Abstract
Sweet potato (Ipomoea batatas) is an important subsistence and famine reserve crop grown in developing countries where Sweet potato chlorotic stunt virus (SPCSV; Closteroviridae), a single-stranded RNA (ssRNA) crinivirus, synergizes unrelated viruses in co-infected sweet potato plants. The most severe disease and yield losses are caused by co-infection with SPCSV and a potyvirus, Sweet potato feathery mottle virus (SPFMV; Potyviridae). Potyviruses synergize unrelated viruses by suppression of RNA silencing with the P1/HC-Pro polyprotein; however, the SPCSV-SPFMV synergism is unusual in that the potyvirus is the beneficiary. Our data show that transformation of an SPFMV-resistant sweet potato variety with the double-stranded RNA (dsRNA)-specific class 1 RNA endoribonuclease III (RNase3) of SPCSV broke down resistance to SPFMV, leading to high accumulation of SPFMV antigen and severe disease symptoms similar to the synergism in plants co-infected with SPCSV and SPFMV. RNase3-transgenic sweet potatoes also accumulated higher concentrations of 2 other unrelated viruses and developed more severe symptoms than non-transgenic plants. In leaves, RNase3 suppressed ssRNA-induced gene silencing (RNAi) in an endonuclease activity-dependent manner. It cleaved synthetic double-stranded small interfering RNAs (siRNAs) of 21, 22, and 24 bp in vitro to products of approximately 14 bp that are inactive in RNAi. It also affected total siRNA isolated from SPFMV-infected sweet potato plants, suggesting a viral mechanism for suppression of RNAi by cleavage of siRNA. Results implicate RNase3 in suppression of antiviral defense in sweet potato plants and reveal RNase3 as a protein that mediates viral synergism with several unrelated viruses, a function previously described only for P1/HC-Pro.
Keywords: plant virus, RNA silencing, suppression of RNAi, viral synergism
Sweet potato (Ipomoea batatas) is one of the most important food crops in the world (annual production 122 million tons; http://faostat.fao.org). In developing countries, it is a famine-reserve crop and consumed by the resource-poor rural populations for subsistence. The only severe disease affecting sweet potatoes and thereby food security is caused by a virus complex that can reduce yields by 90% (1). This sweet potato virus disease (SPVD) characterized by severe leaf malformation, chlorosis, and stunting develops only in plants infected with SPCSV (family Closteroviridae), a single-stranded RNA (ssRNA) virus that causes synergistic diseases with many unrelated viruses (2–4), and for which no resistance has been found in sweet potato germplasm. The worst symptoms and yield losses develop in plants co-infected with SPCSV and an unrelated ssRNA virus, SPFMV (family Potyviridae). Consequently, titers of SPFMV increase by 2–3 orders of magnitude and severe symptoms develop (2, 3, 5). These results indicate a general loss of resistance to viruses in sweet potato plants infected with SPCSV.
Viruses are both inducers and targets of RNA interference (RNAi), a fundamental antiviral defense mechanism in eukaryotic organisms (6). RNAi is a cytoplasmic cell surveillance system to recognize double-stranded RNA (dsRNA) and specifically eliminate by cleavage RNAs homologous to the inducer RNA (7, 8). Cleavage of dsRNA is carried out by Dicer, which is a class 3 RNase III endonuclease (9). Plants encode 4 Dicer-like (DCL) enzymes that recognize and cleave long dsRNA molecules to 21-, 22-, and 24-bp fragments that act as small interfering RNAs (siRNAs) (10–13). A primary silencing siRNA binds to a ribonuclease H–like protein (Argonaute) and is used to detect homologous ssRNA molecules for cleavage (13). In plants, RNAi becomes amplified via transitivity when the cleaved RNA recruits an RNA-directed RNA polymerase to generate dsRNA, which is cleaved by a DCL protein to produce secondary siRNAs (14). Transitivity pathways are essential for certain types of transgene-induced silencing in plants and have a key role in defense against viruses (12, 14–16). In general, RNAi pathways are essential for virus resistance and recovery from virus disease in plants (17, 18).
Many sweet potato cultivars are highly resistant to infection by SPFMV or are able to recover from initial infection during plant growth (1, 2), which is characteristic of RNAi-based antiviral resistance (17, 18). However, resistance to SPFMV severely diminishes following infection with SPCSV. Plants fail to recover from infection by SPFMV and SPVD develops (2). Viruses express a wide range of dedicated RNAi-suppressor proteins (RSP) to interfere with the different steps of the RNAi pathway (19–22). It is therefore conceivable that in mixed viral infections, the presence of several RSPs might help to overcome RNAi, generating a synergism that allows at least one of the co-infecting viruses to accumulate at higher titers than observed in single-virus infections (21, 23). However, so far this has been shown only for the N-proximal part (P1/HC-Pro) of the potyviral polyprotein that is known as the potent and sufficient mediator of synergism in transgenic plants infected with unrelated viruses (23). The central part of HC-Pro that mediates viral synergism is involved in suppression of RNAi (24, 25). However, the unidirectional synergy of SPCSV and SPFMV is unusual in that the concentration of the potyviral component increases by 2–3 orders of magnitude, whereas the titer of the non-potyviral component remains constant or slightly decreases (3, 5). In addition, SPCSV synergizes several unrelated viruses, such as Sweet potato mild mottle virus (SPMMV, genus Ipomovirus) (3, 4) and Sweet potato chlorotic fleck virus (SPCFV, genus Carlavirus) (4), which enhances the concentrations of these viruses and the severity of symptoms. These data suggest that SPCSV encodes a suppressor of a basic antiviral defense system of sweet potato that normally protects the plants. The aim of the present study was to address the question how SPCSV causes the dramatic general loss of sweet potato resistance to SPFMV.
Results
RNase3 Alone Predisposes Sweet Potato Plants to SPVD.
We found previously that SPCSV encodes a unique class 1 RNase III enzyme (RNase3) whose endonuclease activity enhances RNAi suppression caused by another protein (p22) that also is encoded by SPCSV (26). Recent studies have revealed that many SPCSV isolates that lack p22 still synergize with unrelated viruses (4, 5), indicating that p22 is dispensable for synergy between SPCSV and other viruses. Therefore, the aim of the current study was to elucidate the possible involvement of RNase3 in the synergism and development of SPVD.
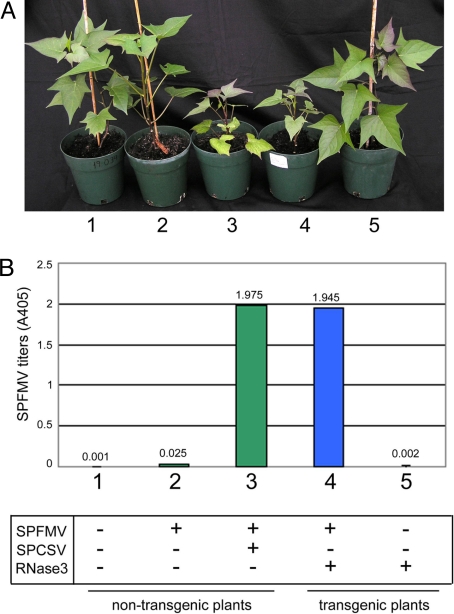
We chose sweet potato variety ‘Huachano’ for the study because it is extremely resistant to SPFMV. It accumulates detectable titers of SPFMV only temporarily at 3 weeks post-inoculation but subsequently recovers from infection. This cultivar was genetically transformed with RNase3. The transgenic plants grew normally and were indistinguishable from the wild-type non-transgenic plants (Fig. 1A and Fig. S1A). They accumulated detectable amounts of the RNase3 protein (Fig. S1B). Following inoculation with SPFMV, the RNase3-transgenic plants accumulated high titers of SPFMV (Figs. 1B and Fig. S2), developed very severe symptoms of SPVD (chlorosis, heavily stunted growth) and did not recover. In contrast, the non-transgenic plants (Fig. 1 A and B) and plants transformed to express the p22 protein recovered from the initial systemic infection with SPFMV (Fig. S2). Disease symptoms and SPFMV titers in RNase3-transgenic plants were similar to non-transgenic plants co-infected with SPCSV and SPFMV (Fig. 1). Therefore, RNase3 alone was sufficient to eliminate resistance to SPFMV and predispose the plant to SPVD. Furthermore, the transgenic plants accumulated higher, readily detectable amounts of virus when inoculated with SPMMV and SPCFV and also displayed discernible symptoms, which was not observed with the non-transgenic plants inoculated with these viruses (Fig. S3).
Fig. 1.
Symptoms of sweet potato virus disease (SPVD) and accumulation of Sweet potato feathery mottle virus (SPFMV) in non-transgenic and transgenic sweet potato expressing the RNase3 protein of Sweet potato chlorotic stunt virus (SPCSV). Sweet potato plants were grown from cuttings taken from graft-inoculated plants. (A) Plants infected with SPCSV and/or SPFMV or transgenic for RNase3 and infected with SPFMV were photographed 6 weeks after planting in soil. (B) SPFMV titers were measured by double antibody sandwich ELISA in the uppermost fully opened leaves of plants 7 weeks after graft inoculation.
RNase3 Suppresses ssRNA-Mediated RNAi.
The ability of RNase3 to suppress plant antiviral defense and increase accumulation of SPFMV suggested a role for RNase3 in suppression of RNAi. However, our previous studies indicated that RNase3 could not suppress RNAi induced by overexpression of ‘hairpin’ dsRNA in leaves (26), which has been reported for the class 1 RNase III of Escherichia coli (27) that is homologous to RNase3 (Fig. S4). Therefore, we tested whether RNase3 could interfere with ssRNA-induced silencing (co-suppression), the originally described form of posttranscriptional silencing of endogenous genes (28) and for which a predominant role in plant defense against viruses has been suggested (12, 15).
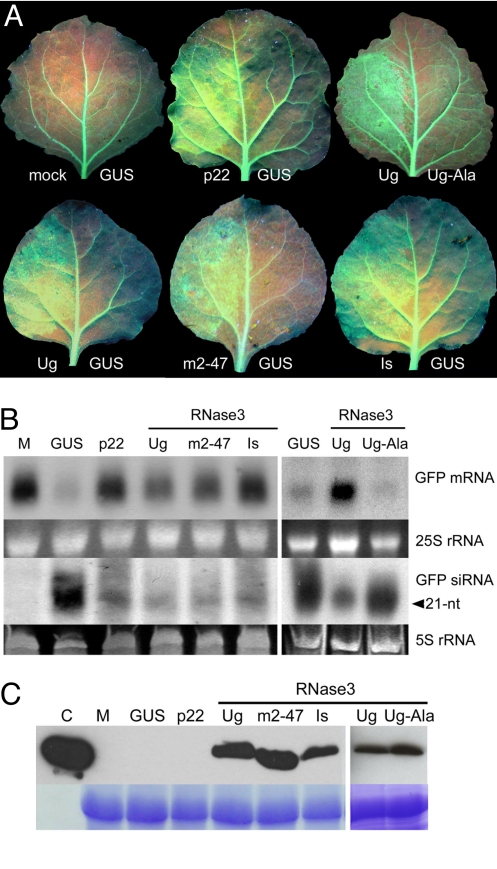
Leaves of transgenic Nicotiana benthamiana that constitutively express the jellyfish green fluorescent protein (GFP) (20) were infiltrated with Agrobacterium tumefaciens engineered to overexpress gfp mRNA. Consequently, GFP fluorescence initially increased, but then decreased substantially by 3 days post-infiltration (d.p.i) (Fig. 2A). Similarly, gfp mRNA levels were substantially reduced, whereas the gfp-homologous siRNA accumulated to high levels, consistent with co-suppression (Fig. 2B). However, when leaf tissues were co-infiltrated with Agrobacterium strains for expression of GFP and RNase3, GFP fluorescence (Fig. 2A) and mRNA expression (Fig. 2B) remained high at 3 d.p.i., and only small amounts of gfp-homologous siRNA were detected (Fig. 2B).
Fig. 2.
RNase3 from SPCSV suppresses RNA silencing. (A) The left sides of Nicotiana benthamiana leaves were mock-infiltrated with buffer or agroinfiltrated with an Agrobacterium tumefaciens strain expressing gfp and a strain expressing p22 of a Ugandan isolate of SPCSV (Ug) (a positive control) or RNase3 of isolate Ug, M2–47 (Peru) or Is (Israel). The right sides of the leaves were agroinfiltrated for expressing gfp and β-glucuronidase (GUS) as a negative control or the RNase3-Ala mutant defective for endonuclease activity (Ug-Ala). The transgenic N. benthamiana plants (line 16c) constitutively express gfp (green fluorescence in veins of the leaf at the upper left corner and the right sides of the leaves). Leaves were photographed and analyzed 3 days post-infiltration. (B) Northern blot of gfp mRNA and siRNA in the leaf tissues illustrated in A. M indicates leaf tissue of the 16c line mock-infiltrated with buffer. Upper shows the accumulation of gfp mRNA in the respective infiltrated regions. Lower shows accumulation of gfp-derived siRNA. Ethidium bromide-stained gels of rRNA were used as loading controls. (C) Western blot of the RNase3 protein in the infiltrated tissues. C indicates purified recombinant RNase3 protein (positive control). (Lower) Coomassie blue–stained gel (control for equal loading of samples).
Endonuclease Activity of RNase3 Is Needed for Suppression of RNAi.
We have shown previously (26) that 2 point mutations introduced into the endonuclease signature motif of RNase3 (Fig. S4) abolish RNase III activity, which was reconfirmed in the present study (Fig. S5). When this mutated protein (RNase3-Ala) was expressed in leaf tissue, only constitutive expression levels of GFP were observed, suggesting that without endonuclease activity RNase3 was not able to protect gfp mRNA from co-suppression (Fig. 2 A and B). Hence, the siRNA-binding ability retained by RNase3-Ala (26) was not sufficient to suppress silencing (Fig. 2B). These data show that RNase3 interferes with RNAi initiated by ssRNA and that the endonuclease activity of RNase3 is required for this interference.
RNase3 Cleaves siRNA.
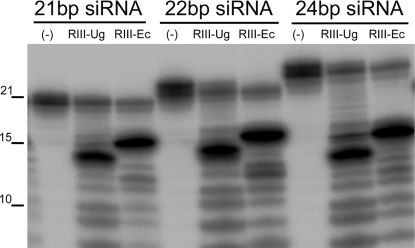
Because the endonuclease activity of RNase3 was crucial for suppression of RNAi, we tested whether RNase3 could target and modify the double-stranded siRNAs essential for RNAi (12). Using purified recombinant RNase3 protein and the RNase3-Ala mutant protein, we observed that synthetic siRNAs of 21, 22, and 24 bp were all cleaved to products of approximately 14 bp by RNase3 (Fig. 3) but not RNase3-Ala (Fig. S5). The cleavage products were smaller than those produced by the E. coli RNase III (≈15 nucleotides) (Fig. 3). It has been shown in many previous studies that RNAs smaller than 20 nucleotides are inefficient triggers of RNAi (29–31). Treatment of siRNA isolated from SPFMV-affected sweetpotato plants (Fig. S5C) with RNase3 caused a clear reduction of the siRNA pool of 21–25 nucleotides, in contrast to the treatment with RNase3-Ala (Fig. S5C). Detection with an SPFMV-specific probe revealed that the amounts of virus-specific siRNAs following treatment with RNase3 were only slightly less than with RNase3-Ala (Fig. S5C). These data indicated that the majority of siRNA cleaved by RNase3 was of host origin, and SPFMV-derived double-stranded siRNA might be in a minority in the pool of total SPFMV-derived siRNA. To reveal the proportions of SPFMV-derived double-stranded and single-stranded siRNA in SPFMV-infected sweet potato leaves, small RNAs of 20–30 nucleotides were isolated by polyacrylamide gel purification and sequenced on the Illumina Genome Analyzer. The resulting sequences were mapped to the complete sequence of SPFMV-Piu. Data showed that the proportion of SPFMV-derived double-stranded siRNA (containing 2-nucleotide 3′ overhangs) was only 3.95% of the total pool of SPFMV-specific siRNA.
Fig. 3.
RNase3 of SPCSV cleaves synthetic double-stranded small interfering RNAs (siRNAs). 32P-labeled double-stranded siRNAs of the indicated sizes were incubated for 1 h with purified recombinant RNase3 proteins of SPCSV (RIII-Ug) or E. coli (RIII-Ec). Numbers at the left of the figure indicate the sizes in nucleotides based on a carbonate-treated ssRNA oligonucleotide. Samples were analyzed by electrophoresis in a TBE-UREA gel as described in SI Materials and Methods. (−), untreated double-stranded siRNA.
Discussion
The RNase III enzyme of SPCSV is the second viral protein directly implicated in viral synergism in plants. The first was the P1/HC-Pro polyprotein of an unrelated virus family (Potyviridae) (23, 24) whose ability to mediate synergism suggested interference with RNAi. Suppression of RNAi was subsequently shown for HC-Pro (19–21) and many other RSPs from a wide range of plant and animal viruses (22), but no other RSPs besides P1/HC-Pro were reported as causal agents of synergistic viral diseases. The synergism involving RNase3 is unusual. While P1/HC-Pro mediates the typical synergism in which potyviruses enhance the titers of unrelated viruses, RNase3-mediated synergism represents a viral interaction in which the potyvirus is the beneficiary. Our data show that RNase3 also mediates synergism with viruses of other taxa known to synergize with SPCSV. These data suggest that SPCSV encodes a suppressor of a basal antiviral defense system of sweet potato that normally protects the plants.
Viral RSPs may bind siRNA (32), affect their methylation status and stability (33), and interfere with formation of the effector complexes required for RNAi (12, 22). These modes of action are known or suggested for RSPs encoded by many viral taxa (22), including those related to the sweet potato viruses that cause synergistic diseases in co-infection with SPCSV (3, 4). However, the viral RNase III endonuclease enzyme of SPCSV is the first viral RSP found to destroy siRNAs. A search of sequence databases revealed animal DNA viruses (family Iridoviridae) that contain genes predicted to encode class 1 RNase III enzymes (Fig. S4). To our knowledge, these RNases also suppress RNAi. These data, along with the fact that some DNA viruses that infect algae encode class 1 RNase III enzymes (Fig. S4), suggest that these endonucleases might play a role in suppression of RNAi in unrelated viruses infecting many types of host organisms. The viral RNase III endonuclease enzymes are not completely without precedent as RSPs because an exonuclease of Caenorhabditis elegans (ERI-1) and its human orthologue degrade siRNAs in vitro (34). Mutation of eri-1 gene results in higher accumulation of siRNA (34). Other exonucleases degrading small regulatory RNAs are also involved in downregulation of the silencing pathways (35).
The different and possibly complementary actions of RSPs encoded by SPCSV and other viruses such as SPFMV may be the ultimate cause of major failure of antiviral defense and the subsequent development of unusually severe diseases in sweet potato plants. The RNase3 protein is different from the HC-Pro protein and suppresses silencing by a mechanism that is unrelated to HC-Pro. HC-Pro inhibits the siRNA-initiated RNA silencing complex assembly pathway, possibly by binding siRNA and preventing RNA silencing initiator complex formation (36). Expression of HC-Pro also greatly enhances the accumulation of endogenous miRNAs in plants, which suggests that potyviruses control plant gene expression to suppress defense and/or facilitate utilization of host resources required during the infection cycle (37). RNA-binding is not sufficient for suppression of RNAi by RNase3 but its endonuclease activity is required. Furthermore, because RNase3 cannot efficiently suppress RNAi that is induced by overexpressed dsRNA (26), it is possible that the endonuclease activity of RNase3 interferes with components of the ssRNA-induced silencing pathway, RNA-directed RNA polymerase dependent transitivity, production of secondary siRNAs and amplification of silencing (12, 14, 15).
Our results from an RNase3 cleavage assay on the pool of siRNA isolated from sweet potato leaves demonstrated that RNase3 can act on host-derived siRNA and/or miRNA, and obviously the double-stranded forms of them since RNase3 does not cleave long or short ssRNA strands (26). The analyzed pool of siRNA included SPFMV-derived double-stranded siRNA but the proportion of them in the total siRNA derived from SPFMV was only 3.95%. These results suggest that RNase3 may synergize SPFMV and other viruses by targeting a specific host component via interference with small-RNA biogenesis in a manner which other viruses are unable to do and which releases a key obstacle that prevents other viruses from getting an upper hand on silencing. An alternative hypothesis is that the phloem-limited SPCSV (2) targets SPFMV-derived double-stranded siRNA in vascular tissue, prevents systemic signaling for silencing and, hence, makes antiviral defense inefficient. Consequently, SPFMV accumulates in high titers in the course of systemic infection and suppresses silencing with its own RSP, HC-Pro. Identification of the specific target of RNase3 remains as an interesting topic for further study.
The results of this study provide a mechanistic understanding of synergism that is addressed to an important disease of a subsistence crop in developing countries. Identification of a viral class 1 RNase III enzyme as a key factor behind the severe disease and yield losses to which SPCSV predisposes sweetpotato plants suggests possibilities for disease control. This is important because extensive screening of sweet potato germplasm for sources of resistance, and conventional approaches of engineered, pathogen-derived resistance used in sweet potato varieties (38) have rendered little progress possible toward durable resistance to SPCSV and SPVD.
Materials and Methods
Transgenic Sweet Potato Lines Expressing SPCSV RNase3.
Pathogen-free in vitro plants of the SPFMV-resistant Peruvian sweet potato landrace ‘Huachano’ (accession no. CIP420065) (38) were obtained from the germplasm collection of The International Potato Center (CIP). The RNase3 gene of SPCSV-Ug (26) was placed under the Cauliflower mosaic virus 35S promoter in the binary vector pKOH200, as described (26), and used to transform sweet potato leaf explants with A. tumefaciens strain EHA105. Plants were regenerated following a somatic embryogenesis protocol (38).
Plant Inoculation and Virus Detection.
Sweet potato plants were graft-inoculated with the East African strains of SPFMV-Piu and SPCSV-M2–47 (4, 38) using scions from the virus propagation host, Ipomoea setosa, in an insect-proof greenhouse at CIP. SPFMV was detected in 150 mg of the tissue sampled from the youngest fully opened leaves by double antibody sandwich ELISA (DAS-ELISA) (5).
Western Blot Analysis.
Proteins were isolated from 200 mg sweet potato leaf tissue, separated in a denaturing 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Hybond-P) by electroblotting. RNase3 protein was detected with a specific rabbit antiserum raised against SPCSV RNase3, as described (26). Anti-rabbit monoclonal mouse antibodies conjugated with horseradish peroxidase (Amersham), the Supersignal West Pico chemiluminescent substrate (Pierce Biotechnology), and exposure to X-ray film were used to detect signals by the ECL method according to the manufacturer's instructions (Amersham).
Agroinfiltration Assay.
The cloning strategy and vector plasmids used in the agroinfiltration assays in this study have been described (26). Sequences of the RNase3 genes of SPCSV-Ug, SPCSV-M2–47, and SPCSV-Is have been described and encode the most different RNase3 protein sequences currently known (amino acid sequence identity 80–97%) (5). In the mutated RNase3 of SPCSV-Ug (designated as RNase3-Ala),2 substitutions (E37A and D44A) were made in the highly conserved RNase III signature motif required for the dsRNA endonuclease activity of RNase III enzymes (Fig. S4). pA-GUS expresses the ß-glucuronidase (GUS) gene with a plant intron to prevent GUS expression in Agrobacterium. pBIN35S-GFP expressed the “cycle 3” GFP gene. Constructs were verified by sequencing. Agroinfiltration was done as described (39) using different A. tumefaciens cultures that were combined before infiltration (26). For co-infiltration treatments that included fewer constructs than others, the missing volume was replaced by the Agrobacterium strain expressing GUS. Infiltrations were carried out on the N. benthamiana line 16c genetically transformed to constitutively express the jellyfish (Aequoria victoriae) GFP (20) (seeds kindly provided by Prof. D. Baulcombe) in a controlled growth chamber. Infiltrated tissues were monitored daily for GFP fluorescence using a hand-held UV lamp and photographed.
RNA Isolation and Northern Blot Hybridization.
Total RNA was isolated from 400 mg fresh leaf material using TRIzol (Invitrogen) following the manufacturer's instructions. Low molecular weight (LMW) RNA was obtained by LiCl preciptation and used to detect siRNA, whereas high molecular weight (HMW) RNA was used to detect gfp mRNA accumulation as described (10, 26). A probe complementary to gfp was prepared and labeled with [γ-32P]UTP (Amersham) by in vitro transcription of gfp cloned into pCR-Blunt (Invitrogen) behind the T7 promoter. After hybridization and washing, membranes were exposed to X-ray film (Kodak) for 4, 16, and 48 h and developed using an X-Omat 1000 automated developer (Kodak).
RNA Cleavage Assays with RNase3.
The genes RNase3 and RNase3-Ala were cloned in pET11d (Stratagene) to transform E. coli BL21(DE3) cells for expression and purification of the RNase3 and RNase3-Ala proteins, respectively, following induction with isopropyl-β-d-thiogalactoside. Purification of the 6× His-tagged proteins was accomplished using Ni-NTA agarose columns according to the manufacturer's instructions (Expressionist, Qiagen). Protein concentrations were measured using the Bradford assay including known amounts of BSA for comparison. The siRNAs were ordered as single-stranded sense and antisense RNA oligonucleotides (21, 22, and 24 nucleotides). The sense oligonucleotides (10 pmol) were labeled by phosphorylation with [γ-32P]ATP for 30 min using T4 polynucleotide kinase (Fermentas) and purified by gel extraction. Duplexes of the oligonucleotides (dsRNA) were obtained by heating a mixture of labeled sense and unlabeled antisense oligonucleotides at 98 °C for 4 min and cooling at room temperature for 1 h. The double-stranded siRNA substrates were incubated in a reaction mix containing RNase3, RNase3-Ala, or E. coli RNase III (New England Biolabs). The reaction was stopped by boiling, analyzed by electrophoresis on a 20% polyacrylamide gel, and the labeled RNAs visualized using a PhosphorImager (Fuji FLA-5010).
For testing cleavage of siRNA isolated from sweet potato plants, LMW RNA was isolated using TRIzol and LiCl precipitation (as described above), 5 μg was heated to 95 °C for 10 min, and then let to cool for 2 h at room temperature. The LMW RNA samples were treated with RNase3 and RNase3-Ala in the presence of 10 mM MgCl2 for 2 h and separated in a 20% polyacrylamide gel stained with ethidium bromide, and visualized using an Epichemi3-Darkroom (UVP Bioimaging System) gel documentation equipment. SPFMV-specific siRNA was detected using a specific probe labeled with [γ-32P]UTP (see SI Materials and Methods).
To reveal the proportions of SPFMV-derived double-stranded and single-stranded siRNA in SPFMV-infected sweet potato, small RNAs of 20–30 nucleotides in size were isolated by polyacrylamide gel purification, and sequenced on the Illumina Genome Analyzer at Fasteris Life Sciences SA according to the service provider's recommendations. The resulting sequences 21–24 nucleotides in length (> 95% of all sequences) were mapped to the complete sequence of SPFMV-Piu (GenBank accession no. FJ155666) using the program MAQ (http://maq.sourceforge.net). The mapping tables were then imported into Microsoft Excel. Sequence were sorted according to size and polarity and further tabulated into histograms, for each size class and strand polarity, in bins corresponding to the siRNA mapping positions for each nucleotide across the SPFMV genome. The exact number of putative siRNAs could then be extracted by comparing the positive and negative polarity histograms for each size class.
SI.
Detailed experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank J. Turunen, M. Frilander, and M. Björklund for expert advice in small RNA analysis, Ana Perez for help with virus inoculation, M. Saarma for critical comments on the manuscript, and M. Ghislain and I. Barker for supporting the study. This work was supported by Academy of Finland Grants 1102134 and 1110797 and the Howard Buffet Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806042106/DCSupplemental.
References
- 1.Gibson RW, et al. Symptoms, aetiology, and serological analysis of sweet potato virus disease in Uganda. Plant Pathol. 1998;47:95–102. [Google Scholar]
- 2.Karyeija RF, Kreuze JF, Gibson RW, Valkonen JPT. Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweet potato plants. Virology. 2000;269:26–36. doi: 10.1006/viro.1999.0169. [DOI] [PubMed] [Google Scholar]
- 3.Mukasa SB, Rubaihayo PR, Valkonen JPT. Interactions between a crinivirus, an ipomovirus and a potyvirus in co-infected sweetpotato plants. Plant Pathol. 2006;55:458–467. [Google Scholar]
- 4.Untiveros M, Fuentes S, Salazar LF. Synergistic interaction of sweet potato chlorotic stunt virus (Crinivirus) with carla-, cucumo-, ipomo-, and potyviruses infecting sweet potato. Plant Dis. 2007;91:669–676. doi: 10.1094/PDIS-91-6-0669. [DOI] [PubMed] [Google Scholar]
- 5.Cuellar WJ, Tairo F, Kreuze JF, Valkonen JPT. Analysis of gene content in sweet potato chlorotic stunt virus RNA1 reveals the presence of the p22 RNA silencing suppressor in only a few isolates: implications for viral evolution and synergism. J Gen Virol. 2008;89:573–582. doi: 10.1099/vir.0.83471-0. [DOI] [PubMed] [Google Scholar]
- 6.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: Strike and counterstrike. Nature Biotech. 2007;12:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 11.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 12.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 14.Baulcombe DC. Amplified silencing. Science. 2007;315:199–200. doi: 10.1126/science.1138030. [DOI] [PubMed] [Google Scholar]
- 15.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantaleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Covey SN, Al-Kaff NS, Langara A, Turner DS. Plants combat infection by gene silencing. Nature. 1997;285:781–782. [Google Scholar]
- 18.Ratcliff F, Harrison BD, Baulcombe DC. A similarity between virus defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 19.Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 20.Brigneti G, et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Anandalakshmi R, et al. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruss G, Ge X, Shi XM, Carrington JC, Vance VB. Plant viral synergism: The potyviral genome encodes a broad range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell. 1997;9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi XM, Miller H, Verchot J, Carrington JC, Vance VB. Mutations in the region encoding the central domain of helper component proteinase (HC-Pro) eliminate potato virus X/potyviral synergism. Virology. 1997;231:35–42. doi: 10.1006/viro.1997.8488. [DOI] [PubMed] [Google Scholar]
- 25.Kasschau KD, Carrington JC. Long-distance movement and replication maintenance functions correlate with silencing-suppression activity of potyviral HC-Pro. Virology. 2001;285:71–81. doi: 10.1006/viro.2001.0901. [DOI] [PubMed] [Google Scholar]
- 26.Kreuze JF, Savenkov EI, Cuellar WJ, Li X, Valkonen JPT. Viral class 1 RNase III involved in suppression of RNA silencing. J Virol. 2005;79:7227–7238. doi: 10.1128/JVI.79.11.7227-7238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichner Z, Silhavy D, Burgyan J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J Gen Virol. 2003;84:975–980. doi: 10.1099/vir.0.18987-0. [DOI] [PubMed] [Google Scholar]
- 28.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang D, et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogler H, et al. Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J Virol. 2007;81:10379–10388. doi: 10.1128/JVI.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in. C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 35.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakatos L, et al. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallory AC, et al. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA. 2002;99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreuze JF, et al. RNA silencing mediated resistance to a crinivirus (Closteroviridae) in cultivated sweetpotato (Ipomoea batatas) and development of sweetpotato virus disease following co-infection with a potyvirus. Mol Plant Pathol. 2008;9:589–598. doi: 10.1111/j.1364-3703.2008.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansen LK, Carrington JC. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126:930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.