Abstract
Mutations in the gene encoding tripartite motif protein 32 (TRIM32) cause two seemingly diverse diseases: limb-girdle muscular dystrophy type 2H (LGMD2H) or sarcotubular myopathy (STM) and Bardet–Biedl syndrome type 11(BBS11). Although TRIM32 is involved in protein ubiquitination, its substrates and the molecular consequences of disease-causing mutations are poorly understood. In this paper, we show that TRIM32 is a widely expressed ubiquitin ligase that is localized to the Z-line in skeletal muscle. Using the yeast two-hybrid system, we found that TRIM32 binds and ubiquitinates dysbindin, a protein implicated in the genetic aetiology of schizophrenia, augmenting its degradation. Small-interfering RNA-mediated knock-down of TRIM32 in myoblasts resulted in elevated levels of dysbindin. Importantly, the LGMD2H/STM-associated TRIM32 mutations, D487N and R394H impair ubiquitin ligase activity towards dysbindin and were mislocalized in heterologous cells. These mutants were able to self-associate and also co-immunoprecipitated with wild-type TRIM32 in transfected cells. Furthermore, the D487N mutant could bind to both dysbindin and its E2 enzyme but was defective in monoubiquitination. In contrast, the BBS11 mutant P130S did not show any biochemical differences compared with the wild-type protein. Our data identify TRIM32 as a regulator of dysbindin and demonstrate that the LGMD2H/STM mutations may impair substrate ubiquitination.
INTRODUCTION
The ubiquitin–proteasome system (UPS) is the major pathway responsible for protein degradation in eukaryotic cells (1). Lysine residues within target proteins are modified by covalent attachment of the 8.5 kDa regulatory protein ubiquitin through the sequential action of three ubiquitin ligases: E1, E2 and E3 (2). Ubiquitin can itself serve as an acceptor for further rounds of ubiquitination, forming a polyubiquitin chain. Lysine 48 (K48)-linked polyubiquitin chains are recognized by the proteasome, which rapidly degrades the modified protein. Through its ability to selectively target proteins for destruction, the UPS is involved in all aspects of eukaryotic cell biology and has an important role in degrading misfolded proteins (3).
In humans, there are 68 members of the TRIM protein family that contain the eponymous tripartite motif domains: RING, BBOX and a coiled-coil (4,5). Many TRIM proteins have additional and variable C-terminal domains, including the SP1a and the RYanodine receptor (SPRY), NCL-1, HT2A and LIN-41 (NHL) and plant homeodomain (PHD) domains (4). Despite the prevalence of TRIM genes in the genome, most members of this protein family have poorly defined biological functions. A small number of TRIM family members have been shown to possess E3 ubiquitin ligase activity and have been implicated in different diseases states, such as cancer (6–8), retroviral restriction (5) and inherited genetic disease (9–12), indicating that they have important cellular roles.
In 2002, Frosk et al. (9) described a mutation, D487N, in the gene encoding tripartite motif protein 32 (TRIM32) as the SPRY cause of limb-girdle muscular dystrophy type 2H (LGMD2H). LGMD2H is a mild, autosomal recessive muscle disease that was originally described in the Manitoba Hutterite population (13). Historically, these patients were described as having sarcotubular myopathy (STM). Clinically, the disease is characterized by moderately elevated serum creatine kinase levels, slowly progressive weakness and wasting of the proximal limb musculature and a relatively late onset in the second or third decade of life. Similarly, Müller-Felber et al. (14) described two German patients with a congenital, non-progressive muscle disease with segmental vacuolation of some muscle fibres diagnosed as STM. The muscle biopsies of these patients showed evidence of Z-line streaming, dilation of the sarcotubular system, autophagic double-membraned vacuoles and an increased number of centrally located myonuclei. Although STM has an earlier onset and is clinically more severe than LGMD2H, the same D487N mutation has been shown to cause both disorders suggesting that the clinical phenotype is heterogeneous (15). This mutation resides in the third NHL repeat of TRIM32 (9). These repeats are thought to fold into a six-bladed β-propeller structure (16,17). Studies on proteins such as Brat and malin have shown that the NHL repeats mediate protein–protein interactions (16–18). Three additional mutations in the first (R394H), fourth (T520TfsX13) and fifth (D588del) NHL repeats of TRIM32 were also found to cause LGMD2H in non-Hutterite populations (19). Interestingly, this study also identified one patient with LGMD2H attributable to heterozygous mutation R394H/+ in the TRIM32 gene (19). Recently, a mouse model of LGMD2H/STM has been described that recapitulates many of the features in the LGMD2H/STM in humans (20). However, in addition to muscular dystrophy, these mice also have evidence of neurological involvement that may contribute to some of the myopathic changes observed in their muscles. These changes have been attributed in part to decreased levels of neurofilaments in Trim32 knockout mice concomitant with a reduction in the diameter of myelinated motor axons (20).
In contrast, a missense mutation P130S, in the BBOX domain of TRIM32, causes Bardet–Biedl syndrome type 11 (BBS11) (21). BBS is a complex condition associated with obesity, retinal degeneration, genito-urinary tract malformations and cognitive impairment (22). Furthermore, BBS appears to be associated with an increased incidence of psychiatric and behavioural abnormalities (23,24). In mammals, the majority of the BBS proteins participate in the formation of the primary cilium and exist in a 450 kDa multiprotein complex, termed the BBSsome (25). In addition to their role in ciliogenesis, BBS proteins participate in a number of signalling pathways and may have a more general role in vesicular trafficking and movement of intracellular organelles such as melanosomes (26). Intriguingly, no muscle abnormality is found in BBS patients, suggesting different aspects of TRIM32 function are disrupted by the LGMD2H mutations.
Although TRIM32 is thought to be involved in protein ubiquitination, its precise physiological role in muscle is unknown (27). A recent report investigating the role of TRIM32 in muscle demonstrated that TRIM32 binds to myosin IIA and ubiquitinates actin in vitro (27). This study also concluded that TRIM32–D487N behaved like the wild-type protein with respect to its ability to monoubiquitinate actin. In a separate study, Saccone et al. (19) have reported that the D487N mutation impairs both self-association and E2-binding in yeast. Paradoxically, this defect would be predicted to drastically impair substrate ubiquitination.
In addition to its role in genetic disease, TRIM32 is up-regulated in epidermal carcinogenesis, where it is thought to increase keratinocyte survival by blocking ultraviolet light B (UVB)-induced tumour necrosis factor-α (TNFα) apoptotic signalling (28). This effect was later shown to be mediated through TRIM32 ubiquitination and destabilization of a pro-apoptotic SUMO-E3 ubiquitin ligase protein inhibitor of activated STAT Y (PIASy) which exhibited altered intracellular distribution in response to UVB/TNFα-treatment (29). Whether PIASy is a substrate of TRIM32 in muscle under normal conditions has yet to be determined.
The aim of this study was to elucidate the cellular role of TRIM32 and to investigate how TRIM32 mutations might alter the function of the protein. We have found that TRIM32 is able to bind and ubiquitinate dysbindin, a protein involved in endosomal–lysosomal trafficking and the genetic aetiology of schizophrenia (30). The LGMD2H/STM mutants have impaired ubiquitin ligase activity, were mislocalized in heterologous cells but were able to self-associate. Furthermore, the common D487N mutant was able to bind to its E2 enzyme but was not monoubiquitinated under basal conditions. In contrast, no biochemical differences between wild-type TRIM32 and the BBS11-associated P130S mutant were found. Our data suggest that TRIM32 may be a regulator of dysbindin and highlight important functional differences between wild-type TRIM32 and the LGMD2H/STM mutants D487N and R394H.
RESULTS
TRIM32 can ubiquitinate dysbindin in heterologous cells
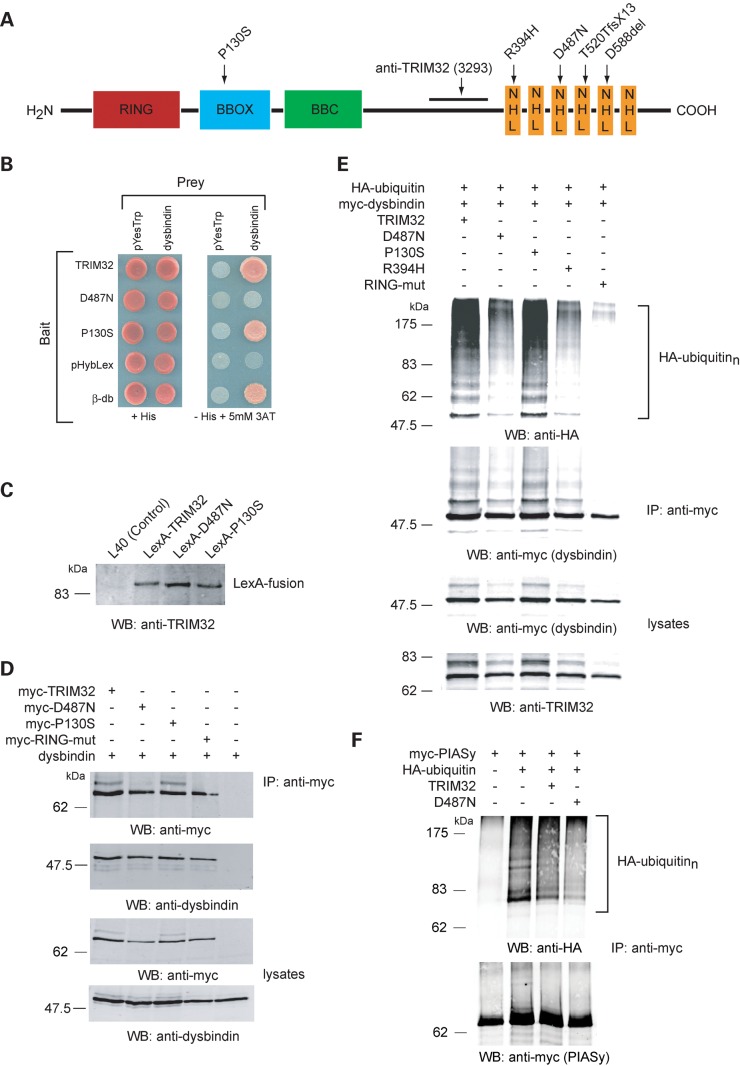
TRIM32 is an E3 ubiquitin ligase composed of several distinct domains (Fig. 1A). Mutations associated with muscle disease cluster within the NHL repeats, whereas the BBS11 mutation is found in the BBOX domain (Fig. 1A). The yeast two-hybrid system (Y2H) was used to identify potential TRIM32-interacing proteins from a mouse skeletal muscle cDNA library. This screen isolated three independent full-length clones encoding the protein dysbindin. Subsequently, we used the Y2H system to determine whether the D487N (LGMD2H) and P130S (BBS11) mutants, generated by site-directed mutagenesis, could interact with dysbindin. In yeast, the LGMD2H mutant TRIM32–D487N failed to interact with dysbindin, whereas a robust interaction between TRIM32–P130S and dysbindin was found (Fig. 1B). To ensure that the TRIM32-, D487N- and P130S-LexA fusion proteins were being expressed at comparable levels, protein extracts were prepared from each yeast strain and analysed by western blot using the anti-TRIM32 antibody (Fig. 1C). Characterization of the anti-TRIM32 antibody is shown in Supplementary Material, Fig. S1A. Each protein was produced at similar levels in yeast suggesting that the lack of interaction between the LGMD mutant and dysbindin did not result from reduced expression of the LGMD mutant bait plasmid.
Figure 1.
Dysbindin interacts with TRIM32 in yeast and in vivo. TRIM32 and P130S interact with dysbindin in the Y2H system. (A) Schematic representation of the domain organization of TRIM32 and location of the disease-associated mutations and the region used for polyclonal rabbit anti-sera generation. (B) TRIM32, D487N and P130S bait strains were co-transformed with empty prey plasmid (pYesTrp2) or dysbindin prey plasmid. The double transformants were tested for histidine auxotrophy. The previously characterized β-dystrobrevin (β-db) bait is used as a positive control for the dysbindin interaction. (C) Bait strain protein lysates were western blotted with the TRIM32 3293 antibody, showing all three DNA-binding LexA-fusions are synthesized at comparable levels in yeast. (D) TRIM32 and dysbindin interact in mammalian cells. HEK293T cells were transfected with dysbindin and myc-TRIM32, myc-D487N, myc-P130S, myc-RING-mut. Dysbindin co-immunoprecipitated only in the presence of myc-TRIM32 (or any of the mutants), indicating they form a complex in transfected cells. (E) Dysbindin is polyubiquitinated by TRIM32. Myc-dysbindin was immunoprecipitated from HEK293T cells expressing HA-ubiquitin and either TRIM32, D487N, P130S, R394H or RING-mut. Immunoprecipitated dysbindin was probed for the presence of conjugated HA-ubiquitin by western blot. A high molecular weight smear is seen on the HA blot indicative of polyubiquitination. As expected, the RING mutant is unable to ubiquitinate dysbindin. Interestingly, D487N and R394H have impaired ubiquitin ligase activity towards dysbindin, exemplified by the weaker smearing on the HA blot. (F) Effect of TRIM32 on the ubiquitination of PIASy. The ubiquitination of PIASy was assessed under identical condition to those for dysbindin as shown in (E). Co-expression of TRIM32 or TRIM32–D487N had no apparent effect upon the ubiquitination of PIASy. Note that PIASy is modified by endogenous ubiquitin ligases when expressed in HEK293T cells.
To confirm the interaction between TRIM32 and dysbindin in mammalian cells, myc-tagged TRIM32 (and mutants thereof) and dysbindin were transiently transfected into HEK293T cells. TRIM32 was immunoprecipitated from cell extracts using the anti-myc 9E10 antibody. The immune complexes were immunoblotted and probed for the presence of dysbindin using the anti-dysbindin antibody PA3111A (31). Figure 1D shows that wild-type TRIM32 and all of the mutants [D487N, P130S and RING mutant (RING-mut)] could co-immunoprecipitate with dysbindin from HEK293T cell extracts.
To determine whether TRIM32 could act as an E3 ubiquitin ligase for dysbindin, various combinations of expression constructs encoding TRIM32, TRIM32–D487N, TRIM–P130S, TRIM32–RING-mut, myc-dysbindin and haemaglutinin (HA)-ubiquitin were transfected in to HEK293T cells. In addition to the aforementioned mutants, we also included a second LGMD2H mutant R394H, which was recently found in two patients from Southern Italy (19). The RING-mut construct was produced by site-directed mutagenesis changing three conserved Zn2+-co-ordinating residues (C21, C40 and H42) in the RING domain to alanine. These mutations were predicted to impair the activity of TRIM32 as an ubiquitin ligase. Transfected cells were lysed under denaturing and reducing conditions (1% SDS, 10 mm DTT) to prevent polyubiquitinated proteins co-immunoprecipitating with dysbindin. Myc-dysbindin was immunoprecipitated from the cell extracts using the 9E10 antibody. Ubiquitinated dysbindin was identified by immunoblotting immunoprecipitated myc-dysbindin with the anti-HA antibody. A band corresponding to monoubiquitinated dysbindin is detected with both the anti-myc and the anti-HA antibodies when wild-type TRIM32 is co-expressed (Fig. 1E). The anti-HA-ubiquitin blot also detects high molecular weight smearing indicative of polyubiquitination. This smearing is present in immunoprecipitates from cells expressing both wild-type TRIM32 and the P130S mutant. Interestingly, the intensity of the polyubiquitin smearing was markedly reduced for the LGMD2H/STM-associated D487N and R394H mutants, suggesting that these mutations are associated with impaired ubiquitin ligase activity. As expected, mutation of the RING domain abolished ubiquitination of dysbindin demonstrating that this ubiquitination was solely due to the function of TRIM32 and not endogenous E3 ubiquitin ligases. To negate the possibility that the impaired ubiquitin ligase activity of the D487N and RING-mut was due to the inherent instability of the protein in transfected cells, we determined the half-lives of each protein. Wild-type TRIM32 and the D487N mutant have very similar half-lives (17.7 and 18.5 h, respectively), whereas P130S and the RING-mut were slightly more stable (Supplementary Material, Table S1).
In control experiments, we performed the identical ubiquitination assays using myc-PIASy instead of myc-dysbindin. PIASy is polyubiquitinated by endogenous E3 ligases in HEK293T cells in the absence of TRIM32. This ubiquitination does not increase in the presence of TRIM32 expression (Fig. 1F). As suggested by Albor et al. (29), one possible explanation for this finding is that PIASy translocation from the nucleus occurs only under UVB/TNFα/MG132 treatment, which then permits TRIM32 to modify PIASy. Without eliciting a stress response, PIASy and TRIM32 are in different subcellular compartments, where interaction and ubiquitination would be unlikely to occur (see below and Fig. 2A).
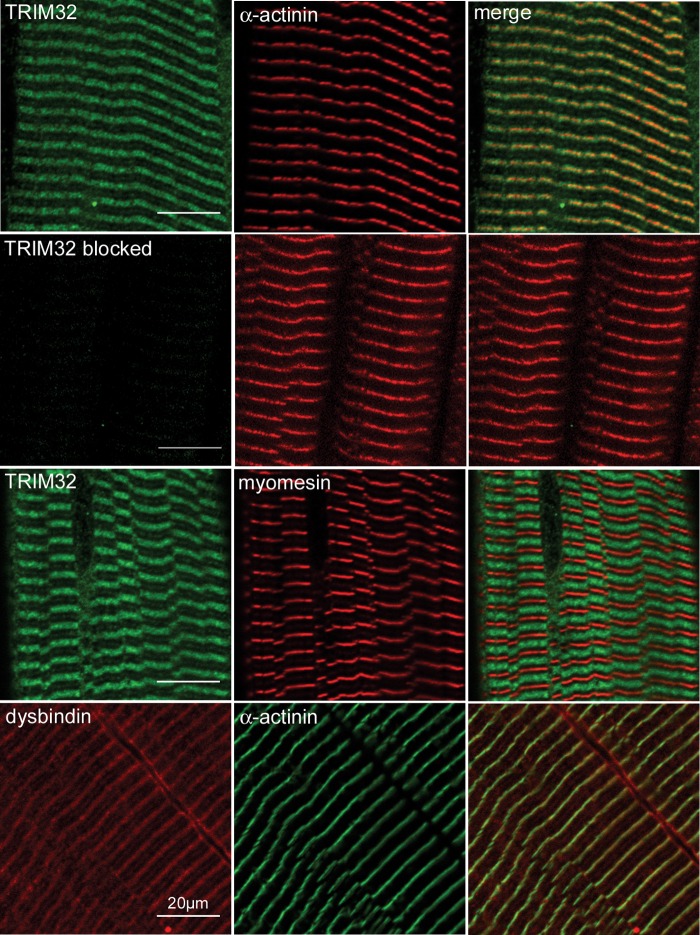
Figure 2.
Localization of TRIM32 and dysbindin in skeletal muscle. Longitudinal sections of guinea pig tibialis anterior muscle were stained with the antibodies 3293 (TRIM32), EA-53 (α-actinin), B4 (myomesin) and PA3111A (dysbindin). Primary antibodies were detected using species-specific fluorescent antibody conjugates (Alexa-488 or rhodamine Red-X). TRIM32 and dysbindin colocalize with α-actinin, suggesting that each protein is found on and around the periphery of the Z-line. In addition to the Z-line labelling, dysbindin is also found at the sarcolemma and M-band. Pre-incubation of the anti-TRIM32 antibody with the fusion protein used for immunization blocked labelling in muscle sections but did not interfere with immunoreactivity of α-actinin on the same tissue section. Scale bar is 20 µm.
Synthesis and localization of TRIM32 and dysbindin in muscle
Western and northern blot analyses of TRIM32 and dysbindin have shown that each protein is expressed at relatively low levels in skeletal muscle compared with other tissues such as the brain (9,20,31) (Supplementary Material, Fig. S1B). To determine the subcellular localization of TRIM32 and dysbindin in skeletal muscle, cryosections of guinea pig skeletal muscle were labelled with rabbit polyclonal antibodies raised against each protein. Dysbindin immunoreactivity, detected with PA3111A, could be seen around the Z-line and sarcolemma of all fibres, as previously reported (Fig. 2) (31). TRIM32 immunoreactivity, detected with the 3293 polyclonal antibody (Supplementary Material, Fig. S1), was also found to be concentrated around the Z-line but was not found at the sarcolemma (Fig. 2). Both antigens extended beyond the boundary of the Z-line. Furthermore, TRIM32 and dysbindin are also found at the M-line, although the labelling is considerably weaker compared with the Z-line (Fig. 2). The location of the Z-line and M-band in each experiment was identified using monoclonal antibodies that detect α-actinin (Z-line) and myomesin (M-band). Pre-absorption of the anti-TRIM32 polyclonal antibody with the immunising fusion protein abolished labelling on muscle sections but had no effect on the detection of α-actinin (Fig. 2). Similarly, pre-absorption of the TRIM32 antibody with the immunogen blocked specific labelling in transfected cells over-expressing TRIM32–EYFP (Supplementary Material, Fig. S2A). Western blotting confirmed the presence of TRIM32 in muscle extracts prepared from different guinea pig muscles (Supplementary Material, Fig. S3A).
We also determined the levels of TRIM32 and dysbindin during C2C12 myoblast differentiation. Using quantitative western blotting, we found that TRIM32 and dysbindin remained at relatively constant levels during myoblast differentiation over a 12 days period (Supplementary Material, Fig. S3B). As a positive control for differentiation, identical cell extracts were probed with the previously characterized anti-α-dystrobrevin-1 antibody (α1CT-FP), which detects an α-dystrobrevin-1 isoform known to be induced during myogenesis (α-dystrobrevin+vr3) (32). These data suggest that the temporal and spatial distributions of TRIM32 and dysbindin are similar in skeletal muscle (Supplementary Material, Fig. S3B).
Immunolocalization of TRIM32, its mutants and dysbindin in transfected cells
To determine whether TRIM32 and dysbindin are expressed in the same subcellular compartments, TRIM32–EYFP and dysbindin were transfected into COS-7 cells. Following fixation, the cells were stained for dysbindin and analysed by confocal microscopy. In agreement with previous reports, TRIM32–EYFP is seen as cytoplasmic speckles (sometimes referred to as cytoplasmic bodies), often located around the nucleus (Fig. 3A). The size and number of the TRIM32 speckles were dependent on the expression level of the protein, similar to TRIM5α cytoplasmic bodies (33). Cells expressing low levels of TRIM32 had a greater number of cytoplasmic bodies with lower fluorescence intensity, whereas in the cells expressing higher amounts of TRIM32, the cytoplasmic speckles appear to coalesce to form larger aggregates that were fewer in number. These cytoplasmic speckles were also immunoreactive for ubiquitinated proteins detected with the anti-FK1 monoclonal antibody (Fig. 3B). Dysbindin exhibited a diffuse cytoplasmic staining pattern, as previously reported (31). In cells co-expressing dysbindin and TRIM32, dysbindin immunoreactivity was found in aggregates that also contained TRIM32–EYFP, suggesting that they are capable of interacting in the cytoplasm of mammalian cells (Fig. 3A). In control experiments, we confirmed that the apparent co-localization of dysbindin and TRIM32 in aggregates in co-transfected cells was not the result of bleed-through from the different channels following sequential scans on the confocal microscope (Supplementary Material, Fig. S2B).
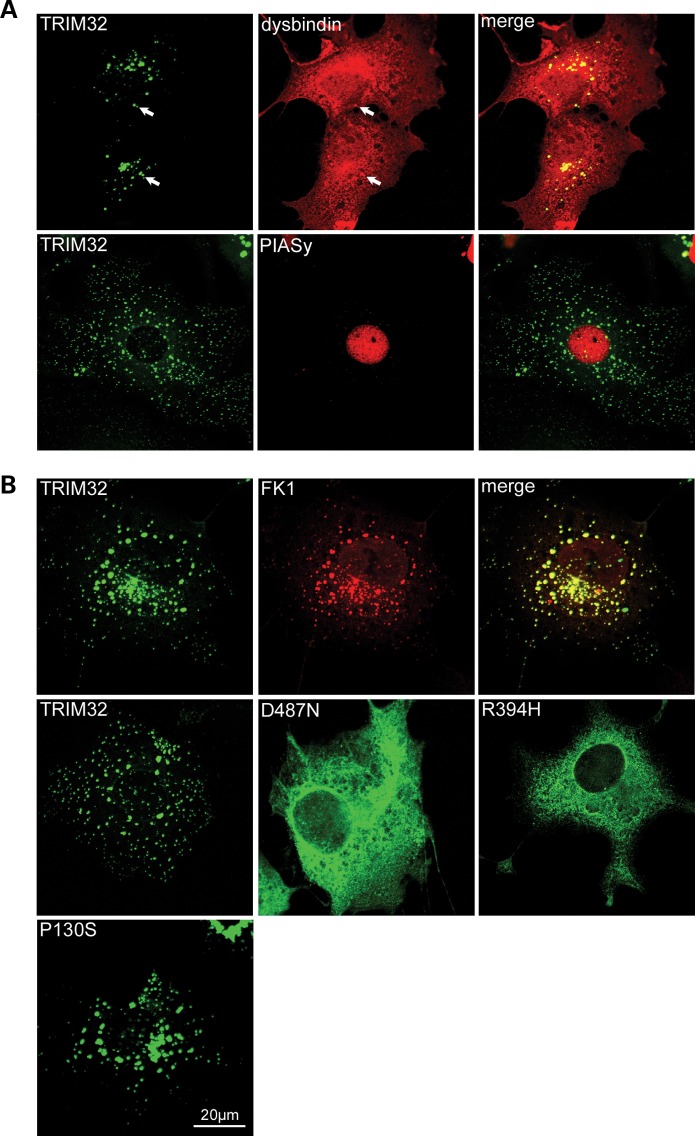
Figure 3.
Subcellular distribution of TRIM32, dysbindin and PIASy in COS-7 cells. (A) Co-localization of TRIM32–EYFP and dysbindin in COS-7 cells. TRIM32–EYFP and dysbindin were transfected into COS-7 cells and stained for dysbindin using PA3111A (red). Dysbindin is diffusely expressed in the cytoplasm and nucleus. In addition, dysbindin can be found in aggregates that co-localize with the TRIM32–EYFP fluorescence (arrows), which appear yellow in the merged image. Myc-tagged PIASy and TRIM32–EYFP were transfected into COS-7 cells and stained for myc using 9E10 (red). PIASy exhibited a nuclear localization, with TRIM32 residing in cytoplasmic speckles. (B) Localization of TRIM32–EYFP and mutants in COS-7 cells. COS-7 cells were transfected with TRIM32–EYFP, D487N–EYFP, R394H–EYFP and P130S–EYFP. TRIM32 and P130S form discrete cytoplasmic speckles, whereas D487N and R394H are predominantly cytoplasmic. TRIM32–EYFP transfected COS-7 cells were stained with FK1, which detects polyubiquitinated proteins (red). Merging the two images shows that the two antigens predominantly co-localize indicating that TRIM32–EYFP cytoplasmic speckles are associated with polyubiquitinated proteins.
For comparative purposes, we co-transfected TRIM32–EYFP and myc-PIASy, a putative TRIM32 substrate. As previously reported, in the absence of proteasome inhibition and irradiation PIASy had a nuclear localization that is clearly distinct from TRIM32 in cytoplasmic speckles (Fig. 3A) (29). We also examined the subcellular localization of the TRIM32 mutants in COS-7 cells (Fig. 3B). P130S–EYFP displayed a similar distribution to the wild-type protein, forming distinct cytoplasmic speckles. In contrast, the LGMD2H/STM-associated D487N–EYFP and R394H–EYFP mutants exhibited diffuse cytoplasmic staining, with loss of the characteristic aggregates, which were only visible in a minority of cells expressing high levels of the chimera. Similar results were found in C2C12 myoblasts (Supplementary Material, Fig. S4). The differences in the immunolocalization of the LGMD2H/STM mutants compare with the wild-type protein are unlikely to be a result of expression difference because each EYFP–chimera was synthesised at similar levels (data not shown).
TRIM32 targets dysbindin for degradation
Polyubiquitination is commonly associated with the proteasomal degradation of certain proteins. Therefore, we examined whether polyubiquitination of dysbindin by TRIM32 had any effect on its turnover. Dysbindin was transfected into COS-7 cells with either wild-type TRIM32 or the RING-mut. Protein synthesis was blocked with cycloheximide and samples were taken every 5 h. The levels of protein remaining at each time point were determined by quantitative western blotting. Figure 4 shows that the presence of TRIM32 had a destabilising effect on dysbindin compared with the ubiquitination-defective RING-mut (Fig. 4B). The western blots show a ladder of lower molecular weight bands < 50 kDa, which are only present when dysbindin is co-expressed with TRIM32, but not with the RING-mut (asterisk in Fig. 4A). It is possible that these bands represent dysbindin breakdown products. Since the RING-mut is able to bind (Fig. 1C), but not ubiquitinate dysbindin, we conclude that polyubiquitination of dysbindin by wild-type TRIM32 augments its degradation.
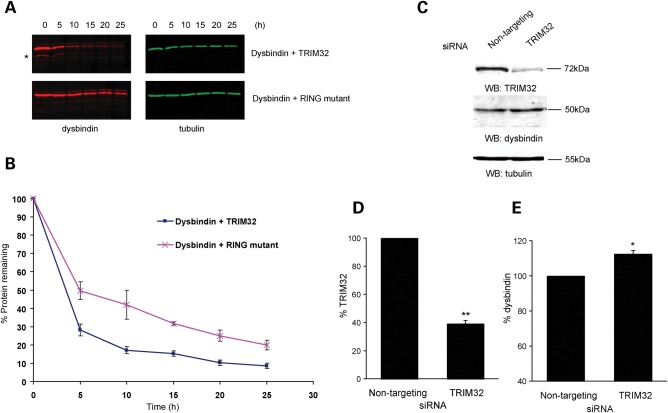
Figure 4.
TRIM32 targets dysbindin for degradation. (A) COS-7 cells were transfected with dysbindin (red) and TRIM32 (green), or dysbindin and the RING mutant (RING-mut) (all untagged in pCIneo). Protein synthesis was blocked by cycloheximide treatment (50 µg/ml) and samples taken every 5 h. The levels of dysbindin in lysates was analysed by quantitative western and plotted graphically (B). The presence of TRIM32 destabilizes dysbindin, increasing its turnover relative to co-expression the RING-mut. Additional bands that might be possible breakdown products of dysbindin can be seen below the main dysbindin band in the presence of TRIM32, but not the RING-mut (asterisk in A). The error bars show the standard error of the mean from three independent experiments. (C) Knockdown of TRIM32 in C2C12s by siRNA increases dysbindin levels. C2C12 cells were transfected on consecutive days with either control siRNA (non-targeting pool) or TRIM32 siRNA. Seventy-two hours after the first transfection lysates were made and levels of TRIM32, dysbindin and tubulin were analysed by quantitative western blot (WB). TRIM32 and dysbindin levels were normalized to tubulin and plotted as a chart (D). The error bars show the standard error of the mean from three independent experiments. The TRIM32-specific J-12 duplex achieved ∼60% knockdown relative to the control siRNA (**P < 0.01 in a one-sample two-tailed t-test). (E) Knockdown of TRIM32 results in a significant increase in dysbindin levels relative to control (*P < 0.05 in a one-sample two-tailed t-test).
Our data presented herein point to a potential loss of function for the LGMD2H/STM TRIM32 mutations D487N and R394H. To determine whether TRIM32 could regulate dysbindin levels in vivo, we used small-interfering RNAs (siRNAs) to ‘knockdown’ TRIM32 in C2C12 myoblasts. We first determined the half-life of endogenous TRIM32 in C2C12 myoblasts. Endogenous TRIM32 appeared to be a relatively stable protein, with a half-life of ∼33 h (Supplementary Material, Table S1). C2C12 myoblasts were transfected with either a TRIM32-specific siRNA or non-targeting control siRNAs. Whole cell lysates taken 72 h post-transfection were analysed by quantitative immunoblotting. The TRIM32 siRNA achieved ∼60% knockdown compared with the control siRNA duplex (Fig. 4C and D). Conversely, dysbindin levels increased by ∼12% in response to depleted TRIM32 (Fig. 4C and E). The increase in dysbindin levels following TRIM32 knockdown was statistically significant, with a P-value of < 0.05 in a one-sample two-tailed t-test. Each experiment was performed in quadruplicate. Tubulin levels were unaffected by TRIM32 knockdown. The moderate increase in endogenous dysbindin levels following siRNA-mediated knockdown of TRIM32 may be attributable to the partial co-localization of TRIM32 and dysbindin in transfected cells (Fig. 3A) or the stability and degree of knockdown of TRIM32 in C2C12 myoblasts.
TRIM32 mutants are not defective in self-association
Having determined that the LGMD2H/STM mutations D487N and R394H impaired dysbindin ubiquitination, we preceded to examine the ability of TRIM32 to self-associate. In addition to dysbindin, TRIM32 was also identified as TRIM32-binding protein in our Y2H screens of muscle and brain cDNA libraries (data not shown) indicating the propensity to self-associate. Furthermore, members of the TRIM family can form dimeric or trimeric complexes mediated by the tripartite motif (4). Previous reports have used the Y2H system to show that mutations in the NHL repeats of TRIM32 impaired self-association (19). Given the previous dichotomy between results obtained in yeast and mammalian cells (Fig. 1B and D), we examined the self-association properties of TRIM32 and its mutants in both systems. Bait and prey constructs were produced for each mutant and co-transformed into the reporter yeast strain L40. In the Y2H system, wild-type TRIM32 and P130S co-transformants could grow in the absence of histidine, indicating that they self-associate (Fig. 5A). In contrast, and in agreement with previous reports, D487N and R394H failed to grow on histidine-deficient media (19).
Figure 5.
Self-association properties of TRIM32 and mutants. (A) Y2H-mediated analysis of TRIM32 self-association. Co-transformants containing TRIM32, D487N, P130S, R394H bait and prey plasmids or empty bait, and prey plasmids were tested for histidine auxotrophy on media lacking histidine containing 5 mm 3-AT. Only wild-type TRIM32 and P130S could self-associate in yeast. In each case, selective and non-selective media are compared. (B) Wild-type TRIM32, D487N and P130S self-associate in mammalian cells. HEK293T cells were transfected with myc-tagged TRIM32, D487N or P130S and their EYFP-tagged counterparts. EYFP-tagged proteins are only immunoprecipitated in the presence of the myc-tagged protein, indicating that the myc-tagged and EYFP-tagged proteins interact. (C) The LGMD2H/STM mutants failed to interact with wild-type TRIM32 in yeast. Bait and preys were co-transformed into yeast as indicated. TRIM32 is able to self-associate, but failed to interact with either of the LGMD2H/STM mutants. In each case, selective and non-selective media are compared. (D) Co-immunoprecipitation of wild-type TRIM32 with the LGMD2H/STM mutants, D487N and R394H. HEK293T cells were transfected with the constructed as indicated. Proteins were immunoprecipitated from the cell extracts using the anti-myc antibody. The anti-GFP antibody was used to detect the co-immunoprecipitated EYFP-tagged TRIM32. In each case, TRIM32 and the each of the mutants co-immunoprecipitated with myc-tagged wild-type TRIM32. The EYFP-tagged proteins did not immunoprecipitate in the absence of myc-tagged wild-type TRIM32 demonstrating that non-specific binding to the beads was not a confounding factor in this experiment. (E) Dysbindin ubiquitination in cells co-expressing wild-type and mutant TRIM32s. Ubiquitination assays were performed as described above (Fig. 1E). Myc-tagged dysbindin was immunoprecipitated from cells expressing the designated combination of plasmids. In each case, wild-type TRIM32 was co-expressed with either 0.5 µg of TRIM32–EYFP, D487N–EYFP, R394H–EYFP or RING-mut–EYFP. Dysbindin ubiquitination was assessed with the anti-HA antibody as described above. In each reaction, dysbindin is clearly polyubiquitinated by each combination of TRIM32 and mutant. However in extracts from cells expressing myc-TRIM32 and R394H–EYFP, we consistently found a reduction in the levels of the monoubiquitinated form of dysbindin (asterisk). This change was also detected with the anti-dysbindin antibody (asterisk). Inspection of the lysates revealed that similar levels of myc-TRIM32, dysbindin and the EYFP-tagged mutants were produced in each transfection.
In complimentary experiments, co-immunoprecipitation was used to examine the self-association properties of TRIM32 and mutants thereof expressed in mammalian cells. The 9E10 antibody was used to co-immunoprecipitate the corresponding EYFP-tagged proteins from HEK293T cells co-transfected with myc-tagged TRIM32 or mutants thereof. In contrast to the data obtained in yeast, we found that myc-tagged TRIM32 and each of the mutants co-immunoprecipitated with their cognate EYFP-tagged construct indicating the propensity for each protein to self-associate (Fig. 5B). On the basis of the studies in mammalian cells, it appears that neither the P130S nor D487N mutations affect the ability of TRIM32 to self-associate. However, in light of the data from the Y2H system, it is equally possible that the interaction between dysbindin and TRIM32 (and the mutants) in HEK cells is mediated by a ‘bridging’ protein or post-translational modification that is not present in yeast.
In light of the interesting findings of Saccone et al. (19), showing that an LGMD2H patient was heterozygous for the R394H allele, we tested the ability of the LGMD2H/STM mutants to associate with the wild-type protein in yeast and in mammalian cells. Neither of the mutants was able to interact with wild-type TRIM32 in the Y2H system (Fig. 5C). In contrast to the results obtained using yeast, both LGMD2H/STM mutants immunoprecipitated with wild-type TRIM32 when co-expressed in HEK293T cells (Fig. 5D). Having found that a wild-type protein was able to interact with a mutant albeit in heterologous cells, we tested the ability of the D487N and R394H mutants to ubiquitinate dysbindin in the presence of wild-type TRIM32. These experiments were performed as described above; however, each transfection was spiked with 0.5 µg of a plasmid encoding TRIM32–EYFP, D487N–EYFP, R394H–EYFP or RING-mut–EYFP. Each condition was compared with the ubiquitination-defective RING-mut, since this would represent a mixture of an active and inactive ubiquitin ligase (Fig. 5E). Although no gross differences in dysbindin ubiquitination were observed, it is notable that the levels of monoubiquitinated dysbindin were reduced when TRIM32 was co-expressed with the R394H mutant (asterisks in Fig. 5E). This result was found in three separate experiments and may reflect the ability of the R394H mutant to attenuate the activity of wild-type TRIM32. Finally, we co-expressed the EYFP-tagged LGMD2H/STM mutants and wild-type TRIM32 in COS-7 cells to determine whether co-expression of TRIM32 could alter the cytoplasmic distribution of the mutants that was reported above (Fig. 3B). In cells expressing myc-TRIM32 and the EYFP-tagged mutants, both proteins co-localized in cytoplasmic bodies that were reminiscent of wild-type TRIM32 (Supplementary Material, Fig. S5).
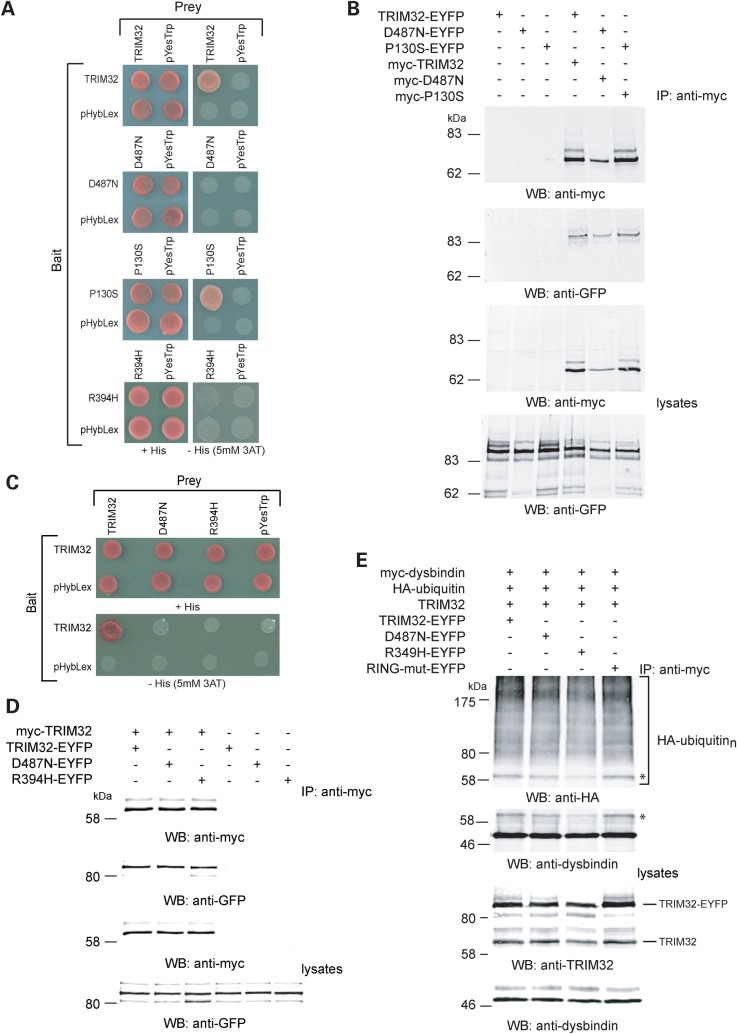
TRIM32 mutants can bind to their E2-enzyme in mammalian cells
The RING domain is thought to be responsible for recruiting E2 ubiquitin-conjugating enzymes to RING E3 ligases. It has recently been suggested that the D487N mutation abolished the interaction of TRIM32 with the E2 enzyme, UBE2N in yeast cells (19). Since D487N, and the RING-mut, appeared to have impaired ubiquitin ligase activity towards dysbindin, we tested whether these proteins could bind to the E2 enzyme UbcM3 in mammalian cells. UbcM3 is the mouse orthologue of human UbcH6, which has previously been shown to be compatible with TRIM32 in in vitro ubiquitination assays (27). TRIM32 and each of the mutants co-immunoprecipitate with HA-tagged UbcM3 from mammalian cells (Fig. 6). These data, and those of Gentry et al. (18), highlight potential limitations in the utility of the Y2H system to examine the interaction between mutant ubiquitin ligases and their associated proteins.
Figure 6.
TRIM32 mutants bind to the E2 ubiquitin-conjugating enzyme UbcM3. Myc-tagged TRIM32 (and mutants) were immunoprecipitated from mammalian cells in low stringency RIPA buffer and probed for the presence of co-immunoprecipitated HA-UbcM3 by western blot using the HA antibody. All of the TRIM32 variants could immunoprecipitate UbcM3, with the RING mutant having the most robust interaction.
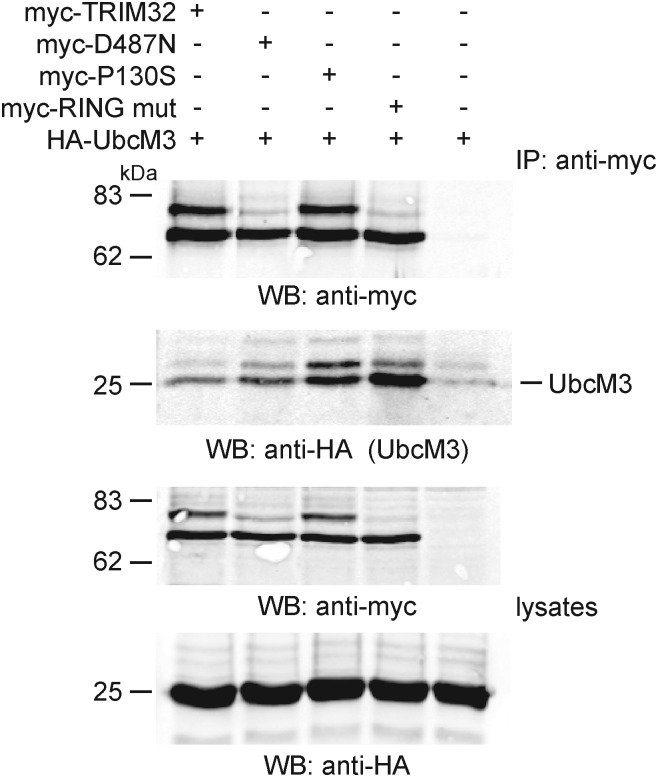
Impaired monoubiquitination of the D487N mutant
TRIM proteins are thought to function as dimers or trimers (4) and self-association can lead to autoubiquitination, whereby an E3 ubiquitin ligase can catalyse ubiquitin conjugation to its reciprocally bound partner. A higher molecular weight TRIM32 cross-reactive protein was present in both transfected cells (see, for example, Figs 1D and 5D) and endogenously in tissues synthesising the highest levels of the protein (Supplementary Material, Fig. S1B, testes). We postulated that this protein might correspond to endogenously monoubiquitinated TRIM32. We used a direct biochemical approach to assay the basal ubiquitination status of TRIM32 and each of the mutants. Myc-TRIM32 and each of the mutants were transiently transfected into HEK293T cells. The cells were lysed under denaturing and reducing conditions, and myc-TRIM32 was purified using immunoaffinity chromatography from cell lysates using 9E10-conjugated Protein G Sepharose beads. Affinity purified myc-TRIM32 was separated by gel electrophoresis and stained with colloidal Coomassie blue. A strong 72 kDa band corresponding to TRIM32 is present at similar amounts in all lanes (Fig. 7). A higher molecular weight band is also present for wild-type TRIM32 and the P130S mutant (Fig. 7). This upper band is not apparent for D487N and RING-mut. The upper and lower bands were excised from the Coomassie stained gel and processed for Fourier transform ion cyclon resonance mass spectrometry. Numerous tryptic peptides corresponding TRIM32 were obtained from bands A1 and A3. In addition to TRIM32, band A1 yielded multiple peptides obtained from ubiquitin (Fig. 7). We therefore concluded that the upper band represents monoubiquitinated TRIM32 and that D487N and the RING-mut fail to gain this modification. These data are consistent with our finding that the TRIM32 speckles found in transfected cells also contain ubiquitin. In fact, it appears that ubiquitin is conjugated directly to TRIM32.
Figure 7.
Impaired monoubiquitination of D487N. HEK293T cells were transfected with myc-tagged constructs encoding, TRIM32, TRIM32–D487N, TRIM32–RING-mut and TRIM32–P130S as indicated. After 24 h, each protein was immunoprecipitated from cell extracts using the 9E10 antibody. Immune complexes were separated by gel electrophoresis on a 4–20% gradient gel. The gel was stained with colloidal Coomassie blue to visualize protein bands. Bands A1 and A3 were excised and subjected to FTICR-MS. The table shows the percentage of their amino acid sequence covered by peptides identified in MS and the number of unique peptides that match this sequence.
DISCUSSION
The aim of this study was to investigate the cellular function of TRIM32 and learn more about the molecular defects that underlie LGMD2H/STM. We used an Y2H approach to identify potential substrates and regulators of TRIM32. We found that TRIM32 could associate with and ubiquitinate dysbindin augmenting its degradation. Dysbindin is part of a multiprotein complex called biogenesis of lysosome-related organelles complex-1 (BLOC-1) that is involved in lysosomal–endosomal trafficking (34–36). Dysbindin and TRIM32 were both found at the Z-line in skeletal muscle (Fig. 2). Interestingly, Z-line streaming is a pathological feature of many muscle diseases including LGMD2H/STM and was also a found in the mouse Trim32 knockout (20). Although no direct link between dysbindin, BLOC-1 and muscle disease has been established, dysbindin is known to participate in the trafficking of proteins to lysosomes and lysosome-related organelles including some lysosome-associated membrane proteins (LAMPs) (37,38). Interestingly, mutations in LAMP-2 cause Danon disease, a complex lysosomal myopathy associated with the accumulation of autophagic vacuoles in myofibres and impaired lysosomal function (39). Our data could implicate TRIM32 in the regulation of trafficking in the lysosomal–endosomal pathway in muscle through the ubiquitination of proteins such as dysbindin.
Mutations in the human dysbindin gene, DTNBP1, cause a complex, hypo-pigmentation and bleeding disorder characterized by defects in the formation of lysosome-related organelles, such as melanosomes and platelet-dense granules (34). A defect in lysosomal–endosomal trafficking may explain recent evidence linking dysbindin to the psychiatric disorder schizophrenia (30). A plethora of genetic studies have linked DTNBP1 variants to an increased susceptibility to develop schizophrenia (30,40). Furthermore, Talbot et al. (36) found that dysbindin protein levels are reduced in the hippocampal formation of schizophrenics relative to age-matched control. In light of these studies, it is important to consider that TRIM32-mediated dysbindin ubiquitination may have a regulatory role, affecting protein trafficking to endosomes or recycling. These processes are often regulated by monoubiquitination and K63-linked polyubiquitination (41). A role for TRIM32 in atypical ubiquitination is further supported by the study of Saccone et al. (19) who showed that the ubiquitin ligase E2N, which normally participates in the formation of lysine-63 linked polyubiquitin chains, may act as an E2 enzyme for TRIM32.
Using several complimentary techniques, we have demonstrated that the LGMD2H/STM-associated D487N and R394H mutations abrogate the ubiquitin ligase activity of TRIM32. In common with wild-type TRIM32, the D487N could bind to UbcM3 consistent with current evidence showing that the RING domain is the main determinant in E2 recruitment (42). Since D487N is able to bind dysbindin, it is possible that the reduced ubiquitin ligase activity of D487N results from disruption in protein interactions with other components of the ubiquitin–proteasome pathway, and suggests that the NHL domain is not essential for substrate binding in this instance. This defect could also be linked to the altered distribution seen for D487N–EYFP and R394H–EYFP in transfected cells (Fig. 3 and Supplementary Material, Fig. S4) and the impaired monoubiquitination of D487N (Fig. 7). Since mutations of the RING domain also resulted in impaired monoubiquitination, it is likely that this represents an ‘auto-monoubiquitination’ event that may have a regulatory role.
Molecular modelling of the NHL repeats of TRIM32 suggested that LGMD2H mutations significantly disrupt the β-propeller structure of the NHL domain (19). This structural malformation is likely to have a substantial role in the altered subcellular distribution and function of the LGMD mutant. In mammalian cells, we found that wild-type and mutant TRIM32 could self-associate, whereas in yeast, the D487N and R394H mutants were unable to transactivate expression of the reporter genes. Since the D487N and R394H mutants can ubiquitinate dysbindin, albeit to a lesser extent, it follows that D487N and R394H should be able to bind to dysbindin. Although we have not used formal biophysical methods to show direct binding between dysbindin and TRIM32 in mammalian cells, our data indicate that the mutants D487N and R394H are able to associate with dysbindin either directly or via an intermediate protein (another TRIM or TRIM-related protein) co-immunoprecipitating from transfected cells. It should be noted that neither TRIM32 nor dysbindin is present in yeast cells suggesting that their interaction in the Y2H is direct and not mediated by a third protein. Our findings are similar to those of Gentry et al. (18), who found that mutations in the NHL domain of the E3 ubiquitin ligase malin (which causes a form of myoclonus epilepsy) completely abolished its interaction with laforin (the substrate of malin) in the Y2H system. Despite this, both proteins co-immunoprecipitated from mammalian cells, with the NHL mutations significantly impairing the ubiquitin ligase activity of malin towards laforin (18).
In addition to the self-association properties of TRIM32, we have also found that wild-type TRIM32 and the LGMD2H/STM mutants co-localized and co-immunoprecipitate from cells expressing both proteins (Supplementary Material, Fig. S4 and Fig. 5D). In the case of the R394H mutant, co-expression with wild-type TRIM32 reduced the level of dysbindin monoubiquitination potentially indicating that this mutation may compromise the function of the wild-type protein (Fig. 5E). Interestingly, Saccone et al. (19) found one LGMD2H patient that was heterozygous for the R394H mutation. It is therefore tempting to speculate that an interaction between wild-type and mutant TRIM32 may cause late onset LGMD2H in this patient.
It is striking that all four LGMD2H/STM mutations cluster within the NHL repeats of TRIM32, yet none of the patients presented with symptoms of BBS11 and vice versa. BBS11 is a complex disorder that is associated with an increased incidence of neuropsychiatric disease. For example, BBS patients are twice as likely to develop schizophrenia compared with the general population, leading to the suggestion that BBS genes may themselves be candidate schizophrenia susceptibility genes (43,44). Indeed, recent data suggest disrupted-in-schizophrenia 1 forms a complex with BBS4 and pericentriolar material 1 protein at the centrosome, where a number of other BBS proteins have been found to localize (43). Here, BBS proteins are thought to be involved in the assembly and function of primary cilia by regulating intracellular transport (22,26). Although TRIM32 has not been localized to primary cilia, knockdown of TRIM32 in zebrafish results in defective melanosome transport (21). Our findings are therefore of particular interest, since we have characterized a second interaction between a BBS gene and a schizophrenia susceptibility gene, both of which appear to have roles in intracellular trafficking. Some genetic variants in the DTNBP1 gene influence cognitive performance in the general population and are also associated with cognitive decline in schizophrenia (45,46). It is therefore interesting to speculate that the interaction between TRIM32 and dysbindin may play role in the cognitive and neuropsychiatric abnormalities in some BBS patients. Although the BBS11 mutation P130S was not defective in dysbindin ubiquitination, these data may suggest that the P130S mutation disrupts as yet unidentified aspects of the protein function not affected by TRIM32 mutations associated with LGMD2H/STM. In support of this, Chiang et al. (21) found that unlike P130S, D487N could rescue TRIM32 knockdown BBS phenotypes in zebrafish.
In summary, we have identified several functional differences between wild-type and LGMD2H/STM-mutated TRIM32. These differences affect the localization and potential regulation of TRIM32 and lead to impaired ubiquitination of substrates such as dysbindin that we have identified herein.
MATERIALS AND METHODS
Y2H screening
Y2H screening was performed using the Hybrid Hunter system (Invitrogen). Full-length TRIM32 was amplified by PCR and cloned into the EcoRI/SalI sites of pHybLex/Zeo (Invitrogen). The bait strain was created by transforming L40 with TRIM32-pHybLex/Zeo and used to screen a mouse skeletal muscle cDNA library (Invitrogen), as previously described (30). Interacting prey plasmids were isolated from yeast using the RPM yeast plasmid isolation kit (QBiogene) and electroporated into Escherichia coli XL1 Blue (Stratagene). Plasmids were then extracted from E. coli using a Qiagen Miniprep kit and sequenced using a vector primer. For TRIM32 self-association studies, TRIM32, D487N, R394H and P130S were subcloned into Y2H bait (pHybLex/Zeo) and prey (pYesTrp2) plasmids and used to co-transform the yeast strain L40. Co-transformants were spotted on minimal media lacking tryptophan (+His) or lacking histidine and containing 5 mm 3-aminotriazole (-His+5 mm 3-AT). The plates were grown at 30°C and examined after 3–5 days.
Antibodies
A PCR fragment encoding amino acids 240–372 of TRIM32 was cloned into pET32b (Novagen) and used to transform E. coli BL21 (DE3) gold (Stratagene). His-tagged fusion proteins were purified using Talon resin as per manufacturer’s instructions (Clontech), and used to immunize New Zealand white rabbits (Sigma-Aldrich). Anti-sera were pre-absorbed against thioredoxin and affinity purified using proteins covalently coupled to Sulfolink (Pierce). The anti-dysbindin antibody PA3111A has been previously described (30). Commercial antibodies were purchased from Sigma-Aldrich (α-actinin monoclonal), Covance (HA monoclonal, anti-GFP monoclonal B34), Biomol (FK1), Rockland immunochemicals (IR dye 800 anti-mouse IgG), Invitrogen (Alexa Fluor 488 and 680) and Jackson immunochemicals (Rhodamine Red X anti-rabbit and anti-mouse). Monoclonal antibodies against c-myc (9E10), α-tubulin (12G10) and anti-myomesin (B4) were produced from hybridomas and purified and concentrated by Protein G Sepharose chromatography (GE Healthcare). The hybridomas developed by Frankel and Nelsen (12G10) and Perriard (B4) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA.
Expression constructs
Full-length TRIM32, UbcM3 and PIASy were amplified from a skeletal muscle cDNA library by PCR, and sub-cloned into pCIneo, pEYFP-N1, pCMV-myc or pCMV-HA. Clones were verified by full-length sequencing. Mutagenesis was performed using the ‘Quikchange’ mutagenesis kit (Stratagene) as per manufacturer’s instructions. Dysbindin expression constructs have been previously described (30). The HA epitope-tagged ubiquitin construct was provided kindly by Professor Dirk Bohmann.
Cell culture and transfection
COS-7, HEK-293T and C2C12 cells were grown in DMEM supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. For differentiation, C2C12 cells were placed in DMEM supplemented with 2% horse serum when cultures reached 70% confluence. Transfections were carried out using Fugene-6 (Roche) as per manufacturer’s instructions. For siRNA, C2C12s were transfected with Lipofectamine 2000 (Invitrogen) on consecutive days and incubated for 72 h. For time courses, cells were treated with cycloheximide (50 µg/ml Sigma) 24 h post-transfection, and samples taken thereafter. Where appropriate, cells were re-treated with cycloheximide after 24 h.
Immunofluorescence microscopy
Twenty-four hours post-transfection cells grown on coverslips were rinsed twice with PBS and fixed in methanol at −20°C. Coverslips were washed twice with PBS and blocked for 20 min in PBS containing 10% fetal calf serum. Antibodies were added for 45 min at room temperature with agitation. Primary antibody dilutions were 1:100 (anti-TRIM32 3293), 1:1000 (PA3111A), 1:400 (9E10) and 1:200 (FK1). Coverslips were washed twice and incubated with anti-Rabbit Rhodamine Red-X IgG (for PA3111A detection) or anti-rabbit Rhodamine Red-X IgM (for FK1 detection). Following three washes in PBS, the coverslips were mounted with Aqua Poly/Mount (Polysciences Inc.), and analysed using a confocal microscope. For muscle immunocytochemistry, tibialis anterior muscles (longitudinal sections) were stretched and fixed in 4% (w/v) paraformaldehyde and incubated in 10% (w/v) sucrose for 1 h, 20% (w/v) sucrose for 1 h and 30% (w/v) sucrose overnight, followed by embedding. Eight micron sections were prepared and blocked in 10% (v/v) FCS for 30 min followed by incubation with primary antibody in TBS. Antibody dilutions used were 1:25 (TRIM32 3293), 1:1000 (α-actinin), 1:10 (PA3111A), 1:100 (myomesin). After washing with TBS, the sections were incubated with donkey anti-rabbit Alexa 488/Rhodamine Red-X IgG or goat anti-mouse Alexa-488/Rhodamine Red-X IgG. For blocking experiments, the anti-TRIM32 3293 antibody was diluted as described above and pre-absorbed with 50 µg/ml of fusion protein (thioredoxin–TRIM32) for 1 h at room temperature prior to use.
Immunoprecipitation and ubiquitination assays
For TRIM32–dysbindin co-immunoprecipitation experiments, transfected proteins were solubilized from HEK293T cells with ice-cold RIPA buffer (150 mm NaCl, 50 mm Tris pH 8.0, 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate and 1 mm EDTA). Lysates were pre-cleared with Protein G Sepharose (Invitrogen) and incubated with 9E10-conjugated beads for 3 h as described previously (47). Immune complexes were eluted from the beads by boiling in ×2 Laemmli sample buffer/5% β-mercaptoethanol, and analysed by western blotting. For TRIM32–UbcM3 co-immunoprecipitation experiments, immunoprecipitation was performed in low-stringency buffer (150 mm NaCl, 50 mm Tris pH 8.0, 0.5% (v/v) NP-40 and 1 mm EDTA). For ubiquitination assays, immunoprecipitation was performed as above, except transfected cells were lysed in denaturing conditions (1% SDS, 50 mm Tris pH 8.0, 10 mm DTT). For immunoaffinity purification, transfected proteins were solubilized in RIPA buffer and immunoprecipitated with 9E10. Immune complexes were resolved by gel electrophoresis using 4–12% Novex pre-cast gradient gels (Invitrogen). The gel was washed in water to remove SDS and the proteins fixed by incubating the gel in 7% acetic acid/50% methanol for 15 min. Following washing the bands were visualized by incubating the gel in colloidal Coomassie blue (GelCode Blue, Pierce) for 2 h followed by destaining in HPLC-grade water. Mass spectrometry was performed at the Functional Genomics and Proteomics facility at Birmingham University.
Western blotting
Quantitative western blots were performed as described previously (47). Briefly, protein extracts were separated by SDS–PAGE and electroblotted onto nitrocellulose membranes before probing with primary antibody. After washing, the membranes were incubated with IR fluorophore conjugated secondary antibodies, IRDye™ 800 donkey anti-mouse IgG and Alexa Fluor 680 goat anti-rabbit IgG. Membranes were then washed with Tris-buffered saline (TBS), containing 0.1% (v/v) Tween-20 and rinsed in TBS. Simultaneous two-colour detection was performed using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, Nebraska). Quantification of dysbindin and TRIM32 was performed using the Odyssey® imager application v1.2, normalized to α-tubulin fluorescence as an internal reference.
Mass spectrometry
Myc-tagged TRIM32 and mutants thereof were transfected into HEK-293T cells as described above. After 48 h, the cells were lysed in RIPA buffer and TRIM32 was purified by immunoaffinity chromatography using 9E10-conjugated Protein G Sepharose beads. Purified proteins were resolved by polyacrylamide gel electrophoresis on 5–20% NuPAGE gels (Novex), fixed and stained with colloidal Coomassie blue dye (Gelcode, Pierce). Coomassie stained bands were manually excised and processed for mass spectrometry as follows. Proteins in the individual gel slices were digested using either a MWG Roboseq 4204 or a QIAGEN 3000 robotic gel handling system (School of Bioscience, University of Birmingham). Analysis of tryptic digests was performed on a Thermo Finnigan LTQ-FT with a capillary high-pressure liquid chromatography system with a nanospray probe. Five microlitre of sample was run on a long column (15 cm×75 µm C18, 3 µm, 100 A) to separate out peptides. Database searches were performed with MASCOT (www.matrixscience.com) using the following settings: fixed modifications, carbamidomethyl (C); variable modifications, oxidation (M), peptide charge 2+ and 3+. The data format was pkl.
RNA interference
C2C12s were transfected with various RNA duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, C2C12 cells were seeded in 12-well plates (4 × 104 cells per well) in DMEM supplemented with 5% fetal calf serum. The following day, cells were transfected with TRIM32-specific siRNA, 5′-GUGACUACUUUCUAGCGAAUU and 5′-PO4-UUGCGUAGAAAGUAGUCACUU (Dharmacon) or control siRNA (siCONTROL non-targeting siRNA pool, Dharmacon), at a final concentration of 33 nm. The growth media was replaced 5 h later. The transfection protocol was repeated the following day. Twenty-four hours later (72 h after the first transfection), cell lysates were prepared by lysing the cells in 100 µl of treatment buffer (75 mm Tris pH 6.8, 4 m urea, 4% SDS, 5% β-mercaptoethanol).
SUPPLEMENTARY MATERIAL
FUNDING
This work was generously supported by grants from the Wellcome Trust (061225, Wellcome Trust Senior Research Fellowship to D.J.B.) and National Institute of Mental Health (R01 MH072880). M.L. is funded by an Usher Cunningham Scholarship from Exeter College, Oxford and an MRC studentship. Funding to pay the Open Access charge was provided by The Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Professor Derek Terrar for providing guinea pig muscle and Drs Jeff McIlhinney and Anthony Morgan for help with confocal microscopy.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Ravid T., Hochstrasser M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat. Rev. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nisole S., Stoye J.P., Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M., Inaguma Y., Hiai H., Hirose F. Developmentally regulated expression of a human ‘finger’-containing gene encoded by the 5′ half of the ret transforming gene. Mol. Cell Biol. 1988;8:1853–1856. doi: 10.1128/mcb.8.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salomoni P., Pandolfi P.P. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 8.Le Douarin B., Zechel C., Garnier J.M., Lutz Y., Tora L., Pierrat P., Heery D., Gronemeyer H., Chambon P., Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C.R., Morgan K., Fujiwara T.M., Wrogemann K. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 2002;70:663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quaderi N.A., Schweiger S., Gaudenz K., Franco B., Rugarli E.I., Berger W., Feldman G.J., Volta M., Andolfi G., Gilgenkrantz S., et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 11.Avela K., Lipsanen-Nyman M., Idanheimo N., Seemanova E., Rosengren S., Makela T.P., Perheentupa J., Chapelle A.D., Lehesjoki A.E. Gene encoding a new RING-B-box-coiled-coil protein is mutated in mulibrey nanism. Nat. Genet. 2000;25:298–301. doi: 10.1038/77053. [DOI] [PubMed] [Google Scholar]
- 12.A candidate gene for familial Mediterranean fever. The French FMF Consortium. Nat. Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 13.Jerusalem F., Engel A.G., Gomez M.R. Sarcotubular myopathy. A newly recognized, benign, congenital, familial muscle disease. Neurology. 1973;23:897–906. doi: 10.1212/wnl.23.9.897. [DOI] [PubMed] [Google Scholar]
- 14.Muller-Felber W., Schlotter B., Topfer M., Ketelsen U.P., Muller-Hocker J., Pongratz D. Phenotypic variability in two brothers with sarcotubular myopathy. J. Neurol. 1999;246:408–411. doi: 10.1007/s004150050374. [DOI] [PubMed] [Google Scholar]
- 15.Schoser B.G., Frosk P., Engel A.G., Klutzny U., Lochmuller H., Wrogemann K. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann. Neurol. 2005;57:591–595. doi: 10.1002/ana.20441. [DOI] [PubMed] [Google Scholar]
- 16.Slack F.J., Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 17.Edwards T.A., Wilkinson B.D., Wharton R.P., Aggarwal A.K. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry M.S., Worby C.A., Dixon J.E. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl Acad. Sci. USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saccone V., Palmieri M., Passamano L., Piluso G., Meroni G., Politano L., Nigro V. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum. Mutat. 2008;29:240–247. doi: 10.1002/humu.20633. [DOI] [PubMed] [Google Scholar]
- 20.Kudryashova E., Wu J., Havton L.A., Spencer M.J. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 2009;18:1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang A.P., Beck J.S., Yen H.J., Tayeh M.K., Scheetz T.E., Swiderski R.E., Nishimura D.Y., Braun T.A., Kim K.Y., Huang J., et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet–Biedl syndrome gene (BBS11) Proc. Natl Acad. Sci. USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blacque O.E., Leroux M.R. Bardet–Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore S.J., Green J.S., Fan Y., Bhogal A.K., Dicks E., Fernandez B.A., Stefanelli M., Murphy C., Cramer B.C., Dean J.C., et al. Clinical and genetic epidemiology of Bardet–Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am. J. Med. Genet. A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnett S., Reilly S., Carr L., Ojo I., Beales P.L., Charman T. Behavioural phenotype of Bardet–Biedl syndrome. J. Med. Genet. 2002;39:e76. doi: 10.1136/jmg.39.12.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 26.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Bardet–Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 27.Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 28.Horn E.J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G.E., Babcock M., Bowden G.T., Hennings H., Lozano G., Weinberg W.C., et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 29.Albor A., El-Hizawi S., Horn E.J., Laederich M., Frosk P., Wrogemann K., Kulesz-Martin M. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in LGMD2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkB. J. Biol. Chem. 2006;281:25850–25866. doi: 10.1074/jbc.M601655200. [DOI] [PubMed] [Google Scholar]
- 30.Benson M.A., Sillitoe R.V., Blake D.J. Schizophrenia genetics: dysbindin under the microscope. Trends Neurosci. 2004;27:516–519. doi: 10.1016/j.tins.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Benson M.A., Newey S.E., Martin-Rendon E., Hawkes R., Blake D.J. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 2001;276:24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 32.Holzfeind P.J., Ambrose H.J., Newey S.E., Nawrotzki R.A., Blake D.J., Davies K.E. Tissue-selective expression of alpha-dystrobrevin is determined by multiple promoters. J. Biol. Chem. 1999;274:6250–6258. doi: 10.1074/jbc.274.10.6250. [DOI] [PubMed] [Google Scholar]
- 33.Campbell E.M., Dodding M.P., Yap M.W., Wu X., Gallois-Montbrun S., Malim M.H., Stoye J.P., Hope T.J. TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol. Biol. Cell. 2007;18:2102–2111. doi: 10.1091/mbc.E06-12-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Zhang Q., Oiso N., Novak E.K., Gautam R., O’Brien E.P., Tinsley C.L., Blake D.J., Spritz R.A., Copeland N.G., et al. Hermansky–Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat. Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starcevic M., Dell’Angelica E.C. Identification of snapin and three novel proteins (BLOS1, BLOS2 and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J. Biol. Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 36.Talbot K., Eidem W.L., Tinsley C.L., Benson M.A., Thompson E.W., Smith R.J., Hahn C.G., Siegel S.J., Trojanowski J.Q., Gur R.E., et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W., Feng Y., Hao C., Guo X., Cui Y., He M., He X. The BLOC interactomes form a network in endosomal transport. J. Genet. Genomics. 2007;34:669–682. doi: 10.1016/S1673-8527(07)60076-9. [DOI] [PubMed] [Google Scholar]
- 38.Salazar G., Craige B., Styers M.L., Newell-Litwa K.A., Doucette M.M., Wainer B.H., Falcon-Perez J.M., Dell’Angelica E.C., Peden A.A., Werner E., et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol. Biol. Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malicdan M.C., Noguchi S., Nonaka I., Saftig P., Nishino I. Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul. Disord. 2008;18:521–529. doi: 10.1016/j.nmd.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Norton N., Williams H.J., Owen M.J. An update on the genetics of schizophrenia. Curr. Opin. Psychiatry. 2006;19:158–164. doi: 10.1097/01.yco.0000214341.52249.59. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 42.Joazeiro C.A., Weissman A.M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 43.Kamiya A., Tan P.L., Kubo K., Engelhard C., Ishizuka K., Kubo A., Tsukita S., Pulver A.E., Nakajima K., Cascella N.G., et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch. Gen. Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta S.R., McQuillin A., Rizig M., Blaveri E., Thirumalai S., Kalsi G., Lawrence J., Bass N.J., Puri V., Choudhury K., et al. A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Mol. Psychiatry. 2008 doi: 10.1038/mp.2008.128. In press. [DOI] [PubMed] [Google Scholar]
- 45.Burdick K.E., Goldberg T.E., Funke B., Bates J.A., Lencz T., Kucherlapati R., Malhotra A.K. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophr. Res. 2007;89:169–172. doi: 10.1016/j.schres.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burdick K.E., Lencz T., Funke B., Finn C.T., Szeszko P.R., Kane J.M., Kucherlapati R., Malhotra A.K. Genetic variation in DTNBP1 influences general cognitive ability. Hum. Mol. Genet. 2006;15:1563–1568. doi: 10.1093/hmg/ddi481. [DOI] [PubMed] [Google Scholar]
- 47.Esapa C.T., Waite A., Locke M., Benson M.A., Kraus M., McIlhinney R.A., Sillitoe R.V., Beesley P.W., Blake D.J. SGCE missense mutations that cause myoclonus–dystonia syndrome impair epsilon–sarcoglycan trafficking to the plasma membrane: modulation by ubiquitination and torsinA. Hum. Mol. Genet. 2007;16:327–342. doi: 10.1093/hmg/ddl472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.