Abstract
The molecular complexity of mammalian proteomes demands new methods for mapping the organization of multiprotein complexes. Here, we combine mouse genetics and proteomics to characterize synapse protein complexes and interaction networks. New tandem affinity purification (TAP) tags were fused to the carboxyl terminus of PSD-95 using gene targeting in mice. Homozygous mice showed no detectable abnormalities in PSD-95 expression, subcellular localization or synaptic electrophysiological function. Analysis of multiprotein complexes purified under native conditions by mass spectrometry defined known and new interactors: 118 proteins comprising crucial functional components of synapses, including glutamate receptors, K+ channels, scaffolding and signaling proteins, were recovered. Network clustering of protein interactions generated five connected clusters, with two clusters containing all the major ionotropic glutamate receptors and one cluster with voltage-dependent K+ channels. Annotation of clusters with human disease associations revealed that multiple disorders map to the network, with a significant correlation of schizophrenia within the glutamate receptor clusters. This targeted TAP tagging strategy is generally applicable to mammalian proteomics and systems biology approaches to disease.
Keywords: gene targeting, postsynaptic complexes, postsynaptic density-95, schizophrenia, tandem affinity purification
Introduction
Synapses are fundamental structural and functional units of the nervous system responsible for information processing. Their principal role is the transmission of electrical activity by the release of neurotransmitter from the presynaptic terminal onto postsynaptic receptors and ion channels. Postsynaptic ionotropic receptors initiate the postsynaptic depolarization that elicits action potential generation in the postsynaptic neuron. The second major role is the detection and processing of information contained in the patterns of electrical activity. This is achieved by the coupling of neurotransmitter receptors to second-messenger signaling pathways that modulate downstream effectors, ranging from modulation of ion channels themselves to structural changes and gene expression.
In recent years, proteomic studies have revealed that mammalian synapses comprise up to 2000 proteins in the presynaptic and postsynaptic terminals (Husi et al, 2000; Walikonis et al, 2000; Husi and Grant, 2001; Sheng and Kim, 2002; Peng et al, 2004; Takamori et al, 2006; Trinidad et al, 2008). To understand the macromolecular organization of complexes and substructures, isolation of complexes by antibody, peptide and ligand affinity methods was used to recover smaller sets of proteins (Husi et al, 2000; Farr et al, 2004; Collins et al, 2006; Dosemeci et al, 2007; Klemmer et al, 2009; Paulo et al, 2009). These methods generally involve a single purification step, which is limited by the specificity of the affinity reagent and potentially recovers more contaminants than those with multiple steps. Furthermore, these protocols are not generally suitable for recovery of native complexes in solution, which could be used for enzymatic and structural studies.
A potential solution to this major problem has been achieved in yeast through genetic modification of the endogenous protein by fusion with a Tandem Affinity Purification (TAP) tag into the C- or N-terminus of the protein of interest (Rigaut et al, 1999). This tagged protein can be isolated (with its associated proteins) in a tandem procedure, overcoming many of the inherent specificity and sensitivity limitations of traditional fractionation methods, as well as antibody, ligand and peptide affinity purification methods. In mammalian tissues, where developmental and cell-type control of regulation is more complex, the targeting of the TAP tag into the endogenous gene provides advantages over transgenic random integration or cDNA overexpression systems (Knuesel et al, 2003; Bouwmeester et al, 2004; Brajenovic et al, 2004; Drakas et al, 2005; Wang et al, 2005, 2006; Angrand et al, 2006; Burckstummer et al, 2006). For those reasons we chose to explore TAP tagging in mammals using gene targeting in mouse.
Our first aim was to test TAP tagging using a gene-targeting approach in mice, with the specific objective of purifying signaling complexes from the synapse. We generated knockin mice in which TAP tags were inserted into the endogenous locus of post synaptic density-95 (PSD-95), which is one of the most abundant scaffold proteins at excitatory brain synapses (Nourry et al, 2003; Peng et al, 2004). PSD-95 is localized to the postsynaptic compartment in which it interacts with neurotransmitter receptors and ion channels to assemble signaling complexes (Kornau et al, 1995; Hunt et al, 1996; Tu et al, 1999; Husi et al, 2000; Nehring et al, 2000; Dosemeci et al, 2007) controlling neuronal plasticity (Migaud et al, 1998; Carlisle et al, 2008; Cuthbert et al, 2007) underlying learning and memory (Migaud et al, 1998), pain (Garry et al, 2003) and drug addiction (Yao et al, 2004). Our second aim was to integrate TAP tag proteomic data with systems biology approaches to analyze the organization and function of complexes.
We show the first example of gene-targeted TAP tagging in mice and show that the tagging did not introduce a mutation or alter the expression or localization of the protein. Clear advantages of two-step purification methods over the existing single-step methods were found. Mass spectrometry analysis of four replicates of the purification revealed that PSD-95-associated complexes comprise the principal ionotropic glutamate receptors and K+ channels in addition to important signaling proteins. Text mining and systematic annotation together with clustering of proteins using protein interaction data revealed the network substructure with a core subnetwork involved in schizophrenia.
Results
A strategy for purification of in vivo multiprotein complexes
We used a TAP tag consisting of a poly-histidine affinity tag (HAT) and a triple FLAG tag (Terpe, 2003) in tandem, separated by a unique TEV-protease cleavage site (Figure 1A). This 5-kDa tag is considerably smaller than the tag first applied in yeast (20 kDa) (Rigaut et al, 1999) and exploits the specificity of both FLAG and HAT-tag binding. Targeting the endogenous gene allows a thorough testing of the potential mutant phenotype by breeding to homozygosity and comparing with the existing mutant mice.
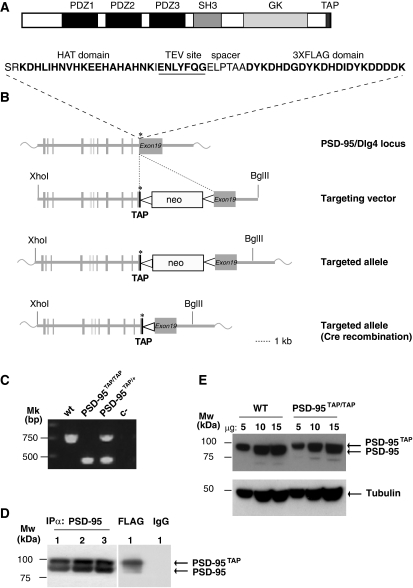
Figure 1.
Generation of Tandem Affinity Purification (TAP)-tagged PSD-95 knockin mice. (A) Domain structure of TAP modified PSD-95. PSD-95 domains, including three PDZ (PSD-95/discs large/zona occludens), a SH3 (Src homology 3), a GK (guanylate kinase) and C-terminal TAP-tag domain. Amino-acid sequence of the TAP tag comprising a histidine affinity tag (HAT)-domain (bold), a TEV site (underlined) and a 3XFLAG domain (bold) separated by a spacer. (B) Scheme of the targeted genomic PSD-95/Dlg4 locus. The Dlg4 allele was targeted with the TAP sequence inserted before the stop codon. Crossing the transgenic Cre-recombinase-expressing mice deleted the neomycin resistance cassette (neo) between loxP sites (bottom). Asterisk: stop codon of the coding sequence; black thick lane: TAP tag sequence; triangle: loxP site. (C) PCR genotyping of TAP-tagged PSD-95 mice, using a common forward primer PSD-95 F5 and two reverse primers PSD-95 R6 and pneoR4, which amplify the wild type (upper band) and targeted allele (lower band), respectively. (D) Immunoblot with PSD-95 antibody for immunoprecipitations. Three different heterozygous mice are shown (PSD-95TAP/+, left panel). PSD-95TAP/+ forebrain was also affinity purified with a FLAG antibody (right panel). (E) PSD-95 protein expression in wt and PSD-95TAP/TAP mouse forebrains. Brain lysates of 5, 10 and 15 μg were loaded and immunoblotted with antibodies against PSD-95 (upper panel) and tubulin (lower panel), which is a loading control. Wt, wild type; PSD-95TAP/TAP, homozygous TAP-tagged PSD-95 mice; c-, PCR water; IgG, mouse total IgG used as a negative control of the immunoprecipitation.
Generation of a TAP-tagged PSD-95 knockin mouse line
We chose to test TAP tagging in mice, with a focus on PSD-95 as a first model gene for the following reasons: (i) PSD-95 has discrete expression in the postsynaptic compartment of excitatory synapses of the brain, (ii) PSD-95 mutant mice are well characterized and show robust phenotypes in electrophysiological studies of synapses and behavior (Migaud et al, 1998; El-Husseini et al, 2000; Yao et al, 2004; Beique et al, 2006), and (iii) this protein has been extensively studied using methods that identify binary interaction partners (Kim and Sheng, 2004).
PSD-95 is a scaffold protein with three PDZ domains, an SH3 and a guanylate kinase domain that mediate protein interactions (Figure 1A). As mouse PSD-95 is known to have multiple isoforms generated by multiple promoters and all forms utilize a common C-terminus (Bence et al, 2005), the TAP tag was inserted into the open reading frame in the 3′-end before the stop codon of exon 19, using Escherichia coli recombineering-based methods (Zhou et al, 2004) (Figure 1B). The final targeting vector, containing a 5′-end homology arm of 6.3 kb and a 3′-end homology arm of 2.9 kb, was transfected into ES cells and integration was detected using standard methods. PCR of neomycin-resistant ES-cell DNA confirmed the expected 3388 bp band in 16 clones (targeting efficiency was 5.6%), and germline transmission of the TAP-tag insertion was established (Figure 1C). This line of mice is referred to herein as PSD-95TAP.
Normal expression and synaptic localization of TAP-tagged PSD-95
We first intercrossed PSD-95TAP heterozygous mice (PSD-95TAP/+) and found no distortion of transmission frequency in the offspring of PSD-95TAP/+ intercrosses (data not shown). We next examined protein expression of TAP-tagged PSD-95 to ensure that introduction of the tag into the gene did not affect the expression and localization of the tagged protein. The solubilization conditions used here have been reported as the best conditions to mostly purify N-methyl-D-aspartate (NMDA) receptors and PSD-95 from adult mouse brain (Husi and Grant, 2001). The forebrain tissue was solubilized from heterozygous (PSD-95TAP/+) mice and PSD-95 was immunoprecipitated and immunoblotted using an anti-PSD-95 antibody (Figure 1D). Two bands of similar intensity were observed, where the upper band corresponded to the TAP-tagged PSD-95 (confirmed by immunoprecipitation using anti-FLAG antibody, right panel) and the lower band to the endogenous PSD-95. Comparison of extracts (5, 10, 15 μg) from wild-type (wt) and homozygous (PSD-95TAP/TAP) mice showed similar amounts of PSD-95 on immunoblots compared with an internal control immunoblot using anti-tubulin antibody (Figure 1E).
We next carried out immunohistochemistry with an anti-PSD-95 antibody on sagittal brain sections to examine the expression pattern of TAP-tagged PSD-95. As shown in Figure 2A, the expression pattern of PSD-95 in PSD-95TAP/TAP brain was the same as in the brains of wt animals, with the highest expression in the CA1 area, dentate gyrus, cortex, cerebellum and lower expression in striatum and brainstem (Figure 2A). There was no detectable abnormality of brain morphology in the PSD-95TAP/TAP mice. As shown in Figure 2B, the expression of PSD-95 in the hippocampal subfields was unaffected by the genetic manipulation and particularly in the stratum radiatum, where the electrophysiological experiments were carried out (described below), was normal.
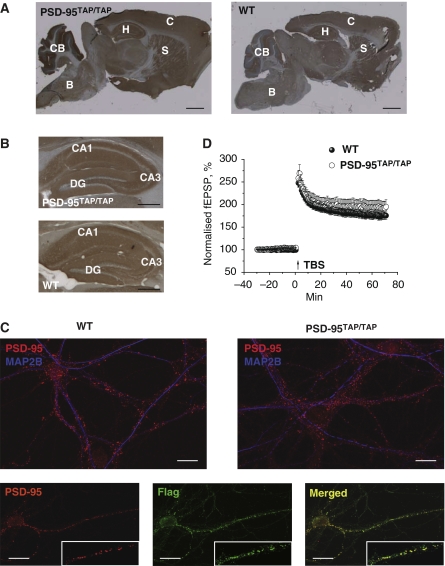
Figure 2.
Analysis of TAP-tagged PSD-95 localization and synaptic plasticity in PSD-95TAP/TAP mice. (A) Immunohistochemical staining of PSD-95 in sagittal brain sections from PSD-95TAP/TAP and wt mice. B, brainstem; C, cortex; CB, cerebellum; H, hippocampus; S, striatum. Scale bar=1 mm. (B) Immunohistochemical staining of PSD-95 in sagittal hippocampus sections from PSD-95TAP/TAP and wt mice showing CA1, CA3 and dentate gyrus (DG). Scale bar=1 mm. (C) Synaptic localization of TAP-tagged PSD-95 in primary hippocampus neurons. DIV14 neurons from wt and PSD-95TAP/TAP mice were stained with PSD-95 and MAP2B antibodies (top panels). Three lower panel show PSD-95 and FLAG antibody staining in a culture from PSD-95TAP/TAP mice (bottom panels). Inset panels show higher magnification of synaptic puncta labeling with each antibody and merged image. Scale bar=10 μm. (D) Long-term potentiation of fEPSPs induced by theta-burst stimulation in CA1 area of hippocampal slices is similar in PSD-95TAP/TAP (13 slices from 4 animals) and wild-type mice (15 slices from 4 animals).
To examine the synaptic localization of TAP-tagged PSD-95, we cultured embryonic hippocampal neurons from PSD-95TAP/TAP and wt mice. The specific subcellular localization of PSD-95 to the postsynaptic compartment of synapses (dendritic spines) was monitored using postsynaptic markers for glutamate neurotransmitter receptors (GluR1 or NR1), presynaptic marker (synaptophysin) and dendritic markers (MAP2) (Figure 2B and Supplementary Figure 1). Similar to PSD-95 staining in wt neurons, TAP-tagged PSD-95 was localized to punctate structures along the length of dendrites in PSD-95TAP/TAP neurons (Figure 2C, top panels). FLAG staining also shows the punctate structures in PSD-95TAP/TAP neurons (Figure 2C, bottom panels). Synaptophysin staining shows typical juxtaposition, indicating that TAP-tagged PSD-95 is found at synapses in PSD-95TAP/TAP neurons (Supplementary Figure 1, top panels). Furthermore, the co-localization of GluR1 and NR1 subunits with TAP-tagged PSD-95 (Supplementary Figure 1, middle and bottom panels, respectively) confirms its postsynaptic localization in the excitatory synapses, just as occurs for wt PSD-95.
The TAP tag does not affect the synaptic electrophysiology
Knockout mutations or overexpression of PSD-95 results in striking changes in synaptic physiology (Migaud et al, 1998; El-Husseini et al, 2000; Beique et al, 2006). In particular, long-term potentiation (LTP) of the excitatory synaptic transmission is greatly enhanced in PSD-95 knockout mice (Migaud et al, 1998; Komiyama et al, 2002; Beique et al, 2006). To determine whether TAP tagging of PSD-95 also altered the synaptic physiology, we studied short- and long-term plasticity in hippocampal slices of PSD-95TAP/TAP mice (Figure 2D). A short episode of theta-burst stimulation was used to induce LTP of field extracellular post-synaptic potentials (fEPSPs) in the CA1 area of the hippocampus. In the period of 60–65 min after theta-burst stimulation, amplitudes of fEPSPs in the test pathway normalized relative to control pathway were not different between PSD95TAP/TAP and wt mice (194±15% versus 176±8%; P=0.295) (Figure 2D). Likewise, paired-pulse facilitation, an established measure of short-term plasticity, was similar in wt and PSD-95TAP/TAP animals (Supplementary Figure 2), whereas it is known that this parameter is significantly enhanced in PSD-95 knockout mice (Migaud et al, 1998; Beique et al, 2006). Therefore, we conclude that the engineering of the TAP tag into PSD-95 using the knockin strategy has not altered the synaptic physiological function of PSD-95. Together, the physiological, biochemical, tissue and subcellular localization studies indicate that the presence of the TAP tag did not significantly alter the expression or function of PSD-95.
Optimization of single-step and tandem affinity purification of PSD-95-associated complexes
The following four-stage protocol for isolation of PSD-95 complexes was used (Figure 3A). First, TAP-tagged PSD-95 (from homozygous PSD-95TAP/TAP mice) was captured from brain extracts with an anti-FLAG antibody covalently coupled to Dynal beads. Second, the complex was eluted by cleavage with TEV protease completing the single step of purification. In the third stage, the complex was recovered from solution by Ni2+–NTA–agarose column that binds the HAT-tagged PSD-95. The fourth and final stage was the release of the PSD-95 complex from the column using imidazole, completing the tandem purification.
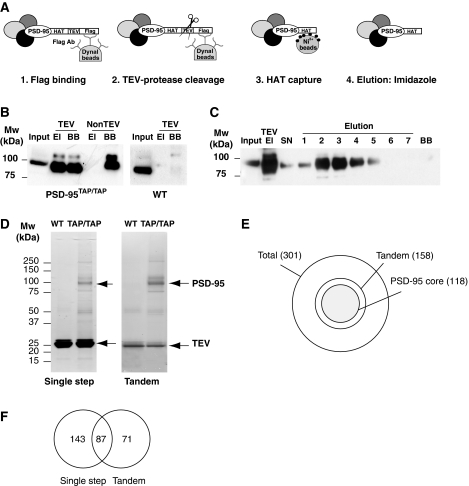
Figure 3.
Tandem affinity purification of PSD-95 complexes. (A) Overview of the TAP protocol. In the first step, the TAP-tagged PSD-95 was captured by FLAG antibody (1) and eluted by TEV cleavage (2). Cleaved TAP-tagged PSD-95 was then captured with Ni2+–NTA–agarose beads (3) and eluted with 250 mM imidazole (4). (B) TAP-tagged PSD-95 was affinity purified with FLAG antibody from forebrain extracts from PSD-95TAP/TAP (left panel) and wt (right panel) mice, then cleaved using TEV protease (TEV) and monitored using immunoblotting with a PSD-95 antibody. The eluted (El) and column retained PSD-95 (BB) are shown. TEV protease was added to the reaction as indicated (TEV) or to control without TEV (non-TEV). Input, total lysate; El, elution after TEV reaction; BB, beads boiled with Laemmli sample buffer after TEV cleavage. (C) HAT-tagged PSD-95 purification monitored using immunoblotting against PSD-95. Following TEV cleavage the eluate (TEV El) was incubated with Ni2+–NTA–agarose, washed and eluted by imidazole 250 mM and collected in 7 fractions. TEV, TEV elution before the Ni2+ column; SN, supernatant remaining after coupling to the Ni2+ column; 1–7, fractions recovered in the imidazole elution. BB, boiled Ni2+–agarose beads after elution. (D) TAP-tagged PSD-95 complex was affinity purified using FLAG antibody (single step, left gel) and tandem (two step, right panel) from wt and PSD-95TAP/TAP forebrain and resolved by SDS–PAGE stained with colloidal Coomasie stain. The lanes were cut for mass spectrometry analysis and the identified proteins listed in Supplementary Table 1. PSD-95 and the TEV enzyme are indicated in both gels. (E) Schematic representation of the total number (301) of proteins identified in the combined single and tandem purifications. In four independent tandem purifications, a total of 158 proteins were identified and 118 appeared in at least three of four replicates (PSD-95 core complexes). (F) Venn diagram with the number of proteins from either single or tandem purifications showing the common proteins (87) and proteins masked (71) in the single-step purification.
We examined the efficiency of the different steps in this protocol by monitoring PSD-95. All the solubilized PSD-95 was captured by FLAG-Dynal beads and >90% was cleaved using TEV protease (Supplementary Figure 3A). In the absence of TEV, there was no spontaneous release of TAP-tagged PSD-95 during incubations (Figure 3B, lane 4, 5 Non-TEV, El lane). TEV incubation released 50–70% of cleaved PSD-95 (Figure 3B, lane 2, El) and 30–50% of cleaved PSD-95 remained on the beads (Figure 3B, lane 3, BB). TEV had efficiently cleaved the retained protein as the size of PSD-95 in the BB lane corresponded to cleaved TAP-tagged PSD-95, and moreover, was not recognized by anti-FLAG antibodies on immunoblotting (Supplementary Figure 3A, comparison of BB lanes). This partial retention of cleaved protein seems to be a protein-specific phenomenon as we have observed this with other proteins studied in a similar manner (data not shown). For other controls, as expected, the FLAG antibody did not precipitate PSD-95 from wt mouse forebrain (Figure 3B, right panel). The recovery of PSD-95 using a Ni2+–NTA–agarose column that binds the HAT-tagged PSD-95 was very efficient (>95%) (Figure 3C, lanes 2 and 3, TEV El, SN). Subsequent elution using imidazole was also highly efficient (>95%) (no detectable retained-PSD-95 in BB lane, Figure 3C). Overall, we estimate that the yield of the protein recovery was 50–60% of the total PSD-95 present in the brain lysate.
Characterization of TAP-tagged PSD-95-associated complexes
We next examined the components of PSD-95 complexes and compared the single-step purification with the tandem purification. Complexes from PSD-95TAP/TAP and wt mice were subjected to SDS–PAGE and stained with colloidal Coomassie for band visualization before gel lanes were cut into slices (Figure 3D). These gels show a strong 95–100 kDa band in the PSD-95TAP/TAP lanes that corresponds to TAP-tagged PSD-95 and was absent in purifications from wt mice. A total of 301 different proteins were identified by LC-MS/MS from PSD-95TAP/TAP in all single-step and tandem purifications (Supplementary Tables 1 and 2).
Toward identifying ‘core' complexes, we found 118 (39% of 301) proteins in three of four independent tandem purifications (a total of 158 proteins were found in all four tandem purification experiments) (Figure 3E and Tables I and II). The tandem purification has a significant technical advantage because abundant proteins (present in single step) ‘mask' less abundant proteins that appear after further purification (Wang et al, 2006). We found 71 (45%) out of the 158 proteins masked in the single purification. This is shown in the Venn diagram (Figure 3F). We created a Web source that provides access to this data and links to physiological and behavioral data from knockout mice for the respective genes (www.genes2cognition.org/TAP-PSD-95).
Table 1.
Functional classification of PSD-95-associated proteins in at least three out of four tandem purifications
| MGI symbol | Protein name | UniProt Acc | Number of peptides | Cluster | |||
|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | ||||
| Adaptor/regulatory | |||||||
| Anks1 | Ankyrin repeat and SAM domain containing 1 | P59672 | 4 | 3 | 3 | ||
| Anks1b | Ankyrin repeat and sterile alpha motif domain containing 1B | Q8BZM2 | 17 | 5 | 10 | 10 | |
| Baiap2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Q8BKX1 | 20 | 12 | 13 | 13 | d |
| Begain | Brain-enriched guanylate kinase-associated | Q68EF6 | 4 | 8 | 12 | 16 | a |
| Dlg1 | Synapse-associated protein 97 | Q3UP61 | 24 | 42 | 34 | 42 | a |
| Dlg2 | Postsynaptic density protein 93 | Q91XM9 | 49 | 67 | 69 | 80 | a |
| Dlg3 | Synapse-associated protein 102 | Q52KF7 | 27 | 17 | 14 | 22 | a |
| Dlg4 | Postsynaptic density protein 95 | Q62108 | 42 | 57 | 64 | 64 | a |
| Dlgap1 | SAP90/PSD-95-associated protein 1 | Q9D415 | 11 | 9 | 14 | 24 | a |
| Dlgap2 | SAP90/PSD-95-associated protein 2 | Q8BJ42 | 18 | 12 | 19 | 24 | a |
| Dlgap3 | SAP90/PSD-95-associated protein 3 | A2A7T7 | 16 | 8 | 7 | 15 | a |
| Dlgap4 | SAP90/PSD-95-associated protein 4 | A2BDU3 | 14 | 9 | 7 | 14 | a |
| Receptor/channels/transporters | |||||||
| Gria1 | Glutamate receptor, ionotropic, AMPA 1 | Q5NBY1 | 2 | 3 | 5 | 13 | b |
| Gria2 | Glutamate receptor, ionotropic, AMPA 2 | P23819 | 4 | 8 | 10 | 20 | b |
| Gria3 | Glutamate receptor, ionotrophic, AMPA 3 | Q9Z2W9 | 2 | 9 | 18 | b | |
| Gria4 | Glutamate receptor, ionotrophic, AMPA 4 | Q9Z2W8 | 2 | 2 | 5 | 9 | b |
| Grik2 | Glutamate receptor, ionotropic, kainate 2 (beta 2) | P39087 | 2 | 4 | 6 | b | |
| Grik5 | Glutamate receptor, ionotropic, kainate 5 (gamma 2) | Q61626 | 2 | 3 | 7 | b | |
| Grin1 | Glutamate receptor, ionotropic, NMDA1 (zeta 1) | A2AI21 | 29 | 40 | 50 | 55 | a |
| Grin2a | Glutamate receptor, ionotropic, NMDA2A (epsilon 1) | P35436 | 24 | 31 | 36 | 46 | a |
| Grin2b | Glutamate receptor, ionotropic, NMDA2B (epsilon 2) | Q01097 | 44 | 54 | 67 | 78 | a |
| Grin2d | Glutamate receptor, ionotropic, NMDA2D (epsilon 4) | Q03391 | 3 | 6 | 9 | 10 | a |
| Gpr123 | G protein-coupled receptor 123 | Q52KJ6 | 2 | 3 | 3 | 3 | |
| Cacng2 | Calcium channel, voltage-dependent, gamma subunit 2 | O88602 | 2 | 2 | 2 | 3 | b |
| Kcna1 | K+ voltage-gated channel, shaker-related subfamily, member 1 | P16388 | 6 | 5 | 5 | 6 | c |
| Kcna2 | K+ voltage-gated channel, shaker-related subfamily, member 2 | P63141 | 4 | 5 | 7 | 6 | c |
| Kcna3 | K+ voltage-gated channel, shaker-related subfamily, member 3 | P16390 | 3 | 4 | 6 | 5 | c |
| Kcna4 | K+ voltage-gated channel, shaker-related subfamily, member 4 | Q8CBF8 | 2 | 3 | 5 | 5 | c |
| Kcnab1 | K+ voltage-gated channel, shaker-related subfamily, beta member 1 | P63143 | 3 | 3 | 4 | 6 | c |
| Kcnab2 | K+ voltage-gated channel, shaker-related subfamily, beta member 2 | P62482 | 5 | 6 | 10 | 11 | c |
| Kcnj10 | K+ inwardly-rectifying channel, subfamily J, member 10 | Q9JM63 | 3 | 3 | 4 | 6 | a |
| Kcnj4 | K+ inwardly-rectifying channel, subfamily J, member 4 | P52189 | 4 | 6 | 6 | 8 | a |
| Vdac1 | Voltage-dependent anion channel 1 | Q60932 | 4 | 4 | 5 | 5 | f |
| Vdac2 | Voltage-dependent anion channel 2 | Q60930 | 4 | 2 | 3 | 4 | |
| Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide1 | P14094 | 3 | 3 | 6 | 3 | |
| Atp6v0d1 | ATPase, H+ transporting, V0 subunit d isoform 1 | P51863 | 3 | 3 | 4 | 2 | |
| Sfxn3 | Sideroflexin 3 | Q91V61 | 3 | 3 | 4 | 3 | |
| Slc1a2 | Solute carrier family, member 2 | P43006 | 3 | 2 | 4 | 4 | |
| Slc25a4 | ADP/ATP translocase 1 | P48962 | 2 | 3 | 6 | g | |
| Slc25a5 | ADP/ATP translocase 2 | P51881 | 2 | 2 | 3 | 6 | g |
| Slc4a4 | Solute carrier family 4 (anion exchanger), member 4 | O88343 | 3 | 6 | 4 | ||
| Cytoskeletal/structural/cell adhesion | |||||||
| Ablim1 | Actin-binding LIM protein 1 | Q8K4G5 | 20 | 10 | 18 | 22 | |
| Adam22 | a disintegrin and metallopeptidase domain 22 | Q9R1V6 | 12 | 16 | 24 | 31 | e |
| Arc | Activity regulated cytoskeletal-associated protein | Q9WV31 | 8 | 13 | 20 | 19 | |
| Arpc4 | Actin related protein 2/3 complex, subunit 4 | P59999 | 2 | 2 | 4 | ||
| Capza2 | Capping protein (actin filament) muscle Z-line, alpha 2 | P47754 | 2 | 2 | 2 | ||
| Cfl1 | Cofilin 1, non-muscle | P18760 | 2 | 2 | 3 | 4 | |
| Dstn | Destrin | Q9R0P5 | 2 | 5 | 6 | 6 | |
| Fscn1 | Fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | Q61553 | 2 | 2 | 3 | f | |
| Lgi1 | Leucine-rich repeat LGI family, member 1 | Q9JIA1 | 6 | 12 | 19 | 15 | e |
| Nefl | Neurofilament, light polypeptide 68kDa | P08551 | 3 | 3 | 6 | a | |
| Nrxn1 | Neurexin 1 | Q9CS84 | 8 | 17 | 12 | 23 | |
| Plp1 | Proteolipid protein (myelin) 1 | P60202 | 2 | 3 | 4 | 5 | |
| Sept11 | Septin 11 | Q8C1B7 | 2 | 2 | 2 | 2 | |
| Sept5 | Septin 5 | Q9Z2Q6 | 2 | 2 | 4 | ||
| Spnb2 | Spectrin beta 2 | Q62261 | 2 | 2 | 5 | a | |
| Tuba1a* | Tubulin, beta polypeptide | P05213, | 11 | 14 | 20 | 20 | |
| Tubb2b* | Tubulin, beta 2b | Q7TMM9 | 19 | 21 | 24 | 27 | |
| Tubb6 | Tubulin, beta 6 | Q922F4 | 8 | 8 | 13 | 13 | |
| Vesicular/trafficking/transport | |||||||
| Arf3 | ADP-ribosylation factor 3 | P61205 | 2 | 2 | 3 | 4 | |
| Cltc | Clathrin, heavy chain (Hc) | Q5SXR6 | 10 | 4 | 12 | a | |
| Cpne4* | Copine IV | Q8BLR2 | 4 | 2 | 3 | ||
| Cpne7 | Copine VII7 | Q0VE82 | 4 | 3 | 2 | 3 | |
| Iqsec1 | IQ motif and Sec7 domain 1 | Q8R0S2 | 11 | 7 | 6 | 15 | |
| Iqsec2 | IQ motif and Sec7 domain 2 | Q5DU25 | 34 | 25 | 24 | 36 | |
| Nsf | N-ethylmaleimide sensitive fusion protein | P46460 | 4 | 4 | 7 | 9 | |
| Stx1b2 | Syntaxin 1B2 | P61264 | 3 | 4 | 3 | 2 | |
| Stxbp1 | Syntaxin binding protein 1 | O08599 | 5 | 3 | 4 | 4 | |
| Syt1 | Synaptotagmin I | P46096 | 5 | 2 | 2 | 2 | |
| Vamp2* | Synaptobrevin 2 | P63024 | 2 | 4 | 2 | ||
| Enzymes | |||||||
| Acat1 | Acetyl-Coenzyme A acetyltransferase 1 | Q8QZT1 | 2 | 3 | 3 | ||
| Aco2 | Aconitase 2, mitochondrial | Q99KI0 | 3 | 3 | 2 | ||
| Acot7 | Acyl-CoA thioesterase 7 | Q91V12 | 3 | 3 | 3 | 4 | |
| Aldoc | Aldolase C, fructose-bisphosphate | P05063 | 4 | 2 | 4 | ||
| Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | Q91VR2 | 3 | 3 | 4 | g | |
| Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit | P56480 | 2 | 6 | 5 | g | |
| Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | Q9DB20 | 2 | 2 | 4 | 4 | g |
| Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 | Q03265 | 5 | 10 | 14 | 11 | g |
| Cnp | 2′,3′-cyclic nucleotide 3′ phosphodiesterase | P16330 | 10 | 3 | 11 | 10 | |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | P16858 | 9 | 8 | 9 | 10 | f |
| Gda | Guanine deaminase | Q9R111 | 29 | 26 | 29 | 31 | a |
| Glul | Glutamate–ammonia ligase (glutamine synthetase) | P15105 | 5 | 17 | 21 | 19 | |
| Gpx4 | Glutathione peroxidase 4 | O70325 | 2 | 2 | 2 | ||
| Msrb2 | Methionine sulfoxide reductase B2 | Q78J03 | 2 | 7 | 4 | ||
| Pdha1 | Pyruvate dehydrogenase E1 alpha 1 | P35486 | 4 | 3 | 8 | 5 | |
| Pdhb | Pyruvate dehydrogenase (lipoamide) beta | Q9D051 | 4 | 4 | 4 | 6 | |
| Pgk1 | Phosphoglycerate kinase 1 | P09411 | 2 | 3 | 13 | 8 | f |
| Pkm2 | Pyruvate kinase, muscle | P52480 | 7 | 6 | 11 | 12 | |
| Ppap2b | Phosphatidic acid phosphatase type 2B | Q99JY8 | 2 | 2 | 5 | ||
| Prdx1 | Peroxiredoxin 1 | P35700 | 4 | 7 | 10 | 14 | |
| Prdx2 | Peroxiredoxin 2 | Q61171 | 2 | 2 | 3 | ||
| Sdha | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | Q8K2B3 | 7 | 6 | 8 | 6 | |
| Sucla2 | Succinate-CoA ligase, ADP-forming, beta subunit | Q9Z2I9 | 4 | 2 | 5 | 6 | |
| Kinases | |||||||
| Camk2a | Calcium/calmodulin-dependent protein kinase II alpha | P11798 | 11 | 3 | 4 | 11 | a |
| Camk2b | Calcium/calmodulin-dependent protein kinase II beta | Q5SVI3 | 10 | 5 | 5 | 7 | a |
| Mapk1 | Mitogen-activated protein kinase 1 | P63085 | 3 | 3 | 4 | 9 | |
| Phosphatases | |||||||
| Ppap2b | Phosphatidic acid phosphatase type 2B | Q99JY8 | 2 | 2 | 5 | ||
| Ppp3ca | Protein phosphatase 3, catalytic subunit, alpha isoform | P63328 | 12 | 6 | 7 | 11 | a |
| Ppp3cb | Protein phosphatase 3, catalytic subunit, beta isoform | P48453 | 7 | 3 | 2 | 4 | |
| G-protein signaling | |||||||
| Abr | Active BCR-related gene | Q6PCY1 | 2 | 2 | 6 | ||
| Gnao1 | Guanine nucleotide binding protein, alpha o | P18872 | 8 | 7 | 15 | 16 | |
| Kalrn | Kalirin, RhoGEF kinase | A2CG52 | 2 | 3 | 2 | 8 | |
| Rac1 | RAS-related C3 botulinum substrate 1 | Q3TLP8 | 2 | 2 | 3 | 3 | d |
| Syngap1 | Synaptic Ras GTPase activating protein 1 homolog (rat) | Q9QUH6 | 21 | 15 | 17 | 38 | a |
| Transcription/translation | |||||||
| Park7 | Parkinson disease (autosomal recessive, early onset) 7 | A2A817 | 3 | 3 | 3 | ||
| Rps14 | Ribosomal protein S14 | P62264 | 2 | 3 | 2 | ||
| Rps3 | Ribosomal protein S3 | P62908 | 2 | 3 | 4 | ||
| Uba52* | Ubiquitin A-52 residue ribosomal protein fusion product 1 | Q66JP1 | 3 | 3 | 5 | 6 | |
| Signal transduction | |||||||
| Btbd11 | BTB (POZ) domain containing 11 | Q6GQW0 | 5 | 3 | 2 | 3 | |
| Phb2 | Prohibitin 2 | O35129 | 2 | 5 | 4 | ||
| Ywhae | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | P62259 | 3 | 2 | 2 | ||
| Pcbp1 | Poly(rC) binding protein 1 | P60335 | 3 | 3 | 5 | ||
| Unclassified | |||||||
| Fam81a | Family with sequence similarity 81, member A | Q3UXZ6 | 7 | 6 | 21 | 18 | |
| AI662250 | Expressed sequence AI662250 | Q3UKV2 | 2 | 2 | 2 | ||
| B630019K06Rik | RIKEN cDNA B630019K06 gene | Q7TNS5 | 6 | 7 | 8 | 9 | |
| Frmpd3 | FERM and PDZ domain containing 3 | Q8BXG0 | 2 | 5 | 5 | ||
| Pgam5 | Phosphoglycerate mutase family member 5 | Q3UK19 | 7 | 7 | 8 | 10 | |
| Prrt1 | Proline-rich transmembrane protein 1 | O35449 | 2 | 2 | 2 | 3 | |
| Slc9a3r1 | Solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | P70441 | 4 | 3 | 7 | 2 | |
MGI approved gene symbols and protein names, and UniProt accession numbers are shown. Numbers of approved peptides for each protein identified by LC-MS/MS in the four tandem purifications are indicated as T1, T2, T3 and T4. More information regarding these proteins is given in Supplementary Table 1. Genes marked with an asterisk represent genes whose peptides are common to other genes.
Cpne4*: Cpne5, Cpne8.
Tuba1a*: Tuba1b, Tuba4c, Tuba1b.
Tubb2b*: Tubb5, Tubb2a, Tubb2c, Tubb4.
Uba52*: Ubc, Ubb.
Vamp2*: Vamp3.
Table 2.
Functional classification of PSD-95-associated proteins in one or two tandem purifications
| MGI Symbol | Protein name | UniProt Acc | Number of peptides | |||
|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | |||
| Adaptor/regulatory | ||||||
| Ap2a1 | Adaptor protein complex AP-2, alpha 1 subunit | P17426 | 3 | |||
| Grb2 | Growth factor receptor bound protein 2 | Q60631 | 2 | 4 | ||
| Receptors/channels/transporters | ||||||
| Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | Q05CJ5 | 2 | 4 | ||
| Grin2c | Glutamate receptor, ionotropic, NMDA2C (epsilon 3) | Q01098 | 5 | |||
| Grm3 | Glutamate receptor, metabotropic 3 | Q9QYS2 | 2 | |||
| Kcnj16 | Potassium inwardly-rectifying channel, subfamily J, member 16 | Q9Z307 | 7 | |||
| Lrp1 | Low density lipoprotein receptor-related protein 1 | Q91ZX7 | 2 | 3 | ||
| Lrrtm1 | Leucine rich repeat transmembrane neuronal 1 | Q8K377 | 2 | |||
| Slc1a3 | Sodium-dependent glutamate/aspartate transporter 1 | P56564 | 3 | 3 | ||
| Slc2a1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | P17809 | 2 | 2 | ||
| Vdac3 | Voltage-dependent anion channel 3 | Q60931 | 2 | 5 | ||
| Cytoeskeletal/structural/cell adhesion | ||||||
| Mtap1a | Microtubule-associated protein 1 A | Q9QYR6 | 3 | 2 | ||
| Nlgn2 | Neuroligin 2 | Q69ZK9 | 2 | |||
| Nlgn3 | Neuroligin 3 | A2AGI2 | 2 | |||
| Syn2 | Synapsin II | Q64332 | 3 | 4 | ||
| Shank1 | SH3/ankyrin domain gene 1 | XP_001474960a | 4 | |||
| Enzymes | ||||||
| Cit | Citron | P49025 | 11 | |||
| Crym | Crystallin, mu | O54983 | 3 | 3 | ||
| Csmd2 | CUB and Sushi multiple domains 2 | A2A8D7 | 5 | |||
| Dusp10 | Dual specificity phosphatase 10 | Q8R3L3 | 3 | 2 | ||
| Jak3 | Janus kinase 3 | Q62137 | 2 | |||
| Mapk3 | Mitogen activated protein kinase 3 | Q63844 | 4 | |||
| Ube2v1 | Ubiquitin-conjugating enzyme E2 variant 1 | Q9CZY3 | 2 | 5 | ||
| Ube2v2 | Ubiquitin-conjugating enzyme E2 variant 2 | Q9D2M8 | 2 | 4 | ||
| G-protein signalling | ||||||
| Gna13 | Guanine nucleotide binding protein, alpha 13 | Q8C5L2 | 2 | 2 | ||
| Gnb1 | Guanine nucleotide binding protein (G protein), beta 1 | P62874 | 3 | 3 | ||
| Rab6 | RAB6, member RAS oncogene family | P35279 | 4 | |||
| Signalling | ||||||
| Fbxo2 | F-box protein 2 | Q80UW2 | 4 | |||
| Fbxo6 | F-box protein 6 | Q9QZN4 | 3 | 4 | ||
| Nxph3 | Neurexophilin 3 | Q91VX5 | 2 | |||
| Pcbp2 | Poly(rC) binding protein 2 | Q61990 | 3 | 3 | ||
| Traf3 | Tnf receptor-associated factor 3 | Q3UHJ1 | 3 | 3 | ||
| Chaperone/protein folding/signalling | ||||||
| Hspa12a | Heat shock protein 12A | Q8K0U4 | 2 | |||
| DNA binding | ||||||
| Hist1h2bj* | Histone 1, H2bb | Q8CGP2 | 3 | 3 | ||
| Transcription/translation | ||||||
| Eef1a1* | Eukaryotic translation elongation factor 1 alpha 1 | P10126 | 2 | 2 | ||
| Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | Q8BUV6 | 2 | 3 | ||
| Unclassified | ||||||
| Clu | Clusterin | Q06890 | 2 | 3 | ||
| Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | O89112 | 4 | |||
| Mog | Myelin oligodendrocyte glycoprotein | Q61885 | 2 | 2 | ||
| Neto1 | Neuropilin (NRP) and tolloid (TLL)-like 1 | Q8R4I7 | 2 | 2 | ||
MGI approved gene symbols and protein names, and UniProt accession numbers are shown. Number of approved peptides for each protein identified by LC-MS/MS in the four tandem purifications is indicated as T1, T2, T3 and T4. More information of these proteins is listed in Supplementary Table 1. Genes marked with an asterisk represent genes whose peptides are common to other genes.
aNCBI accession number.
Eif1a1*: Eif1a2.
Hist1h2bj*: Hist1h2bm, Hist1h2be, Hist1h2bn, Hist1h2bg, Hist1h2bp, Hist1h2bh, Hist1h2bf, Hist1h2bb, Hist3h2bb, Hist1h2bc, Hist1h2bl, Hist2h2bb.
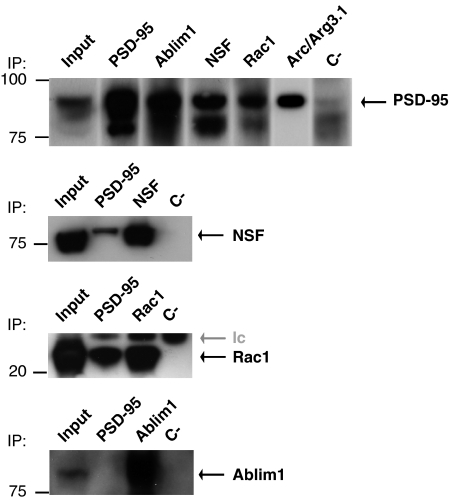
To further explore the advantages of the TAP method, we examined the specific proteins to identify new PSD-95 interactors and also compared the types of proteins remaining in the core set after tandem purification. From the core complex of 118 proteins, 26 (22%) were reported as primary interactors (data present in HPRD, Biogrid, BIND and HOMOMINT databases) and included membrane-associated guanylate kinases (MAGUK or disc large homolog, Dlg family), NMDA receptor subunits, potassium channels and cytoskeletal proteins (Supplementary Table 3). Using immunoblotting we confirmed the presence of 13 PSD-95 interactors (Supplementary Figure 3B). Ten of those interactors were examined by reverse immunoprecipitation and all were validated (Supplementary Figure 3C). We also examined four new interactors using co-immunoprecipitation experiments: Arc/Arg3.1, Rac1, Nsf and Ablim1 (Figure 4 and Supplementary Figure 3B). These proteins are involved in cytoskeletal, vesicular-trafficking and G-protein-mediated signaling pathways.
Figure 4.
Validation of new PSD-95 interaction partners. Immunoprecipitation from forebrain extracts with indicated antibodies (labeled above panels) and immunoblotting with antibodies directed against specific proteins (labeled on the right side of each panel). Antibodies against PSD-95, Nsf, Rac1 and Ablim were used for immunoblotting. Protein molecular weight (kDa) on left. PSD-95 interaction with Arc/Arg3.1 is shown in Supplementary Figure 3B. C-, mouse total IgG was used for immunoprecipitation control; IP, antibodies used for immunoprecipitation; lc, antibody light chain.
We used two methods to examine the types of proteins enriched by the tandem procedure. In the first approach, we grouped all proteins from both single-step and tandem purifications into ten functional categories and graphed the numbers of proteins in each category as a percentage of the dataset (Supplementary Figure 4). The tandem purification was enriched in channels/receptors, cytoskeletal/structural/adhesion and adaptors/regulatory proteins. There was a striking depletion of enzymes in the tandem purification compared with the single-step purification, consistent with the fact that many metabolic enzymes are abundant and can contaminate such purifications (Chen and Gingras, 2007). In the second approach, we considered the emPAI values (a semiquantitative measure of protein abundance based on MS data) for the 301 proteins and divided the dataset into two groups: tandem enriched and tandem depleted (Supplementary Table 4 and Supplementary Figure 5). Analysis of Gene Ontology (GO) terms showed that the tandem depleted set was significantly over-represented with the following GO terms: metabolism (P=1.67e−3), cytoplasm (P=2.10e−5) and mitochondrion (P=7.96e−4). In contrast, the tandem enriched set was significantly enriched with the following GO terms: signal transducer activity (P=6.16e−6), synapse (P=3.52e−7), postsynaptic membrane (P=3.22e−5) and cell communication (P=2.33e−5). Therefore we conclude that the TAP strategy can be used to recover a smaller and more specific subset of proteins than a single immunoaffinity (FLAG antibody) procedure.
We compared the lists of proteins identified in the TAP experiments with earlier studies of synapse proteomes (Supplementary Tables 1 and 5). An earlier report using a single immunoprecipitation with a PSD-95 antibody identified 276 proteins from PSD fractions extracted in the absence of detergent (Dosemeci et al, 2007). The comparison of this list with the PSD-95 core complexes of 118 proteins reported here shows 49 proteins in common. A peptide affinity method for binding PDZ domains of MAGUK proteins (Husi et al, 2000; Husi and Grant, 2001; Collins et al, 2005; Emes et al, 2008) was used in the same extraction conditions reported here and recovered 105 proteins (Collins et al, 2005). This peptide affinity method was not specific to PSD-95 as the peptides are known to bind PSD-93 and SAP102 (Lim et al, 2002; Chung et al, 2004). These 105 proteins and the proteins found by NMDA-receptor immunopurification were used to generate a list of 186 MASC proteins (Collins et al, 2006). Comparison of our 118 PSD-95 TAP list with the 186 proteins from the MASC complex shows 48 proteins in common (Supplementary Tables 1 and 5). An important set of proteins that was recovered using the TAP method consisted of the AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors and K+ channels, which is discussed below. Overall we conclude that the targeted TAP tagging allowed for the enrichment of crucial synaptic proteins.
Composition and organization of PSD-95 interaction networks
To explore the functional organization of the PSD-95 complexes, we reconstructed a network using protein–protein interaction data from high-quality manually-curated interaction data (Pocklington et al, 2006) and the UniHi database (http://www.mdc-berlin.de/unihi). After manual curation, we identified 119 interactions between 50 proteins (excluding self-interactions) from the 118 proteins in the PSD-95 core complex (Table I). No binary interactions were found for the remaining 68 proteins.
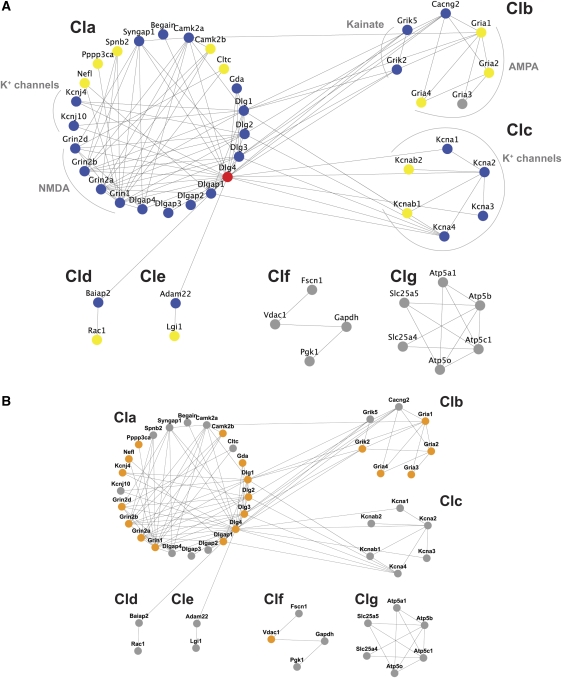
Network clustering of the interacting proteins showed 40 out of the 50 proteins formed a large connected component (major connected component, MCC) and a modular structure that was segregated into five clusters (see Materials and methods), referred to as cluster a (Cla) to cluster e (Cle) (Figure 5A). In addition to the five MCC clusters, two further disconnected clusters (‘Clf' and ‘Clg') were found (see Table I for details).
Figure 5.
Protein interaction network of PSD-95 interacting proteins. (A) 50 proteins of the PSD-95 core complex were connected, with 119 interactions segregated into 5 clusters (Cla–Cle) forming the MCC and two separate small clusters Clf and Clg. PSD-95/Dlg4 is showed in red, primary interactors of PSD-95/Dlg4 are shown in blue and secondary interactors are shown in yellow. The glutamate receptors (NMDA, AMPA and kainate receptors) and potassium channels are bracketed. (B) Schizophrenia susceptibility genes are shown in orange.
It is interesting to note the location and proximity of the receptors and channels responsible for the postsynaptic depolarization and subsequent action potential generation. All NMDA, AMPA and kainate glutamate receptors were restricted to Cla and Clb and the voltage-dependent K+ channels were found in Cla and Clc (entirely comprised of K+ channels). These channels are known to couple to plasticity mechanisms (Watanabe et al, 2002; Chen et al, 2006a; Kim et al, 2007), and we noted that Cla contains important signaling enzymes involved in plasticity, including CamKII (Frankland et al, 2001) and SynGAP (Komiyama et al, 2002). It therefore seems that Cla, Clb and Clc are enriched with membrane proteins responsible for the electrical properties of the postsynaptic terminal.
As PSD-95/Dlg4 was the bait for the biochemical isolation of the complexes, we examined the distribution of its primary interactors (proteins that directly bind PSD-95) and secondary interactors (proteins that do not bind PSD-95 directly, but bind one of its primary interactors) (Figure 5A). Of the 39 MCC proteins (excluding PSD-95), 26 (67%) were primary interactors (blue symbols in Figure 5A) and 12 (31%) were secondary interactors (yellow symbols in Figure 5A) and only one protein, the AMPA receptor subunit Gria3, was a tertiary interactor. The majority of each cluster of the MCC comprised primary interactors: Cla (74%), Clb (43%), Clc (67%), Cld (50%) and Cle (50%). To examine the centrality of each protein in the network the shortest path from each protein to every other protein was counted, and the average shortest path (ASP) calculated. For all proteins, the mean ASP was 2.25. Ranking the ASP of each protein (Supplementary Table 6) showed PSD-95 had the lowest ASP (1.3), consistent with its central role in these networks.
It was of interest to compare the PSD-95 network (MCC of 40 proteins) with the previously published MASC network (MCC of 90 proteins) that was built from proteins co-purified with the NMDA receptor complex and with the PDZ peptide affinity method described in the previous section (Pocklington et al, 2006). The MASC MCC had 13 clusters, with cluster 1 containing PSD-95 (Supplementary Figure 6). A total of 16 proteins were common to PSD-95 MCC and MASC MCC (P<10e−7) and with overlap centered (10/16 proteins, P<10e−3) on Cla and MASC cluster 1. In contrast to MASC MCC, the PSD-95 MCC had Clb (AMPA receptors) and Clc (K+ channels).
Psychiatric disorders and PSD-95 complexes
To explore the potential medical importance of the 118 core PSD-95 interactome we asked (i) which diseases are these proteins involved with, and (ii) is there any relationship between the network clusters and disease types. For each of the proteins in the 118 core set, we manually curated information on their disease involvement from the literature. A total of 49 genes were implicated in multiple diseases: schizophrenia (28), mental retardation (6), bipolar disorder (13) Alzheimer's disease (6) and others (29) (Table III).
Table 3.
Genes associated with neurological and psychiatric diseases
| MGI symbol | Disease | MGI symbol | Disease |
|---|---|---|---|
| Adam22 | Epilepsy1 | Grin2d | Schizophrenia2 |
| Acot7 | Schizophrenia3 | Kcnj4 | Schizophrenia4 |
| Atp1b1 | Rett syndrome5 | Kcna1 | Episodic ataxia, type 16 |
| Neurodegeneration7 | Kcnj10 | Epilepsy8 | |
| Atp5a1 | Alzheimer's9 | Seizure10 | |
| Atp5c1 | Bipolar affective disorder11,12 | Lgi1 | Epilepsy1 |
| Cacng2 | Bipolar disorder13 | Mapk1 | Schizophrenia15,16 |
| CamKIIa | Bipolar disorder14 | Depression18 | |
| CamKIIb | Schizophrenia17 | Msrb2 | Bipolar afective disorder11 |
| Depression17 | |||
| Capza2 | Mental retardation19 | Nefl | CMT120 |
| Cltc | Mental retardation21 | Schizophrenia3 | |
| Cnp1 | Schizophrenia3,22 | Bipolar23 | |
| Dlg1 | Schizophrenia24 | CMT225,26 | |
| Dlg2 | Schizophrenia22 | ALS27,28 | |
| Dlg3 | Schizophrenia23,24 | Nrxn1 | Autism29 |
| Bipolar disorder23 | Schizophrenia22 | ||
| Depression23 | Nsf | Schizophrenia30 | |
| X-Mental retardation31 | Pdha1 | Depression32 | |
| Dlg4 | Schizophrenia33 | Pgk1 | Parkinson's5 |
| Bipolar disorder23 | Mental retardation34 | ||
| Dlgap1 | Schizophrenia35 | Bipolar disorder36 | |
| Gapdh | Alzheimer's39 | Plp1 | Pelizaeus–Merzbacher disease38 |
| Gda | Schizophrenia37 | Depression40 | |
| Gnao1 | Schizophrenia41 | Multiple sclerosis42−45 | |
| Gria1 | Schizophrenia46−50 | Demyelinating disease51−54 | |
| Alzheimer's55–59 | Spastic paraplegia60 | ||
| Epilepsy61,62 | Pppp3ca | Schizophrenia63 | |
| Gria2 | Schizophrenia4,64 | Prdx1 | Alzheimer's65 |
| Epilepsy66–71 | Prdx2 | Parkinson's72 | |
| Gria3 | Schizophrenia47 | Sl1a2 | Schizophrenia22 |
| X-Mental retardation73 | ALS74 | ||
| Gria4 | Schizophrenia22 | Slc25a4 | Bipolar afective disorder12 |
| Grik2 | Mental retardation75 | Ophtalmoplegia76 | |
| Schizophrenia22 | Stxbp1 | Schizophrenia77 | |
| Grin1 | Attention disorder78 | Vdac1 | Alzheimer's9 |
| Bipolar afective disorder80 | Schizophrenia3 | ||
| Schizophrenia81 | Bipolar afective disorder12 | ||
| Seizure82 | Vdac2 | Bipolar afective disorder12 | |
| Grin2a | Alzheimer's83 | Ywhae | Miller–Dieker lissencephaly85 |
| Huntington disease84 | |||
| Schizophrenia33 | |||
| Grin2b | Schizophrenia22,23 | ||
| Bipolar afective disorder86–88 | |||
| Epilepsy11,89 | |||
| Huntington disease84,90 |
Disease association data for proteins in the tandem purification were collected from the Genetic Association Database, CiteXplore and manually curated. References are provided in Supplementary information.
We next analyzed the pair-wise correlation between functional categories (Tables I and II) and disease type. The ‘receptors/channels/transporters' category and ionotropic glutamate receptors were significantly correlated with schizophrenia (P=0.0024 and P<10e−6, respectively). Out of 28 schizophrenia-implicated proteins, 20 were mapped into the network model (orange in Figure 5B). Of those 20 proteins, 70% fell into Cla, which was significantly enriched in schizophrenia-related proteins (P=0.0089). All but one of the remaining schizophrenia-related proteins were found in cluster Clb.
Discussion
Here we report the first isolation of multiprotein complexes from mice using a knockin of a TAP tag fused with the endogenous protein. This allowed expression of the tagged protein to be controlled by its endogenous regulatory elements. A stable and specific complex of 118 proteins associated with PSD-95, containing a range of important synaptic receptors, channels and signaling molecules, including new interactors was isolated.
Previous use of TAP tagging in mammalian cells and tissues was limited to expression of exogenous tagged cDNAs (Knuesel et al, 2003; Bouwmeester et al, 2004; Brajenovic et al, 2004; Drakas et al, 2005; Wang et al, 2005, 2006; Angrand et al, 2006; Burckstummer et al, 2006), which do not recapitulate the natural expression of the protein. A TAP insertion using homologous recombination was published by Chen et al, however, purification of complexes was not reported from mouse tissue (Chen et al, 2006b). In addition to the advantage of recapitulating the natural expression of the protein, and thereby limiting artefactual interactions, the targeting of the endogenous gene allows the breeding of the mice to homozygosity. This permits testing the possibility that the insertion created a mutation. We found no evidence of a mutant phenotype as neither the level, tissue expression pattern, subcellular localization or synaptic physiology of PSD-95 was found in homozygous mice.
The two consecutive steps of purification in the TAP protocol offer advantages over single-step methods such as immunoprecipitation, which is the most commonly used approach. Immunoprecipitation is limited by (i) availability of suitable antibodies and their cross reaction with other proteins, (ii) the possibility that the antibody–protein interaction might be affected by either post-translational modifications or by the binding with other proteins, (iii) the antibody might disrupt interacting partners, (iv) the harsh conditions for the complex elution might result in protein degradation. In addition to overcoming these limitations, the TAP procedure offers an efficient method for isolation of native complexes. Also, we observed that the two-step procedure unmasked core interacting proteins that were not detected by mass spectrometry in the single-step purification: Ten known PSD-95 interactors, Begain, Cit, Grik2, Grik5, Grin2c, Kcna4, Lrp1, Nlgn2, Nlgn3 and Shank1, were present only after the tandem purification. Furthermore, we found 21 new proteins in the PSD-95 core complexes that were not reported in earlier PSP proteomic analysis (Collins et al, 2006 [19]Dosemeci et al, 2007) (Supplementary Table 1), again suggesting that the targeted TAP-tagging strategy produces greater depth and quality of interactors.
The fact that PSD-95-associated complexes contain ionotropic glutamate receptors of the NMDA, AMPA and kainate subtypes as well as major K+ channels is of considerable technical and biological significance. These proteins are the major postsynaptic constituents responsible for synaptic transmission and shaping the postsynaptic electrophysiological response to presynaptic input. We also believe that this is the first method that allows the robust co-purification of these proteins and that the PSD-95TAP mice will be a valuable tool for studying the postsynaptic terminal in vivo. These applications will extend to physiological and behavioral studies of many regions of the brain and disease models.
Although PSD-95 is a known direct binding partner of NMDA receptors, there is conflicting data about physical interactions between PSD-95 and AMPA receptor subunits. PSD-95 expression affects AMPA receptor-mediated excitatory synaptic transmission (Migaud et al, 1998; Beique et al, 2006; Carlisle et al, 2008) and is thought to involve indirect interactions through stargazin, SAP-97, Adam22, Lgi1 and Nsf (Leonard et al, 1998; Osten et al, 1998; Fukata et al, 2006). As we show using reciprocal co-precipitation, the interaction of PSD-95 with N-ethylmaleimide sensitive fusion protein (NSF), a cytosolic ATPase, was required for intracellular membrane fusion, and this reinforces the idea of PSD-95 involvement in synaptic vesicle trafficking and AMPA surface-expression modulation (Luthi et al, 1999; Noel et al, 1999). Other proteins involved in the trafficking and clustering of AMPA receptor are Arc/Arg3.1 (Chowdhury et al, 2006; Shepherd et al, 2006) and Rac1 (Wiens et al, 2005), and these were found within the complexes. The isolation of multiple AMPA-receptor modulators in the PSD-95 complexes underlines the importance of this complex in mediating synaptic plasticity.
We annotated the disease involvement of the proteins in the PSD-95 complexes as a step toward using this proteomic data to drive human genetic studies. We identified 49 of the proteins as involved with human mental disorders, of which there was a high representation of cognitive disorders. Nineteen genes involved in schizophrenia were significantly associated with the clusters Cla and Clb that contain all the glutamate receptors and MAGUK/Dlg proteins. Mapping the primary interactors of these schizophrenia proteins recruited many other proteins found in the other modules of the network. This suggests that the overall network and its various clusters might play a role in schizophrenia, and not simply the glutamate receptors, as was generally considered in the ‘glutamate hypothesis' of schizophrenia (Greene, 2001; Coyle, 2006; Lisman et al, 2008). Proteomic studies are likely to be useful for driving high-throughput sequencing in human diseases and aid in medical systems biology.
Materials and methods
Vector generation and gene targeting
The TAP tag was constructed by assembling two PCR fragments containing histidine affinity tag (HAT), TEV protease and FLAG sequences. The HAT tag was amplified by PCR (PCR1) using 1 ng of the pHAT20 vector (Clontech) as a template with the forward XbaIHATF1 and reverse HATR1 oligos as primers. The 5′-end tail of the forward primer had an XbaI restriction site and the reverse primer had a tail containing the six-amino-acid linker in the 5′-end and the TEV protease sequences. The FLAG tag was amplified by PCR (PCR2) with the forward FLAGF1 and reverse BclIFLAGR1 oligos using as a template 1 ng of the C-terminal p3xFLAG–CMV™14 vector (Sigma). The 5′-end of the forward primer contains the TEV sequence and the six-amino-acid linker sequence, whereas the reverse primer contains a BclI restriction site. A third PCR was then carried out with XbaIHATF1 and BclIFLAGR1 oligos and a mix of PCR1 and PCR2 products as a template. The 3′-end of the PCR1 and the 5′-end of the PCR2 shared the six-amino-acid linker and the TEV sequences, allowing both the fragments to anneal during the third PCR. This fragment (the TAP tag sequence) was cloned into pneoflox vector between the XbaI and BclI restriction sites (TAP tag pneoflox vector). Two homology arms of the genomic PSD-95 sequence were amplified with forward Psd95HAXhoIF and reverse Psd95HAXbaIR primers and forward PSD95HAAcc65IF and reverse Psd95HABglIIR primers using the BAC bMQ239c12 (Adams et al, 2005) as a template. Both homology arms were cloned into the TAP tag pneoflox vector, leaving in between the TAP tag sequence, 2loxP sites, PGK and EM7 promoters, the G418r gene and a SV40 polyadenylation site. The cassette flanked by two homology arms was removed and transformed into EL350 E. coli cells containing a pTargeter vector with the genomic PSD-95 sequence cr11 69851809 to cr11 69861137 (Ensembl release 47). The cassette was inserted into the pTargeter vector by recombination (Knuesel et al, 2003). The final vector containing a 5′-end homolog PSD-95 sequence of 6384 bp and a 3′-end homology arm of 2946 bp (ENSMUSG00000020886) was linearized with PvuI enzyme and electroporated into E14 ES cells. Sixteen neomycin-resistant colonies from 252 were cloned, expanded and frozen. Genomic DNA was extracted from all of them and PCR was carried out using pneoF3 and Psd95R3 to identify TAP-tagged PSD-95 homologous recombinants. One of the ES-cells-positives clones was microinjected into C57BL/6 blastocysts and this generated nine germline chimeras containing 30–70% of targeted cells. These chimeras were crossed to an MF1 genetic background. Tail DNA from the litters was extracted and analyzed by PCR with a 5′ Psd95F5 primer and two 3′ pneoR4 and Psd95R6 primers to distinguish the PSD-95 TAP (+/−) and wild-type alleles (+/+).
Tandem affinity purification
For each independent purification, two forebrains were homogenized in DOC buffer (50 mM Tris pH 9.0, 1% sodium deoxycholate, 50 mM NaF, 20 μM ZnCl, 1 mM Na3VO4, 2 mM Pefabloc SC (Roche) and 1 tablet/10 ml protease inhibitor cocktail tablets (Roche)) and clarified as described earlier (Husi and Grant, 2001). A total of 25 mg of protein was incubated Dynal beads coupled with FLAG antibody for 2 h at 4°C. The resin was washed with three cycles of 15 resin volumes of DOC buffer and twice with TEV-protease cleavage buffer (Invitrogen). The tagged protein was cut from the beads by addition of TEV protease and the protein eluate was collected. The eluate was dialyzed against 2 L of dialysis buffer (50 mM sodium phosphate pH 8.0, 50 mM NaCl) at 4°C with constant agitation. After dialysis the supernatant was collected and added to Ni2+–NTA–agarose resin (Qiagen) pre-washed thrice with the dialysis buffer. The coupling was carried out for 40 min at 4°C with constant agitation in batch, then collected with supernatant and packed into 5 ml plastic columns (Clontech). After sedimentation, the supernatant was collected by gravity flow and the resin was washed thrice with wash buffer containing 0.1% sodium deoxycholate and 1 mM of imidazole. The elution was carried out with 750 μl of elution buffer and fractions were recovered.
All imidazole eluted fractions from the tandem purification that contained PSD-95 were pooled, concentrated in a Vivaspin concentrator (Vivascience, GE), reduced with DTT, alkylated with iodoacetamide and separated by one-dimensional SDS-electrophoresis 4–12% (NUPAGE, Invitrogen, CA). The gel was fixed and stained with colloidal Coomassie and entire gel lanes corresponding to the single-step and tandem purifications from PSD-95TAP/TAP, and wt forebrains were cut into slices and each slice was destained and digested overnight with trypsin (Roche, Trypsin modified, sequencing grade). A solution digest was carried out on the same quantity of starting material as the gel analyzed PSD-95TAP−TAP and wt purifications. Solution digests were carried out using sequencing grade, modified trypsin (Promega) for 4.5 h at 37°C.
LC-MS/MS analysis
Peptides extracted from gel slices were separated on a 40 min RP gradient and solution digests were separated on a 120-min gradient using a PepMap C18 column (75-μm inner diameter × 15 cm; LC Packings). LC-MS/MS analysis was performed on an LTQ-FT (Thermo) mass spectrometer in which the top five most intense ions in a given chromatographic window were subjected to MS/MS sequencing. A total of 102 LC-MS/MS analyses were performed and 59 885 MS/MS spectra were acquired. All data were processed using BioWorks V3.2 (Thermo) and searched using Mascot V2.1 (Matrix science) against mouse IPI sequence database (June, 2007). False discovery rates determined by reverse database searches and empirical analyses of the distributions of mass deviation and Mascot Ion Scores were used to establish score and mass accuracy filters. Only proteins with two or more approved unique peptides were accepted for gel-analysed samples, whereas proteins with only one approved peptide found in a solution digest experiment were required to be present in another replicate analysis of the same solution digest with a different unique peptide. Application of these filters to the PSD-95TAP/TAP datasets resulted in a <1% false discovery rate as assessed by reverse database searching. In general, proteins that were identified in both PSD-95TAP/TAP and control experiments were not accepted. In cases, however, when a protein was found specifically in the tandem TAP-PSD-95 purification but not in the control tandem purification, but was present in both the one-step TAP-PSD-95 purification and in the control single-step purification, then the ratio of approved peptides in the one-step TAP-PSD-95 purification/single-step control purification had to be >3 in order for the protein to be accepted in the tandem purification. Protein hits from all datasets were blast-clustered using a threshold of 95% sequence homology over at least 50% of sequence length.
The data for this manuscript are open access according to the Science Commons CC0 license and can be downloaded from the Tranche network (http://tranche.proteomecommons.org) using the following hash: (J9KSi8FHLDgHFyl2zz1LRq332aRrhVZl/cgPIJAO5WG8tzhAhlrwxvHJOjnre8hIAKLF RTY11dRkXdIEtnkrlqUbg7gAAAAAAAA8fw==). In addition, the raw data are available in Peptide Atlas (http://peptideatlas.org/repository). The protein interaction data have been submitted to the IMEx consortium through the IntAct molecular interaction database (http://www.ebi.ac.uk/intact) and assigned the identifier IM-11694.
emPAI calculation
EmPAI values were calculated for each protein as described (Ishihama et al, 2005) using the following formula: emPAI=10PAI-1, where PAI=number of observed peptides/number of observable peptides. The ratio of protein emPAI vales for proteins in the tandem purification/single-step purification was calculated using emPAI values that were normalized to the total protein emPAI in each set and then normalized to the emPAI value of the bait protein (PSD-95). The tandem-enriched set contained proteins specifically identified in the tandem plus proteins identified in both tandem and single-step purifications with an emPAI ratio (tandem/single step) of >0.5, 126 proteins, and the tandem-depleted set contained proteins specifically identified in the single step plus proteins identified in both tandem and single with an emPAI ratio (tandem/single step) of <0.5, 175 proteins.
Network building and analysis
Proteins appearing in three or more of the replicate experiments were used for the model. Out of these 118, five had gene ambiguity (Cpne4, Tuba1a, Tubb2b, Uba52, and Vamp2) and were removed. A final set of 113 proteins was used. Protein interaction data were sourced from the UniHi database (November 2008) (Chaurasia et al, 2007), with existing curated interactions in the NRC/MASC complex described by Pocklington et al, 2006 and the additional interactions obtained from literature. All interactions were manually re-curated. Interactions sourced from UniHi were traced back to their original database entries and the supporting reference was curated. No high-throughput yeast 2 hybrid data were used unless confirmed by other techniques.
Clustering was performed with the Newman & Girvan algorithm (Newman and Girvan, 2004), using edge betweenness. The modularity coefficient (Q) for the clustering configuration used was 0.37. We found clustering configurations with higher values (up to 0.42); however, these configurations did not reflect the functional organization of the network as well as the one used. With the latter in mind, and also on the basis of the observation that 0.37 was over the average of examined configurations, we decided to use that. A shortest path is defined as a path between two nodes such that the number of its constituent edges is minimized. The average shortest path of a node was calculated as the average of the shortest paths between the node and all the other nodes in the network. Graphical representation of the network was produced by Cytoscape.
Annotation overlap significance was performed according to Pocklington et al (2006). The significance of the overlap between a pair of annotations (e.g. ‘Glutamate Receptors' and ‘Schizophrenia') was evaluated by calculating its probability of occurrence under a random distribution. If within a set of N proteins, na and nb possess the annotations a and b, respectively, and both annotations are distributed randomly in the set the probability of nab proteins possessing both annotations is given by
Given the actual number of proteins possessing the both annotations, mab, we can estimate the significance by calculating the probability P(mab) of an overlap as or less likely under the random distribution.
Additional methods
Detailed description of additional methods is available in the Supplementary information. We described the electrophysiology analysis, immunoprecipitation, immunoblotting, immunocytochemistry and immunohistochemistry. Primers used in this work are summarized in Supplementary Table 7.
Supplementary Material
Supplementary figures S1-6, Supplementary table legends for tables S1-7, Materials & Methods
Supplementary table S1
Supplementary table S2
Supplementary table S3
Supplementary table S4
Supplementary table S5
Supplementary table S6
Supplementary table S7
Acknowledgments
We thank Dr M Pardo for advice on purification conditions and information on cleaved retained proteins, Dr L Yu and S Swamy for support in mass spectrometry analysis, K Porter for tissue perfusion, J Robinson and K Elsegood for mouse colony management, Dr A Enright for protein interaction data, and N Afinowi for the isolation of primary neurons. EF was supported by a Federation of European Biochemistry Societies postdoctoral fellowship; JSC, MOC, MVK, NHK, MDRC and SGNG were supported by the Wellcome Trust; RU was supported by Marie Curie Actions: Research Training Network programs. LZ was supported by the EPSRC/MRC Doctoral Training Centre in Neuroinformatics and Computational Neuroscience.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A (2005) A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758 [DOI] [PubMed] [Google Scholar]
- Angrand PO, Segura I, Volkel P, Ghidelli S, Terry R, Brajenovic M, Vintersten K, Klein R, Superti-Furga G, Drewes G, Kuster B, Bouwmeester T, Acker-Palmer A (2006) Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol Cell Proteomics 5: 2211–2227 [DOI] [PubMed] [Google Scholar]
- Beique JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL (2006) Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci USA 103: 19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence M, Arbuckle MI, Dickson KS, Grant SG (2005) Analyses of murine postsynaptic density-95 identify novel isoforms and potential translational control elements. Brain Res Mol Brain Res 133: 143–152 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G (2004) Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem 279: 12804–12811 [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Fink AE, Grant SG, O'Dell TJ (2008) Opposing effects of PSD-93 and PSD-95 on long-term potentiation and spike timing-dependent plasticity. J Physiol 586: 5885–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia G, Iqbal Y, Hanig C, Herzel H, Wanker EE, Futschik ME (2007) UniHI: an entry gate to the human protein interactome. Nucleic Acids Res 35: D590–D594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GI, Gingras AC (2007) Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42: 298–305 [DOI] [PubMed] [Google Scholar]
- Chen X, Yuan LL, Zhao C, Birnbaum SG, Frick A, Jung WE, Schwarz TL, Sweatt JD, Johnston D (2006a) Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J Neurosci 26: 12143–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YI, Maika SD, Stevens SW (2006b) Epitope tagging of proteins at the native chromosomal loci of genes in mice and in cultured vertebrate cells. J Mol Biol 361: 412–419 [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF (2006) Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Huang YH, Lau LF, Huganir RL (2004) Regulation of the NMDA receptor complex and trafficking by activity-dependent phosphorylation of the NR2B subunit PDZ ligand. J Neurosci 24: 10248–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem 97 (Suppl 1): 16–23 [DOI] [PubMed] [Google Scholar]
- Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, Choudhary JS, Grant SG (2005) Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem 280: 5972–5982 [DOI] [PubMed] [Google Scholar]
- Coyle JT (2006) Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol 26: 365–384 [DOI] [PubMed] [Google Scholar]
- Cuthbert PC, Stanford LE, Coba MP, Ainge JA, Fink AE, Opazo P, Delgado JY, Komiyama NH, O'Dell TJ, Grant SG (2007) Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci 27: 2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosemeci A, Makusky AJ, Jankowska-Stephens E, Yang X, Slotta DJ, Markey SP (2007) Composition of the synaptic PSD-95 complex. Mol Cell Proteomics 6: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakas R, Prisco M, Baserga R (2005) A modified tandem affinity purification tag technique for the purification of protein complexes in mammalian cells. Proteomics 5: 132–137 [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364–1368 [PubMed] [Google Scholar]
- Emes RD, Pocklington AJ, Anderson CN, Bayes A, Collins MO, Vickers CA, Croning MD, Malik BR, Choudhary JS, Armstrong JD, Grant SG (2008) Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat Neurosci 11: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr CD, Gafken PR, Norbeck AD, Doneanu CE, Stapels MD, Barofsky DF, Minami M, Saugstad JA (2004) Proteomic analysis of native metabotropic glutamate receptor 5 protein complexes reveals novel molecular constituents. J Neurochem 91: 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, O'Brien C, Ohno M, Kirkwood A, Silva AJ (2001) Alpha-CaMKII-dependent plasticity in the cortex is required for permanent memory. Nature 411: 309–313 [DOI] [PubMed] [Google Scholar]
- Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M (2006) Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 313: 1792–1795 [DOI] [PubMed] [Google Scholar]
- Garry EM, Moss A, Delaney A, O'Neill F, Blakemore J, Bowen J, Husi H, Mitchell R, Grant SG, Fleetwood-Walker SM (2003) Neuropathic sensitization of behavioral reflexes and spinal NMDA receptor/CaM kinase II interactions are disrupted in PSD-95 mutant mice. Curr Biol 13: 321–328 [DOI] [PubMed] [Google Scholar]
- Greene R (2001) Circuit analysis of NMDAR hypofunction in the hippocampus, in vitro, and psychosis of schizophrenia. Hippocampus 11: 569–577 [DOI] [PubMed] [Google Scholar]
- Hunt CA, Schenker LJ, Kennedy MB (1996) PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J Neurosci 16: 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Grant SG (2001) Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J Neurochem 77: 281–291 [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 3: 661–669 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M (2004) PDZ domain proteins of synapses. Nat Rev Neurosci 5: 771–781 [DOI] [PubMed] [Google Scholar]
- Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA (2007) Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron 54: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemmer P, Smit AB, Li KW (2009) Proteomics analysis of immuno-precipitated synaptic protein complexes. J Proteomics 72: 82–90 [DOI] [PubMed] [Google Scholar]
- Knuesel M, Wan Y, Xiao Z, Holinger E, Lowe N, Wang W, Liu X (2003) Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol Cell Proteomics 2: 1225–1233 [DOI] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, O'Dell TJ, Grant SG (2002) SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci 22: 9721–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH (1995) Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269: 1737–1740 [DOI] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW (1998) SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem 273: 19518–19524 [DOI] [PubMed] [Google Scholar]
- Lim IA, Hall DD, Hell JW (2002) Selectivity and promiscuity of the first and second PDZ domains of PSD-95 and synapse-associated protein 102. J Biol Chem 277: 21697–21711 [DOI] [PubMed] [Google Scholar]
- Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, Grace AA (2008) Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci 31: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi A, Chittajallu R, Duprat F, Palmer MJ, Benke TA, Kidd FL, Henley JM, Isaac JT, Collingridge GL (1999) Hippocampal LTD expression involves a pool of AMPARs regulated by the NSF-GluR2 interaction. Neuron 24: 389–399 [DOI] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG (1998) Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396: 433–439 [DOI] [PubMed] [Google Scholar]
- Nehring RB, Wischmeyer E, Doring F, Veh RW, Sheng M, Karschin A (2000) Neuronal inwardly rectifying K(+) channels differentially couple to PDZ proteins of the PSD-95/SAP90 family. J Neurosci 20: 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys 69: 026113. [DOI] [PubMed] [Google Scholar]
- Noel J, Ralph GS, Pickard L, Williams J, Molnar E, Uney JB, Collingridge GL, Henley JM (1999) Surface expression of AMPA receptors in hippocampal neurons is regulated by an NSF-dependent mechanism. Neuron 23: 365–376 [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP (2003) PDZ domain proteins: plug and play!. Sci STKE 2003: RE7. [DOI] [PubMed] [Google Scholar]
- Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, Ziff EB (1998) The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron 21: 99–110 [DOI] [PubMed] [Google Scholar]
- Paulo J, Brucker W, Hawrot E (2009) Proteomic analysis of an 7 nicotinic acetylcholine receptor interactome. J Proteome Res 8: 1849–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M (2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 279: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Pocklington AJ, Cumiskey M, Armstrong JD, Grant SG (2006) The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behaviour. Mol Syst Biol 2, 2006 0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim MJ (2002) Postsynaptic signaling and plasticity mechanisms. Science 298: 776–780 [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF (2006) Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127: 831–846 [DOI] [PubMed] [Google Scholar]
- Terpe K (2003) Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 60: 523–533 [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics 7: 684–696 [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF (1999) Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23: 583–592 [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW Jr, Mercer JA, Kennedy MB (2000) Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci 20: 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Wang T, Gu S, Ronni T, Du YC, Chen X (2005) In vivo dual-tagging proteomic approach in studying signaling pathways in immune response. J Proteome Res 4: 941–949 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hoffman DA, Migliore M, Johnston D (2002) Dendritic K+ channels contribute to spike-timing dependent long-term potentiation in hippocampal pyramidal neurons. Proc Natl Acad Sci USA 99: 8366–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens KM, Lin H, Liao D (2005) Rac1 induces the clustering of AMPA receptors during spinogenesis. J Neurosci 25: 10627–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG (2004) Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron 41: 625–638 [DOI] [PubMed] [Google Scholar]
- Zhou D, Ren JX, Ryan TM, Higgins NP, Townes TM (2004) Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res 32: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-6, Supplementary table legends for tables S1-7, Materials & Methods
Supplementary table S1
Supplementary table S2
Supplementary table S3
Supplementary table S4
Supplementary table S5
Supplementary table S6
Supplementary table S7