Abstract
Hypoxanthine-guanine phosphoribosyltransferase (HPRT) deficiency results in Lesch-Nyhan disease (LND), where affected individuals exhibit a characteristic neurobehavioral disorder that has been linked with dysfunction of dopaminergic pathways of the basal ganglia. Since the functions of HPRT, a housekeeping enzyme responsible for recycling purines, have no direct relationships with the dopaminergic pathways, the mechanisms whereby HPRT deficiency affect them remain unknown. The current studies demonstrate that HPRT deficiency influences early developmental processes controlling the dopaminergic phenotype, using several different cell models for HPRT deficiency. Microarray methods and quantitative PCR were applied to 10 different HPRT-deficient (HPRT−) sublines derived from the MN9D cell line. Despite the variation inherent in such mutant sublines, several consistent abnormalities were evident. Most notable were increases in the mRNAs for engrailed 1 and 2, transcription factors known to play a key role in the specification and survival of dopamine neurons. The increases in mRNAs were accompanied by increases in engrailed proteins, and restoration of HPRT reverted engrailed expression towards normal levels, demonstrating a functional relationship between HPRT and engrailed. The functional relevance of the abnormal developmental molecular signature of the HPRT− MN9D cells was evident in impoverished neurite outgrowth when the cells were forced to differentiate chemically. To verify that these abnormalities were not idiosyncratic to the MN9D line, HPRT− sublines from the SK-N-BE(2) M17 human neuroblastoma line were evaluated and an increased expression of engrailed mRNAs was also seen. Over-expression of engrailed occurred even in primary fibroblasts from patients with LND in a manner that suggested a correlation with disease severity. These results provide novel evidence that HPRT deficiency may affect dopaminergic neurons by influencing early developmental mechanisms.
INTRODUCTION
Hypoxanthine-guanine phosphoribosyltransferase (HPRT) is a housekeeping enzyme responsible for recycling purines (1). It converts the free purine bases, hypoxanthine and guanine, into utilizable purine nucleotides. It is expressed widely in most organisms from bacteria through mammals. In mammals, it is expressed in virtually all cells throughout the development, beginning at the early blastocyst stage.
Despite nearly ubiquitous expression of HPRT, the consequences of HPRT deficiency in humans predominate in the nervous system. HPRT deficiency is an inborn error of purine metabolism, known as Lesch-Nyhan disease (LND), leading to peculiar neurological and behavioral abnormalities that include compulsive self-injurious behavior, dystonia and cognitive disability (2–4). The neurobehavioral phenotype has been linked with dysfunction of the basal ganglia, and particularly its dopaminergic pathways (5–8). HPRT deficiency is associated with a marked loss of basal ganglia dopamine and dopamine-related enzymes in human autopsy and imaging studies (9–12), and even in HPRT-deficient (HPRT−) mice (13–15). The defects are relatively selective for basal ganglia dopamine pathways. Other brain dopamine pathways are largely spared, and most other neurotransmitter systems are unaffected.
The exact influence of HPRT deficiency on basal ganglia dopamine pathways is unknown, since these pathways have no obvious connection with the known biochemical functions of HPRT. Early speculations that these neural pathways might be particularly dependent upon HPRT because of unusually high levels of enzyme expression were not supported by subsequent studies (16,17). Another early proposal was that the metabolic consequences of HPRT deficiency resulted in degeneration or dysgenesis of dopamine neurons (8,9). This proposal has been undermined by results from both animal and human studies showing the marked loss of dopamine is unaccompanied by any measurable change in the numbers and morphology of dopamine neurons (14,18). It also has been suggested that the metabolic consequences of HPRT deficiency may lead to secondary metabolic defects including oxidative stress or limitation of tetrahydrobiopterin, a cofactor essential for dopamine synthesis. These hypotheses have been refuted too (19–21). The link between HPRT deficiency and dopamine dysfunction therefore remains enigmatic.
Tissue culture models provide powerful tools for exploring pathogenesis in many human diseases (22). They can be exploited to probe the molecular and cellular consequences of specific genetic alterations in a tightly controlled experimental environment. Recent studies with HPRT− sublines of dopaminergic mouse MN9D neuroblastoma showed that HPRT deficiency is associated with a cell-intrinsic loss of dopamine without morphological defects or signs of impaired viability (23). The mutant cells exhibited accompanying abnormalities of nearly all biochemical markers of the dopaminergic phenotype including dopamine metabolites, enzymes involved in synthesis and degradation, and the dopamine transporter.
A novel hypothesis to account for the broad change in the biochemical signature of the dopaminergic phenotype of these cells, with no defect in structure or viability, is that HPRT deficiency influences the developmental programs that regulate their neurochemical phenotype. This hypothesis was explored in the current studies.
RESULTS
Microarray analysis of HPRT-deficient MN9D cells
As a group, the HPRT− sublines displayed a significant reduction in the expression of Hprt mRNA (Table 1). This result was confirmed by qPCR and is consistent with the loss of functional HPRT enzyme activity in these cells (23). The microarray analysis also revealed the HPRT− MN9D sublines to have significantly reduced the expression of the mRNA encoding tyrosine hydroxylase (Th). The reduction in Th also was confirmed by qPCR and is consistent with prior studies showing dysregulation of multiple biochemical markers of the dopamine phenotype, including Th (23).
Table 1.
Microarray significant results for HPRT-deficient MN9D cells
| Gene family | Gene | Protein | Mean fold change |
||
|---|---|---|---|---|---|
| Dye 1 | Dye 2 | qPCR | |||
| Receptors | Adora2a | A2a adenosine receptor | 1.55 | 1.36 | 1.36 |
| Adra2b | Adrenergic receptor alpha2b | 3.17 | 2.73 | 1.36 | |
| Chrnb2 | Cholinergic receptor, nicotinic, beta polypeptide 2 | 1.15 | 1.55 | 2.88 | |
| Grik5 | Glutamate receptor, ionotropic kainate 5 | 1.39 | 1.65 | 1.74 | |
| Chrna6 | Cholinergic receptor, nicotinic, alpha polypeptide 6 | −1.54 | −1.32 | −3.09 | |
| Gria3 | Glutamate receptor, ionotropic, ampa3 | −1.72 | −1.82 | −37.79 | |
| Sstr2 | Somatostatin receptor 2 | −3.23 | −4.17 | −1.27 | |
| Transcription factors | En1 | Engrailed 1 | 3.21 | 18.93 | 19.39 |
| Hoxa5 | Homeo box A5 | 1.47 | 1.86 | 9.67 | |
| Jund1 | Jun proto-oncogene related gene d1 | 2.51 | 2.02 | 2.07 | |
| Pax5 | Paired box gene 5 | 1.61 | 1.91 | 1.88 | |
| Differentiation | Arrb2 | Arrestin-beta2 | 2.47 | 2.72 | 2.06 |
| Vim | Vimentin | 1.61 | 3.47 | 1.43 | |
| Eno2 | Enolase 2 | −1.54 | −3.23 | −0.92 | |
| SynI | Synapsin Ib | −1.23 | −2.27 | ND | |
| Kinases | Camk4 | Calcium/calmodulin dependent protein kinase IV | 1.67 | 3.75 | 2.14 |
| Ntrk3 | Neurotrophic tyrosine kinase receptor type 3 | 1.75 | 3.65 | 1.60 | |
| Camk2b | Calcium/calmodulin dependent protein kinase II beta | −1.64 | −1.30 | −1.55 | |
| Other | Cacna1c | Calcium channel voltage-dependent L type, alpha 1C subunit | 1.88 | 2.51 | 1.54 |
| L1cam | L1 cell adhesion molecule | 2.10 | 2.25 | 1.88 | |
| Nos1 | Nitric oxide synthase 1 neuronal | 2.08 | 3.83 | 2.06 | |
| Prdx3 | Peroxiredoxin 3 | 1.60 | 1.95 | 1.52 | |
| Hprt | Hypoxanthine-guanine phosphoribosyltransferase | −1.85 | −4.76 | −4.72 | |
| Hprt | Hypoxanthine-guanine phosphoribosyltransferase | −3.33 | −7.14 | −4.72 | |
| Th | Tyrosine hydroxylase | −2.63 | −2.78 | −1.92 | |
Dyes were reversed for the duplicate to reduce the effects of dye incorporation bias (Dye 1 and Dye 2). The only significant changes are shown in this table. ND: not detected by qPCR with two different primer pairs.
In addition to abnormalities for Hprt and Th that were anticipated from prior studies, the microarray analysis revealed significant changes in several additional mRNAs (Table 1). The majority (21 of 24) of abnormalities detected via microarray were confirmed by qPCR. Two were not further studied because they could not be detected by qPCR and one gave a contradictory result. A large proportion of the changes involved mRNAs encoding transcription factors known to be involved in neuronal development and differentiation. The largest quantitative change was a 19-fold increase in mRNA transcripts associated with the gene encoding engrailed 1 (En1). The homeobox A5 (HoxA5), Jund1, and paired box gene 5 (Pax5) also were increased. Other transcription factors involved in development such as the dlx gene family were not altered. These results provide support for the hypothesis that the abnormal dopaminergic phenotype of the HPRT− MN9D cells may be due to dysregulation of mechanisms that control early development and differentiation.
Developmental markers in HPRT-deficient MN9D cells
To further address the hypothesis relating HPRT deficiency to a defect in developmental programming in the HPRT− MN9D cells, we performed a more extensive qPCR survey of additional molecular markers known to be involved in dopamine neuron differentiation. Of the 29 developmental mRNAs evaluated, more than half were significantly altered. Among these, 12 mRNAs were significantly increased, 3 were significantly decreased (Table 2) and 2 showed changes of borderline statistical significance (Supplementary Material 1). The largest quantitative changes included an 18-fold increase in En1 mRNA, and a 26-fold increase in mRNA transcripts associated with the gene encoding the closely related transcription factor, engrailed 2 (En2). Only 12 of 29 developmental mRNAS were not significantly affected (Supplementary Material 1). While more than half of the developmental markers were expressed at abnormal levels, multiple control markers evaluated by microarray and/or qPCR were normal: beta-actin, Adsl, Adk.1, App, Aprt, Akt1, Calbindin-D9K, Eefla1.1, Elk1, Elk2, Erg2 Erg3 Gapdh, Glast, Gpx1, Hcn2, Kcnq2, Kir7.1, Kv1.2, Kv1.4, Kv3.1, Kv3.3, Kv33.4a, Lamr1.1, Laptm4a.1, Lrrk2, Mbnac2, Mek1, Myl6, Ppox.1, R-erg, Rps4x.1, Rsk3, Snap25/p140. These qPCR results provide further evidence for a disruption of developmental mechanisms in the HPRT− MN9D cells.
Table 2.
Marked up-regulation of engrailed 1 and engrailed 2 and significant changes of developmental markers in HPRT-deficient MN9D cells
| Gene | AG2 | AG5 | AG6 | AG8 | AG10 | TG1 | TG2 | TG3 | TG4 | TG5 | Overall change | t-test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| En1 | 2156 ± 123 | 2908 ± 270 | 4269 ± 320 | 2350 ± 97 | 1511 ± 89 | 226 ± 23 | 2489 ± 89 | 1351 ± 44 | 1196 ± 36 | 63 ± 5 | 1839 ± 406 | P < 0.0001 |
| En2 | 65 ± 9 | 2335 ± 176 | 5215 ± 233 | 2195 ± 213 | 5208 ± 220 | 1284 ± 200 | 5907 ± 390 | 1780 ± 71 | 135 ± 64 | 2257 ± 161 | 2625 ± 671 | P = 0.0005 |
| Hoxa5 | 627 ± 33 | 1383 ± 86 | 2312 ± 84 | 46 ± 23 | 263 ± 40 | 933 ± 57 | 1377 ± 52 | 258 ± 28 | 783 ± 82 | 685 ± 71 | 867 ± 214 | P = 0.0003 |
| Jund1 | 60 ± 7 | 117 ± 17 | 232 ± 35 | 155 ± 11 | 18 ± 4 | 142 ± 19 | 116 ± 21 | 47 ± 29 | 68 ± 2 | 133 ± 37 | 109 ± 20 | P < 0.0001 |
| Ngn2 | 171 ± 14 | 259 ± 11 | 43 ± 11 | 234 ± 17 | 77 ± 9 | 45 ± 16 | 532 ± 17 | 243 ± 29 | 409 ± 16 | 10 ± 5 | 200 ± 55 | P = 0.0004 |
| Pard3 | 46 ± 14 | 28 ± 7 | 72 ± 8 | 30 ± 4 | −34 ± 1 | 55 ± 8 | 45 ± 7 | 43 ± 5 | 109 ± 10 | 76 ± 8 | 47 ± 12 | P = 0.0008 |
| Pax5 | 90 ± 2 | 5 ± 8 | 79 ± 1 | 121 ± 7 | −25 ± 1 | 174 ± 14 | 88 ± 22 | 95 ± 8 | 89 ± 10 | 160 ± 6 | 88 ± 19 | P < 0.0001 |
| Rac3 | 96 ± 1 | 229 ± 12 | 161 ± 16 | 48 ± 6 | 121 ± 13 | 52 ± 14 | 15 ± 4 | 108 ± 17 | 100 ± 11 | 114 ± 15 | 104 ± 19 | P < 0.0001 |
| Rap1b | 49 ± 10 | 53 ± 11 | 24 ± 4 | 32 ± 8 | 11 ± 2 | 35 ± 6 | 6 ± 3 | 23 ± 4 | 21 ± 1 | 13 ± 2 | 27 ± 5 | P = 0.0007 |
| RhoA | 27 ± 4 | 44 ± 7 | 77 ± 8 | 24 ± 8 | 13 ± 8 | 50 ± 15 | 36 ± 12 | 11 ± 7 | 79 ± 8 | 7 ± 4 | 37 ± 8 | P = 0.0017 |
| Shh | 112 ± 47 | 84 ± 10 | 6 ± 1 | 211 ± 50 | 121 ± 25 | 124 ± 38 | 646 ± 100 | 543 ± 61 | 1321 ± 48 | 169 ± 55 | 317 ± 132 | P < 0.0001 |
| Wnt5 | 108 ± 14 | 82 ± 12 | 94 ± 22 | 64 ± 15 | −38 ± 2 | 39 ± 9 | 37 ± 9 | 78 ± 14 | 192 ± 10 | 100 ± 17 | 76 ± 19 | P = 0.0047 |
| Lmx1b | −57 ± 1 | −89 ± 1 | −65 ± 2 | −21 ± 8 | −73 ± 5 | −77 ± 1 | −38 ± 4 | −68 ± 4.2 | −53 ± 2.3 | −62 ± 5 | −60 ± 6 | P < 0.0001 |
| Msx2 | −68 ± 3 | −98 ± 1 | −86 ± 1 | −661 ± 2 | −47 ± 2 | −88 ± 3 | −44 ± 7 | −42 ± 7 | −18 ± 2 | −73 ± 1 | −63 ± 8 | P < 0.0001 |
| Pitx3 | −69 ± 5 | −68 ± 3 | −62 ± 3 | −66 ± 2 | −61 ± 2 | −84 ± 3 | −54 ± 3 | −56 ± 1 | −17 ± 7 | −66 ± 2 | −60 ± 5 | P < 0.0001 |
The results for each mRNA in each cell line were determined in quadruplicate by qPCR and normalized to a simultaneously processed control mRNA (Myl6). Results from the HPRT− lines are presented as percent changes relative to a simultaneously processed sample of the HPRT+ parent line. The overall change reflects the aggregate results of all the HPRT− lines combined. The results for all 10 HPRT− MN9D cell lines were compared as a group against the parent MN9D line by a single-sample t-test. A Bonferroni correction for the 29 simultaneous comparisons in this Table and Supplementary Material 1 was used to define P < 0.002 as a conservative estimate for statistical significance. Values of 0.002 < P < 0.05 were considered borderline.
Engrailed mRNA and protein in HPRT-deficient MN9D cells
We focused further studies on engrailed 1 and engrailed 2 since these showed the largest quantitative changes in both the microarray and qPCR studies. To determine if the increased mRNA levels were accompanied by increased protein levels, we performed immunoblots using the polyclonal 86/8 antiserum, which recognizes both engrailed protein products (24).
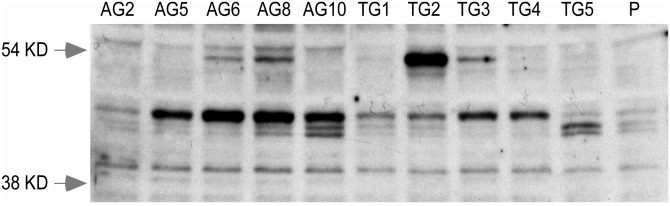
A band corresponding to the expected molecular weight of 54 kDa for engrailed 1 was evident in 4 of the 10 HPRT− MN9D sublines (AG6, AG8, TG2 and TG3), yet only faintly visible in the parental MN9D line (Fig. 1). The bands visible on immunoblots always corresponded to sublines with increased mRNA by qPCR. Even after over-exposure of the immunoblots, a similar band of increased intensity at 54 kDa could not be demonstrated for 6 of the HPRT− sublines (AG2, AG5, AG10, TG4 and TG5). The absence of a visible band on immunoblots for the TG5 subline corresponded to a lack of increased mRNA in this subline (Table 2). However, the other four sublines exhibited a disparity between their immunoblot band intensity and their mRNA levels. Further, the most intense band on immunoblots was seen for the TG2 subline, yet highest mRNA levels were found in subline AG6. These results suggested that the correlation between band intensity on immunoblots and qPCR results was not absolute. The absence of a strict correlation between the immunoblot results and qPCR could not be explained by variations in immunoblot protein loading since the same amount of protein was added in all lanes, re-probing the same blots with an antibody to beta-actin led to roughly equal band intensities, and similar results for all the sublines were obtained in three independently prepared immunoblots. These results confirm that elevated mRNA encoding engrailed 1 can be detected at the protein level in the HPRT− sublines. However, the lack of tight correlation between the immunoblot and qPCR results implied additional variations in the efficiency of engrailed 1 protein translation or post-translational processing among some sublines.
Figure 1.
Increased engrailed protein products in HPRT-deficient MN9D sublines. Immunoblot for engrailed 1 and engrailed 2 protein products in normal and all 10 HPRT− MN9D sublines HPRT, using the 86.6 antiserum, revealed both engrailed 1 (54 kDa) and engrailed 2 (45 kDa). The first 10 lanes show results for the mutant sublines (AG2, AG5, AG6, AG8, AG10, TG1, TG2, TG3, TG4, TG5), while the last lane (P) shows results for the parent line. A similar banding pattern was observed for three independent immunoblots. The locations of molecular weight markers are shown to the left.
The immunoblots also revealed a band corresponding to the expected molecular weight of 45 kDa for engrailed 2 in all HPRT− MN9D sublines (Fig. 1). This band was relatively faint in samples from two of the HPRT− MN9D sublines (AG2 and TG5), and from the parental MN9D line. Similar to the results for engrailed 1, there was only a rough correlation between the band intensity on immunoblot and the qPCR results for En2. For example, the strongest bands on immunoblots were always associated with sublines expressing elevated levels of mRNA (AG5, AG6, AG8, AG10 and TG3). Conversely, a weak band on immunoblots for AG2 corresponded to low mRNA levels (Table 2). However, the TG2 subline had only a modest band on immunoblots yet the highest mRNA levels among all the sublines. The TG5 subline also was unusual with high mRNA levels, a weak immunoblot band signal at the expected molecular weight, but two moderately strong bands at slightly lower molecular weights. These smaller bands were also evident in some of the other subclones, most notably AG10. There were no apparent correlations between the intensities of the bands at 54 kDa for engrailed 1 and at 45 kDa for engrailed 2. These results confirm that the elevated mRNA for En2 also could be detected at the protein level in the HPRT− sublines, together with additional evidence or variations in engrailed 2 protein translation or post-translational processing among the sublines.
Restoration of HPRT reverses engrailed over-expression in MN9D cells
To establish a causal relationship between HPRT deficiency and changes in engrailed mRNAs, we tested whether restoring HPRT expression would reverse the over-expression of engrailed 1 or engrailed 2. Five of the HPRT− MN9D sublines exhibiting robust over-expression of mRNAs encoding En1 or En2 (AG6, AG8, AG10, TG2, TG3) were transfected with a minigene in which human HPRT was driven by the chicken beta-actin promoter and selected for functional expression of HPRT enzyme in HAT medium. Using species-specific primers for HPRT, four of the HAT-resistant lines were determined to be transfectants expressing human HPRT mRNA and one appeared to be a spontaneous revertant expressing mouse HPRT mRNA (AG8).
The restoration of HPRT activity was confirmed by enzymatic assay. The parent MN9D line had readily detectable HPRT enzyme activity, and all five HPRT− MN9D sublines had enzyme levels at or below levels of detection (Table 3). After the transfection procedure and selection in HAT medium, HPRT enzyme activity was restored in all five sublines, with levels ranging from 55 to 940% of the activity seen in the parent MN9D line. For comparison, we examined the activity of the closely related purine salvage enzyme, adenine phosphoribosyltransferase (APRT). APRT activity was detectable in all of the HPRT− MN9D sublines and their HAT-selected transfectants, with levels varying from 40 to 212% of the values in the parent MN9D line. These results confirm the selective loss of HPRT activity in the HPRT− MN9D sublines and its restoration in their HAT-selected transfectants.
Table 3.
HPRT and APRT enzyme activities in HPRT-deficient MN9D sublines and their transfectants
| Cell line | HPRT | % control | APRT | % control |
|---|---|---|---|---|
| MN9D parent | 0.8 | 100 | 7.3 | 100 |
| AG6 | 0 | 0 | 10.7 | 146.6 |
| AG6 transf | 1.5 | 176.5 | 15.5 | 212.3 |
| AG8 | 0 | 0 | 7.4 | 121.9 |
| AG8 revert | 1.2 | 135.3 | 11.9 | 101.4 |
| AG10 | 0 | 0 | 11.9 | 163.0 |
| AG10 transf | 2.6 | 309.4 | 3.0 | 40.5 |
| TG2 | 0 | 0 | 14.9 | 204.1 |
| TG2 transf | 8.0 | 941.2 | 7.3 | 100.0 |
| TG3 | 0.1 | 7.1 | 5.2 | 71.2 |
| TG3 transf | 0.5 | 55.3 | 3.2 | 43.8 |
Results show enzyme activity expressed as nmol/mg protein per min, and percent of control. Each value was determined from a kinetic assay with five time points. Four of the lines in which HPRT was restored were found to be transfectants expressing the human HPRT mRNA (transf), while one was found to be a spontaneous revertant expressing the mouse HPRT mRNA (revert).
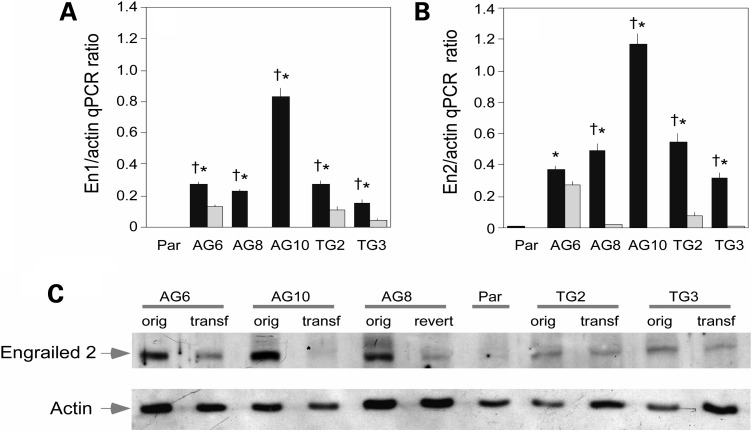
The restoration of HPRT activity attenuated the over-expression of both En1 and En2 mRNAs in all of the MN9D transfectants (Fig. 2A–B). En1 mRNA was reduced to levels indistinguishable from the parent MN9D line in two of the four HPRT transfectants (AG10 and TG3), and the AG8 revertant. En2 mRNA was reduced to control levels in two of the five HPRT transfectants (AG10 and TG3) and in the AG8 revertant.
Figure 2.
Restoration of HPRT normalizes engrailed 1 and engrailed 2 expression. Quantitative PCR for engrailed 1 (A) and engrailed 2 (B) and immunoblot (C) following restoration of HPRT activity in MN9D cells. HPRT+ transfectants (in gray) were compared with HPRT− MN9D sublines (in black). The qPCR data for En1 and En2 were subject separately to one-way ANOVA. The overall ANOVA was significant for both En1 (F = 424.7, P < 0.001) and En2 (F = 64.5, P < 0.001). Post hoc Tukey t-tests revealed significant differences at the P < 0.01 level between the parent (Par) and each HPRT− MN9D subline (*), and between the HPRT− MN9D subline and its corresponding HPRT+ derivative (†) as indicated by the symbols in the figure. The 86.6 antiserum was applied to extracts of the parent MN9D line (Par), five of the HPRT mutant sublines with engrailed overexpression (AG6, AG8, AG10, TG2, TG3; orig) and their corresponding transfectants (AG6 tranf, AG8 revert, AG10 transf, TG2 transf, TG3 transf). The approximate amount of protein loaded was estimated after stripping the blots and reprobing with an antibody to beta-actin.
Corresponding changes in the expression of protein were evaluated via immunoblotting. The 45 kDa band corresponding to engrailed 2 was reduced towards the levels observed in the parent MN9D line in each of the HPRT− MN9D sublines (Fig. 2C). These results show that restoring HPRT activity results in reversal of engrailed over-expression at both the mRNA and protein levels, demonstrating a causal relationship between HPRT deficiency and over-expression of engrailed in MN9D cells.
Morphological evidence of aberrant developmental mechanisms in HPRT-deficient MN9D cells
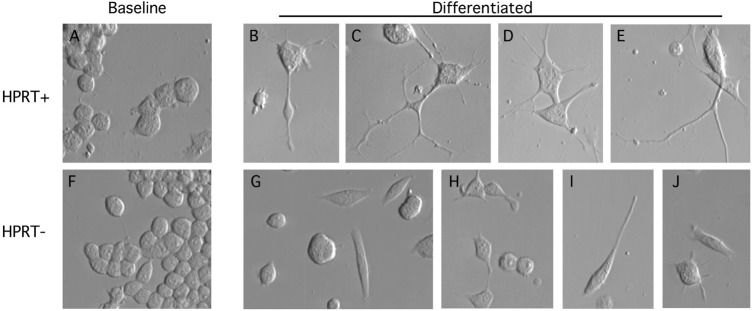
The above studies show HPRT influences the molecular signature of developmental markers in MN9D cells. To explore further the hypothesis that HPRT affects neuronal development, we searched for morphological signs of abnormal development in the HPRT− MN9D cells. At baseline, the HPRT− MN9D sublines had growth characteristics and gross morphological appearances similar to the MN9D parent line (23). Morphologically, cultures were mixed, with most cells appearing as simple round blast-like cells (Fig. 3A and F). Differentiation of the parent line with sodium butyrate led to multiple changes suggestive of differentiation as previously described (25). These changes included cell cycle arrest, enlargement and flattening of the soma, and extension of multiple lengthy neurites (Fig. 3B–E).
Figure 3.
Morphological appearance of parent and HPRT− mouse MN9D sublines. The parent MN9D cells are shown in the top row, and the HPRT− mutants are shown in the bottom row before (A and F) and after (B–E, G–J) sodium butyrate differentiation. Undifferentiated parent MN9D (A) and mutant (F) cultures appeared predominantly as simple round or ovoid cells. Differentiated parent cells demonstrated cell cycle arrest, enlargement and flattening of the soma, and elaboration of multiple complex branching neurites that often extended out of the field of view (B–E). Differentiated mutant cells demonstrated cell cycle arrest with soma changes similar to control cells. However, there was a high proportion of cells with no neurites (G and H), and the neurites tended to be short and simple (H–J).
Differentiation of the HPRT− sublines also led to cell cycle arrest with enlargement and flattening of the soma, but neurite growth was clearly abnormal in all of the lines (Fig. 3G–J). The numbers of neurites per cell were markedly reduced, and neurites were short and simple. These results provide morphological evidence that HPRT deficiency influences developmental processes in the MN9D cell line.
Over-expression of engrailed in other HPRT-deficient cells
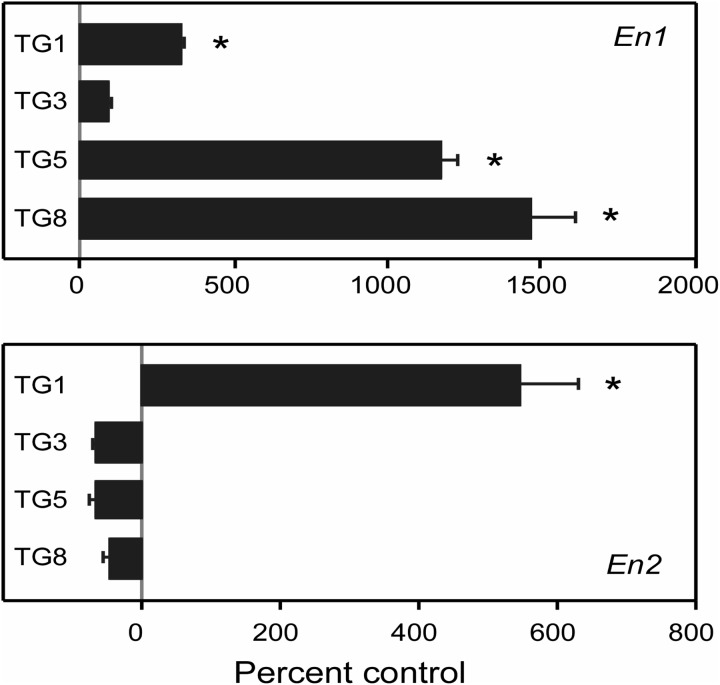
To determine whether over-expression of En1 or En2 might be an idiosyncratic effect of HPRT deficiency limited to MN9D cells, we also examined four HPRT− sublines of the M17 human neuroblastoma line (22) by qPCR. In comparison to the HPRT-competent parent M17 line, En1 mRNA was increased in three of the HPRT− M17 sublines, with levels as high as 15-fold above controls. En2 mRNA was increased in only one of the lines, by more than 5-fold (Fig. 4). Control mRNAs were expressed at normal levels including beta-actin, Gapdh, Myl6. These results revealed that over-expression of the engrailed transcription factors is not limited to HPRT− mouse MN9D cells, but also can be observed in HPRT− human M17 neuroblastoma cells.
Figure 4.
Increased engrailed expression in HPRT-deficient human M17 neuroblastoma cells. Quantitative PCR for En1 and En2 mRNA was done for each mutant subline (TG1, TG3, TG5, TG8), determined in quadruplicate and normalized to a simultaneously processed control mRNA (Myl6), where the raw data showed little variation among the lines. Results from the HPRT− human M17 sublines are presented as percent changes relative to a simultaneously processed sample of the HPRT+ parent line. The overall ANOVA was significant for both En1 (F = 109.2, P < 0.001) and En2 (F = 59.5, P < 0.001). Post hoc Tukey t-tests revealed significant differences at the P < 0.05 level between the parent and each subline as indicated by asterisk.
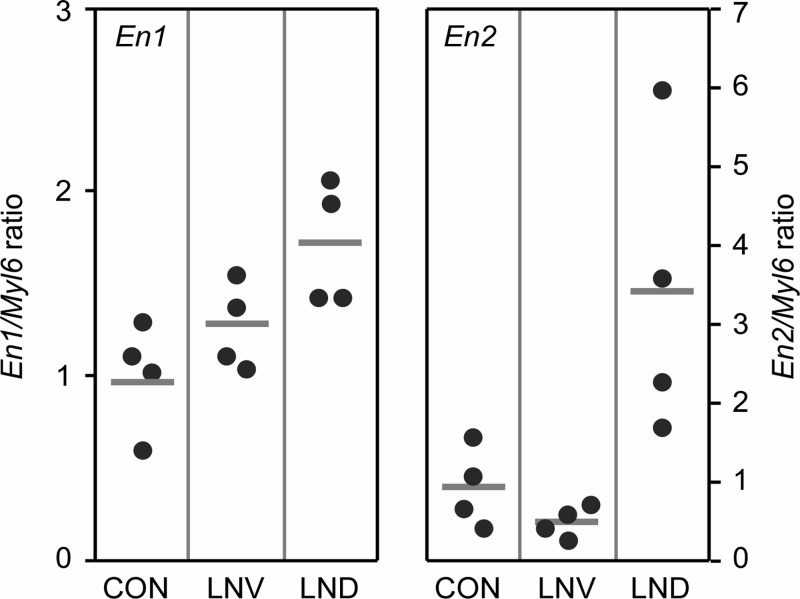
To determine whether the over-expression of En1 or En2 might be a more general property of HPRT deficiency even in non-neural cells, we examined primary skin fibroblasts obtained from eight patients with HPRT deficiency. Half of these fibroblasts came from patients with the classic Lesch-Nyhan phenotype (LND) with hyperuricemia and severe neurobehavioral dysfunction, while half came from Lesch-Nyhan variants (LNV) with hyperuricemia but little or no neurobehavioral dysfunction. Fibroblasts from four patients with unrelated neurobehavioral disorders served as controls. Measurable levels of En1 mRNA were found in all fibroblast samples (Fig. 5). Though there was considerable overlap in the levels among patients with HPRT deficiency and controls, there was a population trend that suggested a correlation between En1 mRNA level and disease severity, with a statistically significant difference between controls and those with the classic LND phenotype (P < 0.02).
Figure 5.
En1 and En2 expression in normal and HPRT-deficient primary human fibroblasts. Cultures came from normal controls (CON), patients with partial HPRT-deficiency syndromes (LNV) and patients with the full classic syndrome (LND). The points show data for individual patients (n = 4 each), normalized to a simultaneously processed control mRNA (Myl6). Horizontal bars mark the group mean. Results for each of the engrailed mRNAs were compared separately by one-way ANOVA. The overall ANOVA was significant for both En1 (F = 6.0, P = 0.022) and En2 (F = 7.5, P = 0.012). Post hoc Tukey t-tests for En1 revealed a significant difference for CON versus LND (P = 0.019), but not for CON versus LNV (P = 0.43), or LNV versus LND (P = 0.14). Post hoc Tukey t-tests for En2 revealed significant differences for CON versus LND (P = 0.03) and LNV versus LND (P = 0.014), but not for CON versus LNV (P = 0.85).
All fibroblasts also expressed En2 mRNA. The four mildly affected Lesch-Nyhan variants had levels indistinguishable from controls. However, En2 was increased in the fibroblasts from patients who had neurobehavioral manifestations typical of the classic LND form. These samples showed no overlap with controls, with an average increase more than 3-fold controls (P < 0.05). These results revealed that over-expression of En2 occurred even in HPRT− human primary fibroblasts, in a manner that suggested a correlation with the manifestation of neurobehavioral dysfunction. While En1 or En2 were increased in the HPRT− fibroblasts, multiple control mRNAs were expressed at normal levels, including Aprt, Adsl, Myl6, Prps1, Prps2 and Comt (not shown).
DISCUSSION
A serious challenge in elucidating the pathogenesis of human HPRT deficiency is its rarity, with so few patients or patient-derived materials available for experimental investigations. A further challenge in elucidating the mechanisms responsible for the neurological abnormalities is the relative inaccessibility of relevant brain pathways to experimental investigations. In view of these challenges, surrogate experimental models have been used for exploring pathogenesis. In the current studies, we took advantage of multiple independent cell models for HPRT deficiency. Because of the central importance of dopamine neuron dysfunction to LND, we focused on the 10 recently developed HPRT− sublines of the dopaminergic mouse MN9D neuroblastoma line (23). These cells replicate the key phenomena of significant dopamine loss without gross baseline morphological defects or impaired viability evident in Lesch-Nyhan human brains and HPRT− mice. A microarray survey of these cell models pointed to abnormalities in pathways relevant to early neuronal development and differentiation. Rigorous and methodical evaluation by qPCR revealed abnormal expression of 15 of 29 mRNAs known to encode proteins relevant to dopamine neuron development and differentiation. The most robust quantitative changes involved overexpression of the engrailed genes (En1 and En2) and their products (engrailed 1 and 2). The changes in the developmental markers do not reflect a global and non-specific abnormality affecting all mRNAs in the HPRT− MN9D sublines, since multiple control mRNAs were normal.
While cell models have some obvious advantages as experimental tools for identifying novel mechanisms of pathogenesis, they also have some important limitations. One limitation of cell models is that individual sublines of established cell lines may exhibit idiosyncratic properties that differ from the parental controls independent of the genetic alteration introduced (22,23). This limitation was first addressed in the current studies by evaluating 10 independently isolated HPRT− MN9D sublines. The statistical approach for the microarray and qPCR studies involved comparing the parental MN9D cell line against all 10 HPRT− MN9D sublines, searching for significant changes across the whole group of mutant sublines, rather than idiosyncratic changes among individual sublines. This approach reduces the risk of finding spurious abnormalities unrelated to HPRT deficiency and revealed highly significant changes in multiple developmentally regulated mRNAs, most notably over-expression of the engrailed genes. The relationship between HPRT deficiency and over-expression of the engrailed genes was further tested by examining the influence of restoring functional HPRT in the HPRT− MN9D sublines in which En1 and/or En2 were over-expressed. The over-expression of the engrailed genes was partly or completely reversed in each case. Thus, the increased expression of engrailed is not a spurious phenomenon in the mutant sublines, but one that appears to depend on the expression of functional HPRT.
Another limitation of cell models is that findings may be idiosyncratic to the cell model studied, with no relevance to other experimental models. This limitation was addressed by evaluating HPRT− cell models from multiple different sources. In addition to the 10 HPRT− MN9D sublines, we examined four HPRT− sublines of human M17 neuroblastoma. All four of the mutant M17 sublines exhibited an increase in En1 expression, while En2 expression was more variable. We also evaluated primary human fibroblasts from eight patients with varying degrees of HPRT deficiency. There was again evidence for increased expression of En1 and/or En2 in the HPRT− fibroblasts relative to controls. Thus, increased expression of one or both engrailed genes appears to be common to several cell models of HPRT deficiency, including those derived from affected patients.
The final limitation of cell models is that the findings may be restricted to the in vitro environment, with little relevance for the in vivo state, especially in the brain. This limitation can be addressed only by examining brain tissue from affected Lesch-Nyhan patients. Because the developmental markers in question have specific temporal windows of expression, it may be necessary to study these brains at various stages of development. Though such studies are not feasible because of the absence of appropriate human brain specimens, it is important to note that the hypothesis regarding a defect in developmental programming of the dopamine neurochemical phenotype is consistent with prior human studies showing a relatively broad disruption of many biochemical markers of the dopamine phenotype (7,9,10,26) with no apparent histological evidence for loss of cells or axonal projections (2,10,27).
A disruption of molecular pathways that control early development provides a novel mechanism that could account for the well-known association between HPRT deficiency and dysfunction of brain dopamine systems. The En1 and En2 genes encode two closely related transcription factors that are part of a large family of homeobox transcription factors responsible for many developmental processes. The engrailed products have specific roles in two events during neural development. Early in embryogenesis, they take part in establishing the size and region of the midbrain in which dopamine neurons later develop. Later they play a role in the specification and survival of dopamine neurons (28–30). They function as nuclear transcription factors that alter the expression of multiple other genes, but they also can be released from cells and internalized by neighbors, where they are thought to exert paracrine-like activities (31,32). The genes have partly overlapping functions, such that one can partly compensate for loss of the other (33). In mutant mice homozygous for null mutations in both genes, dopamine neurons are generated between embryonic days 11 and 14, but the entire population disappears before birth and the pups cannot survive (28,29). Intermediate genotypes with one or more copies of either gene display varying degrees of midbrain dopamine neuron loss (24,34). In our HPRT− MN9D sublines, there were no apparent correlations between the expression of En1 and En2 among the HPRT− cells. Some HPRT− cells exhibited an increase in both engrailed genes, while others exhibited increases in only one. The lack of consistent increases in both mRNA and protein products may be related to the redundant biological functions of these two homeoproteins.
There is good evidence that alterations in the expression of the engrailed genes influence dopamine neuron development and survival. Transcription factors of the homeoprotein family present cell autonomous and non-cell autonomous activities that can influence neurite elongation and guidance. In the case of engrailed, several of its putative transcriptional targets may play such a function. Among the latter targets are EphrinA5, a cell surface protein with collapsing activity (35) and MAP1B a microtubule associated protein that regulates microtubule polymerization in the axon (36). More recently, it was shown that engrailed serves as an axon guidance factor, in a non-cell autonomous way, by regulating mRNA translation within the growth cones (37). It is also of interest that engrailed 1 is present in the dendrites of midbrain dopaminergic neurons (38). Although the latter localization is in want of a function, the ability of engrailed to regulate translation in developing axons leaves open the possibility that a similar mechanism is used to regulate dendrite geometry. Thus, the morphological defects seen in our differentiated HPRT− MN9D cells might represent downstream consequences of abnormal expression of the engrailed.
The mechanisms by which HPRT deficiency influences the expression of the engrailed genes remain to be determined. One possibility is that engrailed expression may be affected directly by HPRT. While there currently is no evidence that HPRT can affect the engrailed or other developmental pathways directly, there is ample precedent for other housekeeping enzymes having such additional ‘moonlighting’ activities in addition to their traditionally ascribed functions (39–41). Thus, HPRT may have a moonlighting function beyond its role in purine recycling. Alternatively, HPRT deficiency may influence engrailed pathways indirectly, through the secondary changes in purine metabolism that occur when HPRT is missing. Indeed, disruption of purine levels is known to have an important influence on neuronal differentiation in other cells (42,43). It also is possible that over-expression of engrailed genes reflects a compensatory developmental response to a defect elsewhere in the developmental programs of these neurons. Whether the effect is direct or indirect, the implication of the current findings is that HPRT-mediated purine recycling has an important role in regulating early developmental programming of dopaminergic neurons. Further studies will be required to more completely delineate the complex cascade of events resulting from HPRT deficiency and their influence on brain development.
MATERIALS AND METHODS
Cell cultures
The initial studies were conducted with the MN9D cell line, which is a hybrid line derived through the somatic fusion of primary midbrain dopaminergic neurons from an embryonic day 14 mouse with the mouse neuroblastoma line N18TG2 (44). A total of 10 spontaneously arising HPRT− MN9D sublines were isolated by selection in 6-thioguanine or 8-azaguanine (23). These cells were grown routinely at 37°C under 5% CO2 and 95% air in DMEM (Sigma, St Louis, MO, USA) supplemented with 15% fetal bovine serum, 100 U/ml penicillin and 50 mg/ml streptomycin. For some studies, the cells were differentiated with sodium butyrate as previously described (25).
Confirmatory studies were conducted with the SK-N-BE(2) M17 line (M17), a human dopaminergic neuroblastoma of peripheral origin (ATCC, Manassas, VA, USA). Instead of selecting spontaneously arising mutants as in the case of the MN9D line, HPRT− M17 sublines were prepared by targeting a psoralen cross-link to a site immediately adjacent to exon 5 with a triple helix-forming oligonucleotide followed by selection in 6-thioguanine (22). These cells were cultured at 37°C with 5% CO2 in RPMI 1640 media supplemented with 15% fetal bovine serum and 2 mm L-glutamine.
Additional studies were conducted with primary cell lines from affected patients and controls. Primary human male fibroblast cultures were available from a tissue bank for four patients who had the full classic phenotype of LND, four mildly affected LND variants in whom some of the clinical features were absent or attenuated and four controls without HPRT deficiency. Fibroblast cultures were prepared following full thickness skin biopsy. Tissue was minced aseptically in phosphate buffered saline free of Ca2+ and Mg2+, then incubated with gentle agitation at 37ºC for 1 h in a sterile collagenase/dipase solution at 1 mg/ml (Roche Diagnostics, Mannheim, Germany). The partially digested material was pelleted at 1000g, then resuspended in DMEM with 20% fetal bovine serum supplemented with 2 mm glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 2.5 µg/ml amphotericin B. The suspension was plated in a 10 cm plate and fibroblasts were allowed to attach and grow out undisturbed for a few days. Cells were split when they reached confluence and maintained in DMEM with 10% fetal bovine serum supplemented with 2 mm glutamine thereafter.
Restoration of HPRT
An expression vector in which human HPRT was driven by the chicken beta-actin promoter (pCX-HPRT) was constructed by subcloning the human HPRT cDNA from pBACE-HPRT (45) into pCX-EGFP (46) in place of the sequences encoding the green fluorescent protein. The HPRT− MN9D sublines were plated at a density of 5 × 104 cells/cm2 and allowed to attach overnight. Cells then were transfected with pCX-HPRT using the Fugene 6 transfection reagent (Roche, Basel, Switzerland) and grown in medium supplemented with HAT (100 µM hypoxanthine, 0.4 µM aminopterin and 16 µM thymidine) for 2–3 weeks to select for HPRT competence. HAT-tolerant sublines of AG6, AG8, AG10, TG2, TG3 and were isolated and expanded in HAT-free media. Stable human transfectants and spontaneous revertants were distinguished by reverse transcriptase PCR with species-specific primers and conditions leading to selective amplification of the human cDNA without amplification of murine cDNA (primers and qPCR conditions in Supplementary Material 2).
HPRT and APRT enzyme activities
These enzymes were measured as previously described (47,48). Briefly, cells were grown to ∼90% confluency in a T75 culture flask, dislodged by trypsinization and pelleted by centrifugation at 1000g for 5 min. Pelleted cells were then frozen and stored at −80°C until assayed. Enzyme activities were assessed in total soluble extracts of cellular lysates by using [14C]-hypoxanthine or [14C]-adenine followed by separation of substrate and product by chromatography. Activity was normalized to total protein quantified in the lysate by the Lowry method.
Microarray procedure
Microarrays encompassing 289 genes directly relevant to neural transmission were used (NeuroTrans NTX, Genescore, Paris, France) to compare matched pairs of HPRT− samples and controls on the same chip. These microarrays directly compare the amount of fluorescence (Cy3/Cy5) of matched pairs on a single slide. Total RNA was extracted from cell pellets containing approximately 2 × 107 cells and treated with DNase using Qiagen's RNeasy Midi Kit (Valencia, CA, USA). The amount of RNA and the quality of each sample was verified using an Agilent 2100 Bioanalyser (Agilent Technologies Inc., Palo Alto, CA, USA). Total RNA (20 µg) from each HPRT− clone and parental cell line was reverse transcribed into cDNA, using random hexanucleotides (RPO, 50 µM), Superscript II polymerase (Invitrogen, Carlsbad, CA, USA) and dNTP mix (dATP, dCTP, dGTP at a final concentration of 125 µM and dTTP at 62.5 µM), dUTP labeled with either Cy5 or Cy3 fluorescent dyes at 25 µM. Matched pairs were analyzed in duplicate. Dyes were reversed for the duplicate to reduce the effects of dye incorporation bias. Labeled cDNAs were hybridized to the microarray overnight at 42°C in 50% formamide, 4X SSC, 0.1% sodium dodecyl sulfate and 5X Denhart's solution. After hybridization, the microarrays were washed in 2X SSC with 0.1% sodium dodecyl sulfate twice for 5 min, followed by 0.2X SSC for 1 min and 0.1X SSC for 2 min. Slides were then dried by centrifugation for 5 min at 120g.
Microarrays were scanned with an Axon 4000B GenePix Scanner (Foster City, CA, USA) at 10 µm resolution. Analysis was performed using the GenePix Pro 5.0 software (Axon instruments, Sunnyvale CA, USA). Areas of the arrays with visible blemishes were manually flagged and excluded from the analyses. The raw data were then submitted to Varan (http://www.bionet.espci.fr) for normalization and differential expression analysis using a Lowess fit (49). Varan provides the scaled fold, which is useful to eliminate intensity-dependent variability of the log (2) (Cy3/Cy5) ratios. These scaled folds were used for statistical analysis using SAM to identify significantly altered genes (http://www.stat.stanford.edu/~tibs/SAM/).
Quantitative PCR
Total RNAs were isolated from each cell sample using TRIzol following the manufacturer's recommendations (Invitrogen). Briefly, 1 ml of TRIzol was added to the cell pellet and homogenized by repetitive pipetting. Then 200 µl of chloroform was added, the sample was shaken vigorously for 20 s and incubated at 4°C for 5 min. The sample was centrifuged at 10 000g for 15 min at 4°C. An aliquot of 500 µl of the upper aqueous phase was removed and mixed with 500 µl of isopropyl alcohol. RNA was allowed to precipitate overnight at −20°C. The next day, RNA was pelleted at 10 000g for 10 min at 4°C and the pellet was washed twice with 1 ml of 75% ethanol and centrifuged at 5000g for 5 min at 4°C. Following the second wash, the pellet was air-dried for 10 min. Total RNA was dissolved in RNAse-free water and adjusted to 1 µg/µl after a spectrophotometric measure of RNA at 260 nm. The samples were stored at −80°C until further analysis.
For reverse transcription of mRNA, 1 µl of RNA at a concentration of 1 µg/µl was incubated overnight at 37°C with 0.5 µl of Reverse-iT RTase blend from ABgene (50 U/µl), 4 µl of 5X First Strand Synthesis Buffer, 1 µl of DTT 0.1 M, 1 µl of dNTPs 10 mM, 2 µl of random hexamers oligonucleotides at 50 µM (GE Heathcare Bioscience, Uppsala, Sweden) and 10.5 µl of RNase-free water to a final volume of 20 µl.
Quantitative PCR (qPCR) was performed using a LightCycler and the FastStart DNA MasterPLUS SYBR Green I kit (Roche Applied Science, Mannheim, Germany) as previously described (23). Primers were designed via Primerbank (pga.mgh.harvard.edu/primerbank). Reactions were conducted with 1 µl of each cDNA, 0.1 µl of each primer at 0.5 µM, 2 µl of 5X FastStart DNA Master SYBR Green containing the FastStart Taq DNA polymerase, MgCl2, the dNTP mix and the fluorescent dye SYBR Green I. RNase-free water was added to reach the final volume of 10 µl. The pre-incubation and denaturation of the samples were performed at 95°C for 10 min. This was followed by amplification of the target cDNA through 45 cycles at 95°C for 10 s, with annealing temperatures adapted according to the Tm of primers (primers and qPCR conditions are described in Supplementary Materials 2 and 3) and elongation at 72°C for 12 s. Finally, samples were cooled to 35°C. Each reaction was performed in quadruplicate for each sample. Expression of Myl6, beta-actin and GAPDH was used to normalize the raw data. The qPCR for the target gene and Myl6 or beta-actin or GAPDH was performed under the same PCR conditions in the same run. Each RT qPCR assay was designed and experimentally validated to amplify a single, gene-specific amplicon of the correct size with uniform PCR efficiency and without non-specific products. To check the specificity of each amplicon, a melting curve analysis was included in each run and showed a single specific peak for each amplicon. For all amplicons tested, the melting temperature was always above 80°C, avoiding detection of primer dimers having a melting temperature at 55–60°C. The quantification was performed by the crossing point method. In order to detect differences in PCR efficiency, standard curves for each analyzed cDNA were constructed employing reverse transcribed control RNA from parental cells as calibrator.
Western blot and antibodies
Protein samples were heated for 10 min at 95°C in sample buffer (0.4 M Tris–HCL; 0.1 M DTT, 2% SDS, 8.7% glycerol), separated on a 10% SDS–PAGE gel and transferred onto Immobilon P membranes (Millipore, Billerica MA, USA). Membranes were blocked for 1 h at room temperature in 5% non-fat milk in Tris-buffered saline containing 0.2% (v/v) Tween 20 and incubated with an anti-engrailed rabbit polyclonal antibody (86.8; 1:4000) at 4°C overnight (24). Membranes were washed five times for 5 min in the same buffer, incubated with horseradish peroxidase-conjugated secondary anti-rabbit antibody (1:4000) for 1 h at room temperature and washed five times for 5 min. Membranes were incubated with ECL+ HRP detection reagent (Amersham, Buckinghamshire, UK) and exposed to x-ray film. To verify equal loading of total protein, membranes were stripped for 30 min at 50°C in 100 mM 2-mercaptoethanol and 2% sodium dodecyl sulfate in 62.5 mM Tris–HCl pH 6.7. Successful stripping was verified with the detection reagent and exposure to x-ray film for 5 min. In the absence of any remaining signal, the membrane was washed two times for 5 min, blocked as described above, exposed to anti-beta actin rabbit polyclonal at 1:500 dilution (sc-1616, Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C and revealed as described above.
Data analysis
The qPCR results for each mRNA were normalized to simultaneously processed control mRNAs (beta-actin, GAPDH and Myl6). The final results for qPCR studies were similar for normalization with each of these control markers. Data for most qPCR experiments therefore are presented with normalization against Myl6 only, except for one experiment where a simultaneous immunoblot using an antibody to beta-actin is presented.
The statistical approach for the microarray analysis compared results for the parental MN9D cell line against all 10 HPRT− MN9D cell lines, searching for significant changes across the whole group of HPRT− lines, rather than idiosyncratic changes among individual lines (23). The same approach was used to compare qPCR results for the control versus the HPRT− MN9D mutants. In other experiments involving fewer samples for comparisons, data were evaluated by one-way ANOVA with post hoc Tukey t-tests. Individual means were compared via Student's t-test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Association Lesch-Nyhan Action, the Fondation Jérôme Lejeune, the Fédération des Maladies Orphelines, the Association Malaury, the Fondation Louis D. Institut de France and National Institutes of Health [grant numbers NS40470, DK082840]. Some of the human fibroblasts were obtained with support from core grant HD24061 from the National Institute for Child Health and Development to the Kennedy-Hopkins Mental Retardation Research Center.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pr. Alain Prochiantz (Collège de France and CNRS UMR 8542, Paris, France) for providing us with the anti-engrailed polyclonal antibody (86.8) and for advice and comments on the results. We thank Dr Alfred Heller for supplying the original MN9D cell line and V. Droin for her technical assistance in enzymatic HPRT determination.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Jinnah H.A., Friedmann T. Lesch-Nyhan disease and its variants. In: Scriver C.R., et al., editors. The Metabolic and Molecular Bases of Inherited Disease. New York, USA: McGraw-Hill; 2000. pp. 2537–2570. [Google Scholar]
- 2.Jinnah H.A., Visser J.E., Harris J.C., Verdu A., Larovere L., Ceballos-Picot I., Gonzalez-Alegre P., Neychev V., Torres R.J., Dulac O., et al. Lesch-Nyhan Disease International Study Group. Delineation of the motor disorder of Lesch-Nyhan disease. Brain. 2006;129:1201–1217. doi: 10.1093/brain/awl056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schretlen D.J., Ward J., Meyer S.M., Yun J., Puig J.G., Nyhan W.L., Jinnah H.A., Harris J.C. Behavioral aspects of Lesch-Nyhan disease and it variants. Dev. Med. Child Neurol. 2005;47:673–677. doi: 10.1017/S0012162205001374. [DOI] [PubMed] [Google Scholar]
- 4.Schretlen D.S., Harris J.C., Park K.S., Jinnah H.A., Del Pozo N.O. Neurocognitive functioning in Lesch-Nyhan disease and partial hypoxanthine-guanine phosphoribosyltransferase deficiency. J. Int. Neuropsychol. Soc. 2001;7:805–812. doi: 10.1017/s135561770177703x. [DOI] [PubMed] [Google Scholar]
- 5.Visser J.E., Baer P.R., Jinnah H.A. Lesch-Nyhan syndrome and the basal ganglia. Brain Res. Rev. 2000;32:449–475. doi: 10.1016/s0165-0173(99)00094-6. [DOI] [PubMed] [Google Scholar]
- 6.Nyhan W.L. Dopamine function in Lesch-Nyhan disease. Environ. Health Persp. 2000;108:409–411. doi: 10.1289/ehp.00108s3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito Y., Takashima S. Neurotransmitter changes in the pathophysiology of Lesch-Nyhan syndrome. Brain Dev. 2000;22:S122–S131. doi: 10.1016/s0387-7604(00)00143-1. [DOI] [PubMed] [Google Scholar]
- 8.Baumeister A.A., Frye G.D. The biochemical basis of the behavioral disorder in the Lesch-Nyhan syndrome. Neurosci. Biobehav. Rev. 1985;9:169–178. doi: 10.1016/0149-7634(85)90043-0. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd K.G., Hornykiewicz O., Davidson L., Shannak K., Farley I., Goldstein M., Shibuya M., Kelley W.N., Fox I.H. Biochemical evidence of dysfunction of brain neurotransmitters in the Lesch-Nyhan syndrome. N. Engl. J. Med. 1981;305:1106–1111. doi: 10.1056/NEJM198111053051902. [DOI] [PubMed] [Google Scholar]
- 10.Saito Y., Ito M., Hanaoka S., Ohama E., Akaboshi S., Takashima S. Dopamine receptor upregulation in Lesch-Nyhan syndrome: a postmortem study. Neuropediatrics. 1999;30:66–71. doi: 10.1055/s-2007-973462. [DOI] [PubMed] [Google Scholar]
- 11.Ernst M., Zametkin A.J., Matochik J.A., Pascualvaca D., Jons P.H., Hardy K., Hankerson J.G., Doudet D.J., Cohen R.M. Presynaptic dopaminergic deficits in Lesch-Nyhan disease. N. Engl. J. Med. 1996;334:1568–1572. doi: 10.1056/NEJM199606133342403. [DOI] [PubMed] [Google Scholar]
- 12.Wong D.F., Harris J.C., Naidu S., Yokoi F., Marenco S., Dannals R.F., Ravert H.T., Yaster M., Evans A., Rousset O., et al. Dopamine transporters are markedly reduced in Lesch-Nyhan disease in vivo. Proc. Natl Acad. Sci. USA. 1996;93:5539–5543. doi: 10.1073/pnas.93.11.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinnah H.A., Langlais P.J., Friedmann T. Functional analysis of brain dopamine systems in a genetic mouse model of Lesch-Nyhan syndrome. J. Pharmacol. Exp. Ther. 1992;263:596–607. [PubMed] [Google Scholar]
- 14.Jinnah H.A., Wojcik B.E., Hunt M., Narang N., Lee K.Y., Goldstein M., Wamsley J.K., Langlais P.J., Friedmann T. Dopamine deficiency in a genetic mouse model of Lesch-Nyhan disease. J. Neurosci. 1994;14:1164–1175. doi: 10.1523/JNEUROSCI.14-03-01164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinnah H.A., Jones M.D., Wojcik B.E., Rothstein J.D., Hess E.J., Friedmann T., Breese G.R. Influence of age and strain on striatal dopamine loss in a genetic mouse model of Lesch-Nyhan disease. J. Neurochem. 1999;72:225–229. doi: 10.1046/j.1471-4159.1999.0720225.x. [DOI] [PubMed] [Google Scholar]
- 16.Jinnah H.A., Hess E.J., Wilson M.C., Gage F.H., Friedman T. Localization of hypoxanthine-guanine phosphoribosyltransferase mRNA in the mouse brain by in situ hybridization. Mol. Cell. Neurosci. 1992;3:64–78. doi: 10.1016/1044-7431(92)90010-y. [DOI] [PubMed] [Google Scholar]
- 17.Jinnah H.A., Page T., Friedmann T. Brain purines in a genetic mouse model of Lesch-Nyhan disease. J. Neurochem. 1993;60:2036–2045. doi: 10.1111/j.1471-4159.1993.tb03488.x. [DOI] [PubMed] [Google Scholar]
- 18.Egami K., Yitta S., Kasim S., Lewers J.C., Roberts R.C., Lehar M., Jinnah H.A. Basal ganglia dopamine loss due to defect in purine recycling. Neurobiol. Dis. 2007;26:396–407. doi: 10.1016/j.nbd.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visser J.E., Smith D.W., Moy S.S., Breese G.R., Friedmann T., Rothstein J.D., Jinnah H.A. Oxidative stress and dopamine deficiency in a genetic mouse model of Lesch-Nyhan disease. Dev. Brain Res. 2002;133:127–139. doi: 10.1016/s0165-3806(02)00280-8. [DOI] [PubMed] [Google Scholar]
- 20.Hyland K., Kasim S., Egami K., Arnold L.A., Jinnah H.A. Tetrahydrobiopterin and brain dopamine loss in a genetic mouse model of Lesch-Nyhan disease. J. Inherit. Metab. Dis. 2004;27:165–178. doi: 10.1023/B:BOLI.0000028728.93113.4d. [DOI] [PubMed] [Google Scholar]
- 21.Smith D.W., Jinnah H.A. Role of neuronal nitric oxide in the dopamine deficit of HPRT-deficent mice. Metab. Brain Dis. 2007;22:39–43. doi: 10.1007/s11011-007-9044-7. [DOI] [PubMed] [Google Scholar]
- 22.Shirley T.L., Lewers J.C., Egami K., Majumdar A., Kelly M., Ceballos-Picot I., Seidman M.M., Jinnah H.A. A human neuronal tissue culture model for Lesch-Nyhan disease. J. Neurochem. 2007;101:841–853. doi: 10.1111/j.1471-4159.2007.04472.x. [DOI] [PubMed] [Google Scholar]
- 23.Lewers J.C., Ceballos-Picot I., Shirley T.L., Mockel L., Egami K., Jinnah H.A. Consequences of impaired purine recycling in dopaminergic neurons. Neuroscience. 2008;152:761–772. doi: 10.1016/j.neuroscience.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnier L., Le Pen G., Hartmann A., Bizot J.C., Trovero F., Krebs M.O., Prochiantz A. Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for engrailed1. J. Neurosci. 2007;27:1063–1071. doi: 10.1523/JNEUROSCI.4583-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kweon G.R., Marks J.D., Krencik R., Leung E.H., Schumacker P.T., Hyland K., Kang U.J. Distinct mechanisms of neurodegeneration induced by chronic complex I inhibition in dopaminergic and non-dopaminergic cells. J. Biol. Chem. 2004;279:51783–51792. doi: 10.1074/jbc.M407336200. [DOI] [PubMed] [Google Scholar]
- 26.Jankovic J., Caskey T.C., Stout J.T., Butler I.J. Lesch-Nyhan syndrome: a study of motor behavior and cerebrospinal fluid neurotransmitters. Ann. Neurol. 1988;23:466–469. doi: 10.1002/ana.410230507. [DOI] [PubMed] [Google Scholar]
- 27.Del Bigio M.R., Halliday W.C. Multifocal atrophy of cerebellar internal granular neurons in Lesch-Nyhan disease: case reports and review. J. Neuropathol. Exp. Neurol. 2007;66:346–353. doi: 10.1097/nen.0b013e3180515319. [DOI] [PubMed] [Google Scholar]
- 28.Abeliovich A., Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev. Biol. 2007;304:447–454. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 29.Alavian K.N., Scholz C., Simon H.H. Transcriptional regulation of mesencephalic dopaminergic neurons: the full circle of life and death. Mov. Disord. 2008;23:319–328. doi: 10.1002/mds.21640. [DOI] [PubMed] [Google Scholar]
- 30.Sgaier S.K., Lao Z., Villanueva M.P., Berenshteyn F., Stephen D., Turnbull R.K., Joyner A.L. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–2335. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prochiantz A., Joliot A. Can transcription factors function as cell–cell signalling molecules? Nat. Rev. Mol. Cell. Biol. 2003;4:814–819. doi: 10.1038/nrm1227. [DOI] [PubMed] [Google Scholar]
- 32.Brunet I., Di Nardo A.A., Sonnier L., Beurdeley M., Prochiantz A. The topological role of homeoproteins in the developing central nervous system. Trends Neurosci. 2007;30:260–267. doi: 10.1016/j.tins.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Hanks M., Wurst W., Anson-Cartwright L., Auerbach A.B., Joyner A.L. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 34.Sgado P., Alberi L., Gherbassi D., Galasso S.L., Ramakers G.M., Alavian K.N., Smidt M.P., Dyck R.H., Simon H.H. Slow progressive degeneration of nigral dopaminergic neurons in postnatal Engrailed mutant mice. Proc. Natl Acad. Sci. USA. 2006;103:15242–15247. doi: 10.1073/pnas.0602116103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Retaux S., Harris W.A. Engrailed and retinotectal topography. Trends Neurosci. 1996;19:542–546. doi: 10.1016/s0166-2236(96)10062-x. [DOI] [PubMed] [Google Scholar]
- 36.Foucher I., Montesinos M.L., Volovitch M., Prochiantz A., Trembleau A. Joint regulation of the MAP1B promoter by HNF3ß/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and Fox transcription factors. Development. 2003;130:1867–1876. doi: 10.1242/dev.00414. [DOI] [PubMed] [Google Scholar]
- 37.Brunet I., Weinl C., Piper M., Trembleau A., Volovitch M., Harris W., Prochiantz A., Holt C. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Nardo A.A., Nedelec S., Trembleau A., Volovitch M., Prochiantz A., Montesinos M.L. Dendritic localization and activity-dependent translation of Engrailed1 transcription factor. Mol. Cell Neurosci. 2007;35:230–236. doi: 10.1016/j.mcn.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Sriram G., Martinez J.A., McCabe E.R., Liao J.C., Dipple K.M. Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffery C.J. Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 2004;14:663–668. doi: 10.1016/j.sbi.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Jeffery C.J. Multifunctional proteins: examples of gene sharing. Ann. Med. 2003;35:28–35. doi: 10.1080/07853890310004101. [DOI] [PubMed] [Google Scholar]
- 42.Messina E., Micheli V., Giacomello A. Guanine nucleotide depletion induces differentiation and aberrant neurite outgrowth in human dopaminergic neuroblastoma lines: a model for basal ganglia dysfunction in Lesch-Nyhan disease. Neurosci. Lett. 2005;375:97–100. doi: 10.1016/j.neulet.2004.10.076. [DOI] [PubMed] [Google Scholar]
- 43.Messina E., Gazzaniga P., Micheli V., Guaglianone M.R., Barbato S., Morrone S., Frati L., Agliano A.M., Giacomello A. Guanine nucleotide depletion triggers cell cycle arrest and apoptosis in human neuroblastoma cell lines. Int. J. Cancer. 2004;108:812–817. doi: 10.1002/ijc.11642. [DOI] [PubMed] [Google Scholar]
- 44.Choi H.K., Won L.A., Kontur P.J., Hammond D.N., Fox A.P., Wainer B.H., Hoffmann P.C., Heller A. Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 1991;552:67–76. doi: 10.1016/0006-8993(91)90661-e. [DOI] [PubMed] [Google Scholar]
- 45.Craig S.P., 3rd, Yuan L., Kuntz D.A., McKerrow J.H., Wang C.C. High level expression in Escherichia coli of soluble, enzymatically active schistosomal hypoxanthine/guanine phosphoribosyltransferase and trypanosomal ornithine decarboxylase. Proc. Natl Acad. Sci. USA. 1991;88:2500–2504. doi: 10.1073/pnas.88.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. Green mice as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 47.Cartier P., Hamet M. Purine phosphoribosyltransferase activity of human erythrocytes: technic of determination. Clin. Chim. Acta. 1968;20:205–214. doi: 10.1016/0009-8981(68)90152-6. [DOI] [PubMed] [Google Scholar]
- 48.van Bogaert P., Ceballos I., Desguerre I., Telvi L., Kamoun P., Ponsot G. Lesch-Nyhan syndrome in a girl. J. Inherit. Metab. Dis. 1992;15:790–791. doi: 10.1007/BF01800022. [DOI] [PubMed] [Google Scholar]
- 49.Golfier G., Dang M.T., Dauphinot L., Graison E., Rossier J., Potier M.C. VARAN: a web server for variability analysis of DNA microarray experiments. Bioinformatics. 2004;20:1641–1643. doi: 10.1093/bioinformatics/bth117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.