35. VPS35 is the core molecule of the retromer complex.
Scherzer et al. [49] compared the gene expression patterns of lymphocytes of AD patients against age-, sex-, and ethnicity-matched controls and found that the fluency intensity ratio (fold change) for sporadic AD patients was significantly higher (1.8-fold) than in controls. Six genes (SORLA/LR11, IFNGR1, STAF50, Pleckstrin, Amylo-[1,4−1,6]-transglycosylase, and Homo sapiens SNC73 mRNA) were differentially expressed in lymphoblasts of two independent groups of patients with probable and autopsy-confirmed late-stage AD. Immunohistochemistry of 13 AD and 7 control brains reconfirmed a reduction of SORL1 expression in histologically normal neurons in AD brains, including neurons in frontal cortex and hippocampus, compared with control brains. One caveat of studies based on lymphoblasts, rather than affected brain tissue, is that such studies cannot answer 1) whether the differential expression of SORL1 in lymphoblasts and immunohistochemistry is in fact specific to AD; and 2) whether it is a cause or consequence of the disease. Thus, mRNA profiling of the physiologically affected brain tissue against a control region over different stages of the disease would be necessary to settle this question.
Interaction of SORL1 with Other Known Genetic Risk Factors
To date, only one study has explored whether known AD genes interact with SORL1 or not. Dodson et al. [50] compared immunohistochemistry and immunoblotting of SORL1 in PS1/APP transgenic and wild-type mice as well as in human frontal cortex of nondemented controls, sporadic AD cases, and familial AD cases. In the PS1/APP transgenic mice model, SORL1 levels were not affected by genotype or accumulation of amyloid pathology. Consistent with this finding, SORL1 immunostaining intensity in human frontal cortex was reduced in sporadic AD cases, but similar between controls and familial AD cases. These findings suggest that the effect of SORL1 is independent of PSEN1 and PSEN2 genotype. In addition, Rogaeva et al. [7••] observed that two genes in the sortilin pathway, SORCS1 and SORSC2, were weakly associated with AD. Naturally, further studies are needed to clarify these complex relations.
Role of SORL1 in Genetic Medicine
Several lines of evidence strongly support that SORL1 plays an important role in AD pathogenesis. Theoretically, SORL1 can be developed as part of a genetic profiling tool for AD, and as a potential novel therapeutic target for treatment of late-onset AD. However, its current utility in medicine is unclear. Several issues must be resolved before SORL1 can be useful in clinical settings. First, to use SORL1 as a genetic risk–profiling tool, the precise putative genetic variants for SORL1 have to be identified and, at the same time, additional risk factor genes need to be known. For common late-onset AD, accurate overall risk can only be estimated when we better understand the putative and protective genetic variants involved, as well as how they interact among them. Without such information, the risk estimates will be inaccurate and will cause a greater harm than good. Second, it is necessary to clarify the exact mechanisms of intracellular transport and processing through which SORL1 acts on APP protein trafficking. Identification of the mechanisms underlying APP sorting will help us to understand the role of SORL1 in the amyloid cascade and could provide targets for effective intervention. Finally, it is necessary to further characterize molecular pathways involving SORL1. Only when these issues are better understood can the medical applications involving SORL1 be devised.
Conclusions
We have shown that SORL1 is involved in Aβ production and that at least one of the two clusters of SNP variants in SORL1 is associated with AD in many, but not all, cases. However, putative variants have not yet been identified. Thus, deep sequencing is needed to identify the putative variant(s). Once identified, it will be necessary to better characterize their impact on phenotypic outcomes, including AD, as well as on endophenotypes (eg, Aβ, cognitive performance, imaging data). Furthermore, it will be biologically insightful to examine the relations among genes in the sortilin pathway. We are currently pursuing these possibilities.
Acknowledgments
Funding for this project was provided by the National Institutes on Aging, National Institutes of Health (R37 AG15473, P50-AG08702, P01-AG07232). In addition, support was provided by the Charles S. Robertson Gift for Research on Alzheimer's Disease from the Banbury fund. We also thank the members of Estudio Familiar de Influencia Genetica en Alzheimer, The Sociedad Dominicana de Geriatria y Gerontologia, The Sociedad Dominicana de Neurologia y Neurocirugia, The Sociedad Dominicana de Psiquiatria and the Associacion Dominicana Alzheimer y Similares, Inc, and The Bioethics National Committee for Research in the Dominican Republic.
Footnotes
Disclosures
No potential conflicts of interest relevant to this article were reported.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1•.Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [• Of importanceThis paper reviews the current state of knowledge for neuropsychiatric disorders] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.St George-Hyslop PH, Petit A. Molecular biology and genetics of Alzheimer's disease. Curr Rev Biol. 2005;328:119–130. doi: 10.1016/j.crvi.2004.10.013. [• Of importanceThis paper reviews the molecular biology and genetics of AD] [DOI] [PubMed] [Google Scholar]
- 3.Bettens K, Brouwers N, Engelborghs S, et al. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 4•.Lee JH, Cheng R, Honig LS, et al. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70:887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [• Of importanceThis study first reported the relation between SORL1 and autopsy-confirmed AD cases] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Cheng R, Schupf N, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng Y, Lee JH, Cheng R, et al. Association between SORL1 and Alzheimer's disease in a genome-wide study. Neuroreport. 2007;18:1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [•• Of major importanceThis study first reported the association between SORL1 and AD] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Seshadri S, DeStefano AL, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [• Of importanceThis paper examined the relation between SORL1 and endophenotypes, rather than AD itself] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan EK, Lee J, Chen CP, et al. SORL1 haplotypes modulate risk of Alzheimer's disease in Chinese. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.10.013. in press. [DOI] [PubMed] [Google Scholar]
- 10.Andersen OM, Reiche J, Schmidt V, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 12.Offe K, Dodson SE, Shoemaker JT, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 14.Masters CL, Simms G, Weinman NA, et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgaber D, Lerman MI, McBride OW, et al. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer's disease. Science. 1987;235:877–880. doi: 10.1126/science.3810169. [DOI] [PubMed] [Google Scholar]
- 16.Kang J, Lemaire HG, Unterbeck A, et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 17.Robakis NK, Ramakrishna N, Wolfe G, et al. Molecular cloning and characterization of a cDNA encoding the cerebrovascular and the neuritic plaque amyloid peptides. Proc Natl Acad Sci U S A. 1987;84:4190–4194. doi: 10.1073/pnas.84.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanzi RE, Gusella JF, Watkins PC, et al. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235:880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 19.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 20.Levy E, Carman MD, Fernandez-Madrid IJ, et al. Mutation of the Alzheimer's disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 21.Haass C, Selkoe DJ. Cellular processing of beta-amyloid precursor protein and the genesis of amyloid beta-peptide. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 22.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 23.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Levy-Lahad E, Wijsman EM, Nemens E, et al. A familial Alzheimer's disease locus on chromosome 1. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 25.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 26.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 27.Duff K, Eckman C, Zehr C, et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 28.Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 29.Burdick D, Soreghan B, Kwon M, et al. Assembly and aggregation properties of synthetic Alzheimer's A4/beta amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 30.Hilbich C, Kisters-Woike B, Reed J, et al. Aggregation and secondary structure of synthetic amyloid beta A4 peptides of Alzheimer's disease. J Mol Biol. 1991;218:149–163. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 31.Sinha S, Anderson JP, Barbour R, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 32.Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 33.Edbauer D, Winkler E, Regula JT, et al. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 34.Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 35.He X, Li F, Chang WP, et al. GGA proteins mediate the recycling pathway of memapsin 2 (BACE). J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- 36•.Small SA, Kent K, Pierce A, et al. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [• Of importanceThis paper provides an extensive review describing the sorting of SORL1 and other transmembrane proteins through the retromer complex and clathrin coats in AD] [DOI] [PubMed] [Google Scholar]
- 37.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Brinkman RR, Dube MP, Rouleau GA, et al. Human monogenic disorders—a source of novel drug targets. Nat Rev Genet. 2006;7:249–260. doi: 10.1038/nrg1828. [DOI] [PubMed] [Google Scholar]
- 39.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet. 2000;66:1310–1327. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Boehnke M, Abecasis GR. Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet. 2005;76:934–949. doi: 10.1086/430277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q, Biernacka JM, Chen MH, et al. Using linkage and association to identify and model genetic effects: summary of GAW15 Group 4. Genet Epidemiol. 2007;31(Suppl 1):S34–S42. doi: 10.1002/gepi.20278. [DOI] [PubMed] [Google Scholar]
- 42•.Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006;22:306–313. doi: 10.1016/j.tig.2006.04.004. [• Of importanceThis article provides guidelines for using endophenotypes in psychiatric genetic research] [DOI] [PubMed] [Google Scholar]
- 43.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Wetten S, Li L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Rowland C, Catanese J, et al. SORL1 variants and risk of late-onset Alzheimer's disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster JA, Myers AJ, Pearson JV, et al. SORL1 as an Alzheimer's disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 47.Reiman EM, Webster JA, Myers AJ, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small SA, Peirce AL, Kent K, et al. Combining functional imaging with microarray; identifying an unexplored cellular pathway implicated in sporadic Alzheimer's disease.. Paper presented at Society for Neuroscience meeting.; New Orleans, LA. Nov, 2003. pp. 8–12. [Google Scholar]
- 49.Scherzer CR, Offe K, Gearing M, et al. Loss of apolipo-protein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 50.Dodson SE, Gearing M, Lippa CF, et al. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]