Abstract
Deletion of a subset of the D4Z4 macrosatellite repeats in the subtelomeric region of chromosome 4q causes facioscapulohumeral muscular dystrophy (FSHD) when occurring on a specific haplotype of 4qter (4qA161). Several genes have been examined as candidates for causing FSHD, including the DUX4 homeobox gene in the D4Z4 repeat, but none have been definitively shown to cause the disease, nor has the full extent of transcripts from the D4Z4 region been carefully characterized. Using strand-specific RT–PCR, we have identified several sense and antisense transcripts originating from the 4q D4Z4 units in wild-type and FSHD muscle cells. Consistent with prior reports, we find that the DUX4 transcript from the last (most telomeric) D4Z4 unit is polyadenylated and has two introns in its 3-prime untranslated region. In addition, we show that this transcript generates (i) small si/miRNA-sized fragments, (ii) uncapped, polyadenylated 3-prime fragments that encode the conserved C-terminal portion of DUX4 and (iii) capped and polyadenylated mRNAs that contain the double-homeobox domain of DUX4 but splice-out the C-terminal portion. Transfection studies demonstrate that translation initiation at an internal methionine can produce the C-terminal polypeptide and developmental studies show that this peptide inhibits myogenesis at a step between MyoD transcription and the activation of MyoD target genes. Together, we have identified new sense and anti-sense RNA transcripts, novel mRNAs and mi/siRNA-sized RNA fragments generated from the D4Z4 units that are new candidates for the pathophysiology of FSHD.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant muscular dystrophy caused by a deletion of D4Z4 macrosatellite repeats in the subtelomeric region of the 4qA161 haplotype of chromosome 4 (1,2). The unaffected population has approximately 11–100 D4Z4 units arranged as a tandem array on 4q and deletions resulting in arrays of 1–10 D4Z4 units are associated with FSHD. Current models of FSHD pathogenesis suggest that the deletion results in aberrant gene expression, either expression of the DUX4 gene in the D4Z4 unit, expression of genes close to the D4Z4 repeat in cis or expression of genes in trans that might be influenced by the D4Z4 region of 4q (reviewed in 2,3). These models are supported by the demonstration of repressive epigenetic modifications of the D4Z4 units that are decreased on the contracted allele (4) and by a small number of FSHD individuals with D4Z4 DNA hypomethylation in the absence of a D4Z4 contraction, referred to as ‘phenotypic’ FSHD. Increased expression of several genes centromeric of the 4q D4Z4 repeats has been reported in some FSHD studies (5), and abnormal expression of the DUX4 transcript from the D4Z4 repeat has also been suggested as a cause of the disease (6,7). Therefore, studies show that the FSHD-associated D4Z4 region has diminished repressive epigenetic markings and aberrant gene expression has been reported for DUX4 and other genes in the subtelomeric region. However, disagreement remains on whether individual genes are reliably mis-expressed or causative for FSHD.
Smaller residual repeat-array sizes roughly correlate with more severe disease, but at least one repeat remains in affected individuals (2), suggesting a necessary role for the D4Z4 sequence. Further, deletions of D4Z4-like repeats present on chromosome 10q do not cause FSHD despite being within a subtelomeric region that is highly similar to 4q, and deletions on only one of the nine 4q haplotypes have been shown to cause FSHD (1). These suggest a specific allelic requirement for a region of D4Z4 in FSHD pathogenesis. Recent studies have shown that the evolutionarily conserved double-homeobox gene DUX4 in the D4Z4 repeat is a transcriptional activator that can induce cellular apoptosis (6,8,9) and activate expression of the PITX1 gene (7). An attractive hypothesis, therefore, is that the loss of epigenetic repression leads to DUX4 expression and cellular damage, and that allele-specificity is due to allele-specific sequence, either an as yet uncharacterized regulatory polymorphism(s) in the D4Z4 region or the 4qA sequence that is telomeric to the last repeat, the pLAM sequence or both.
Although studies have now detected a low abundance of DUX4 protein and transcripts in some FSHD cells and biopsies (7), the full-length RNA and protein have been difficult to identify. This anomaly caused us to investigate whether the full-length DUX4-encoding transcript might be processed to produce regulatory RNA fragments, such as miRNA, or whether alternative transcriptional start sites and open reading frames (ORFs) might encode proteins in addition to the full-length DUX4 that might contribute to FSHD pathology. In this report, we present evidence for processed and possibly alternatively translated transcripts from the DUX4 locus and identify overlapping sense and antisense transcripts in the D4Z4 macrosatellite units that might have additional regulatory or pathologic roles.
RESULTS
Sequence of D4Z4 repeats
The phage clone λ-42 contains two complete residual D4Z4 units cloned from an FSHD individual with the 4qA161 disease-associated haplotype (10,11). We re-sequenced the two residual D4Z4 units in the λ-42 clone to resolve sequence discrepancies and determined the location of potential ORFs. The first repeat contains two major ORFs: a 135 amino acid (aa) ORF beginning with the codons for methionine (M), glutamate (E) and arginine (R) (MER) that has no significant homology to proteins in the Genbank database; and a 689 aa ORF beginning with the MQGR codons that include internal in-frame ORFs: one beginning with MKG and the previously designated DUX4 ORF that begins with the MAL codons (Fig. 1). In addition, there are several ORFs that overlap the 689 MQGR ORF in different reading frames (data not shown). In the last repeat, sequence variations extend the MER ORF to 256 amino acids and truncate the MQGR ORF after 241 aa. MNE initiates a 633-aa ORF immediately after the MER stop codon. This ORF includes the MKG ORF and the MAL DUX4 ORF. Multiple additional predicted sense and antisense ORFs overlap with these major ORFs (data not shown).
Figure 1.
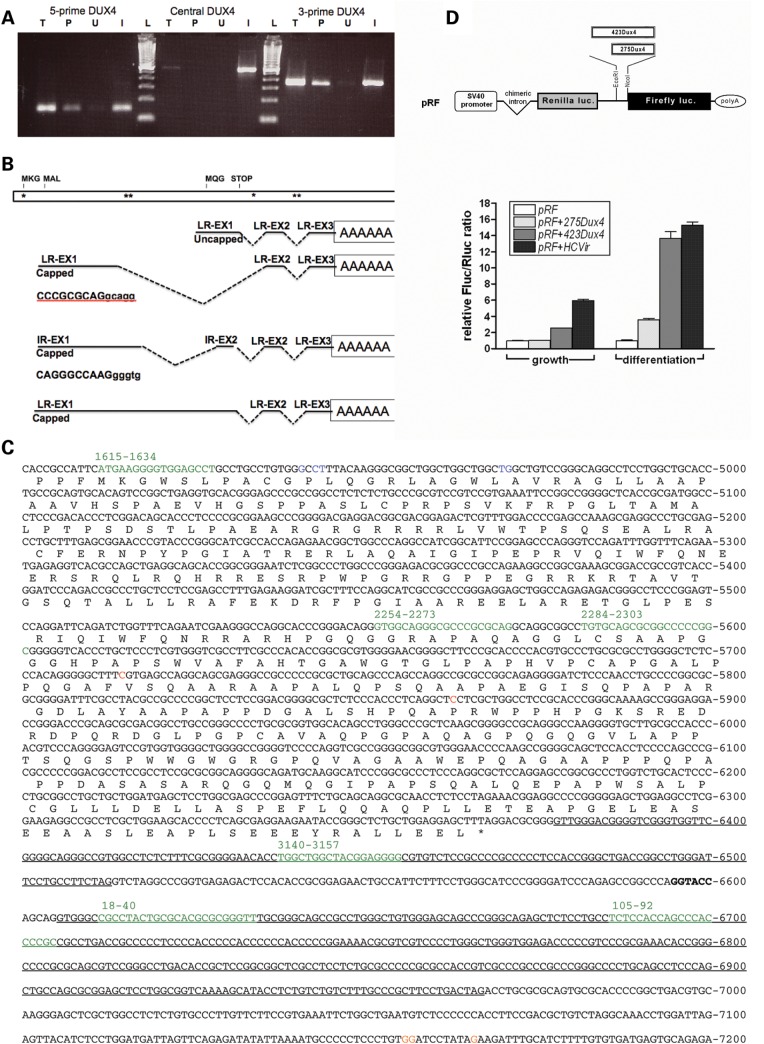
Schematic representation of the FSHD locus, ORFs, transcripts and location of miRNA-sized fragments. The top panel shows the FSHD locus with two full D4Z4 units. The region centromeric to the D4Z4 units is designated as p13E-11. The small triangle represents a partial D4Z4 unit centromeric to the First and Last D4Z4 units (large triangles), followed by another partial D4Z4 unit (partial triangle), then the region designated as pLAM, followed by beta-satellite repeat sequence. The sequence in Supplementary Material, Figure S1 extends from the first D4Z4 unit through the pLAM sequence and is numbered 0 to 7760, as shown schematically. First repeat ORFs: schematic of ORFs in the first D4Z4 unit that extends from position 0 to 3300. Each ORF is predicted from the DNA sequence and denoted by the first few amino acids. MKG, MAL and MQG represent internal in-frame methionines that might also be used for translation initiation. The ORF beginning with MAL is the previously identified DUX4 ORF. STOP indicates a stop-codon in-frame with the upstream ORF. Numbering is from the KPNI site at the beginning of the D4Z4 repeat to the KPNI site at the end of the repeat. Last repeat ORFs: potential ORFs in the last D4Z4 unit differ from first due to sequence variations. Transcripts mapped by strand-specific RT–PCR and small RNAs: map of transcription units determined by strand-specific RT–PCR in wild-type and FSHD fibroblasts and muscle cells. Solid arrows (A–F) represent regions where strand-specific RT–PCR has identified RNA transcripts and dashed lines represent regions where transcripts were not identified. Location of the endpoints of these transcripts in the first/last repeats is as follows: A, 404/3701-1325/4628; B, 1614/4912-2237/5535; C, 2704/6002-3340/6638; D, 1003-1374/4672; E, 1623/4921-2100/5400; F, 2634/5932-3071/6369. Asterisks represent locations of small miRNA-sized fragments confirmed by northern.
Multiple transcripts identified from 4qA D4Z4
To map transcripts from the D4Z4 region, we used primary myoblasts and primary fibroblasts derived from FSHD and control individuals. The myoblasts can be directly differentiated into muscle cells by withdrawing mitogens, whereas the fibroblasts can be induced to differentiate to muscle cells by the expression of the myogenic bHLH transcription factor MyoD. For the fibroblasts, we used retroviral transduction of a fusion of MyoD to the hormone-binding domain of the estrogen receptor such that differentiation can be induced by the addition of beta-estradiol (12). Analysis of the primary muscle cells and the fibroblasts converted to muscle by MyoD gave similar results and are discussed together.
Using RT–PCR, RNA transcripts were detected in the 5-prime and 3-prime regions of DUX4 in fibroblasts and muscle cells from FSHD and control individuals, but not from the central portion of the DUX4 ORF (Fig. 2A and B). For each amplified region, multiple PCR products matched the 4qA sequence from λ-42, and, in regions with informative polymorphisms, the sequences matched the last repeat. Occasional sequences had polymorphisms indicating additional transcripts from other 4q or non-4q D4Z4 repeats. Parallel RT–PCR on in vitro transcribed DUX4 RNA and PCR on DUX4 DNA readily amplify the central portion of DUX4 (Fig. 2B, IVT and DNA samples), indicating that the inability to amplify this region from cellular RNA is not a technical artifact, nor were any splice forms detected that would delete this central region and maintain the 5-prime and 3-prime coding regions (data not shown).
Figure 2.
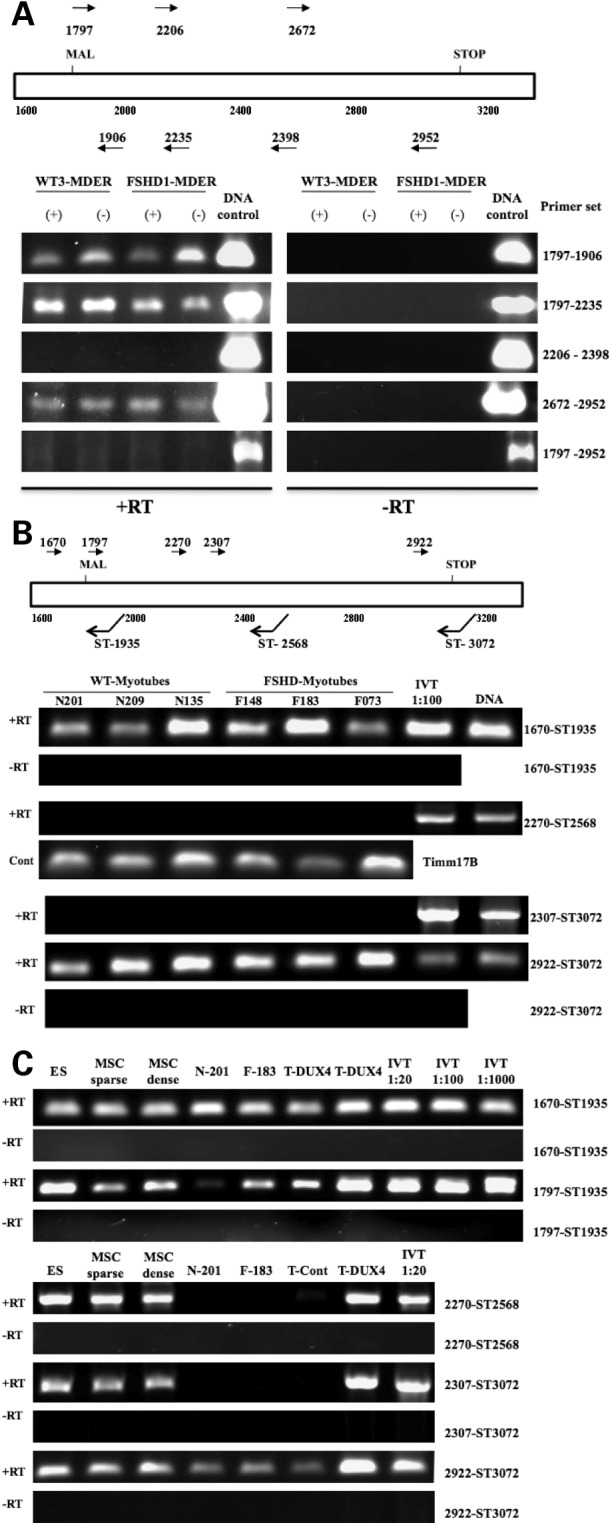
Discontinuous regions of DUX4 transcripts in wild-type and FSHD fibroblasts. (A) RT–PCR of random hexamer primed total RNA from fibroblasts, wild-type or FSHD-derived, amplified with primers for the 5-prime (1797–1906; 1797–2235), the central portion (2270–2398), the 3-prime end (2672–2970) and the full-length DUX4 (1797–2970). The fibroblasts were transduced with a retrovirus expressing MyoD-ER and assayed under differentiation conditions (DMEM with 1 µg/ml insulin and transferrin for 96 h) either without beta-estadiol (−) or with beta-estradiol (+) to induce MyoD activity; however, no consistent differences were noted in RNA obtained from MyoD induced and non-induced cells. The presence of amplification products from DNA controls shows that all primer sets can amplify the sequence from DUX4 cDNA templates. (B) Strand-specific RT–PCR amplification products using total RNA from control and FSHD muscle cells. Primer pairs are designed to amplify the 5-prime (1670-ST1936), middle (2270-ST2568) and 3-prime (2922-ST3072) regions of DUX4. IVT is a dilution of RNA from an in vitro transcribed DUX4 cDNA to demonstrate that the RT reaction and PCR can amplify the central portion of DUX4 from an RNA template. (C) Strand-specific RT–PCR amplification products using total RNA from human embryonic stem cells (ES), mesenchymal stem cells (MSC) and muscle cells from control (N-201) and FSHD (F-183) individuals. In contrast to the wild-type and FSHD muscle cells, the central portion of DUX4 can be amplified from the ES and MSC, but not from wild-type or FSHD muscle cells. Template controls include: IVT, dilutions of RNA from in vitro transcribed DUX4 cDNA; T-DUX4 and T-Control, RNA from mouse C2C12 muscle cells transfected with a DUX4 expression vector and the empty expression vector, respectively. In both (B) and (C), three independent wild-type and FSHD-derived myoblasts cultures were grown to confluence and induced to differentiate for 96 h. Similar results were obtained from undifferentiated wild-type and FSHD myoblasts (data not shown). Note that the size of the PCR product can be calculated from the position of the PCR primers.
DUX4 protein can be readily detected in undifferentiated human embryonic stem cells and mesenchymal stem cells (S. Winokur, personal observation). In contrast to cultured fibroblasts and muscle cells, all of the DUX4 regions can be amplified from human embryonic and mesenchymal stem cell RNA (Fig. 2C, ES and MSC), whereas the central portion of DUX4 remains difficult to detect in both wild-type and FSHD muscle cells (Fig. 2C, N-201 and F-183). To determine the relative abundance of each region of DUX4, we performed quantitative real-time RT–PCR on DUX4 RNA from three wild-type and three FSHD muscle-derived cultures and normalized the values to an internal control and to the comparable region amplified from human ES cells (Fig. 3). The 5-prime end of the DUX4 transcript was slightly more abundant in the muscle cells compared with the ES cells, whereas the middle region was present in muscle cells at one-tenth or less the levels in ES cells (note discontinuity of Y-axis). The 3-prime region was detected at varying levels in the muscle cells and, in some muscle samples approached the abundance in the ES cells. The FSHD-derived cells showed a higher abundance of transcripts of all DUX4 regions compared with the wild-type cells, although transcript abundance did not correlate with the number of residual D4Z4 units. Finally, sequencing individual PCR products from the FSHD samples confirmed that each of the regions had transcripts with a sequence that matched the 4qA161 sequence from the λ42 clone, usually the last repeat but occasionally the first repeat. It is difficult to interpret sequences without a perfect match because of high sequence error rates in GC rich regions. Therefore, the data suggest that the full-length DUX4 transcript is present in ES cells, but that the central portion is deficient in fibroblasts and muscle cells, possibly due to RNA processing, and that the DUX4 transcribed regions are slightly more abundant in FSHD muscle cells compared with controls.
Figure 3.
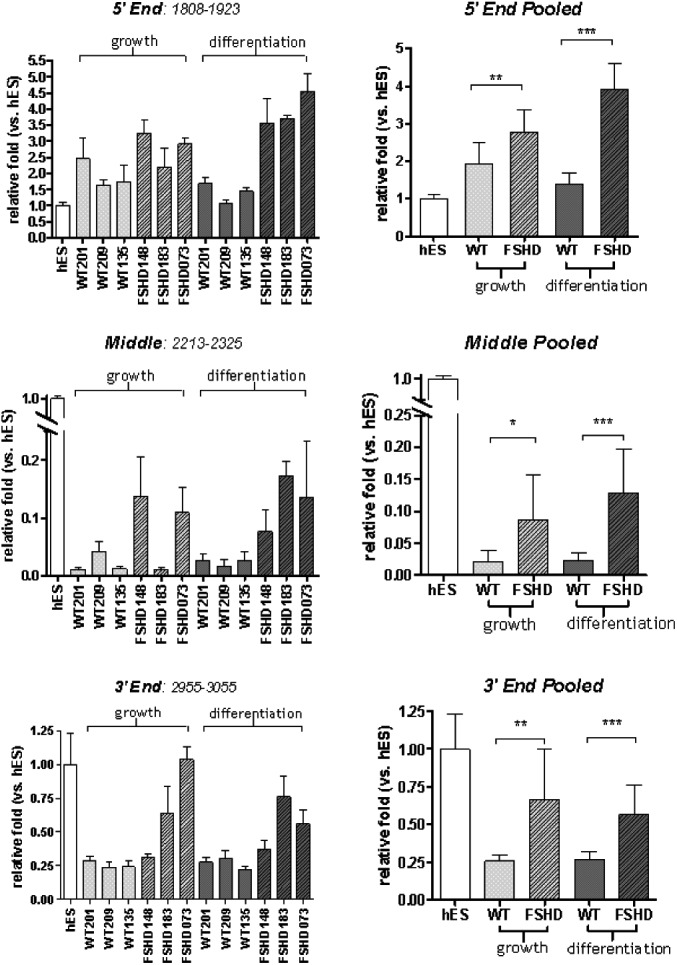
Quantification of amplification products from various DUX4 regions using real-time RT–PCR. RNA sources were cultured myoblasts and myotubes from control and FSHD-affected individuals, and human ES cells. In each grouping, the left panel shows the average of triplicate data from six independent muscle-derived cell cultures (three wild-type and three FSHD) under growth (myoblast) and differentiation (myotube) conditions and the right panel shows the combined average values for wild-type and FSHD with values expressed as the amount relative to the RNA abundance in ES cells. Note that amplification products from the 5-prime region of the DUX4 transcript are slightly more abundant in muscle cells when compared with ES cells; however, amplification products from the middle region of DUX4 are much less abundant in muscle cells. Amplification products from the 3-prime region of DUX4 are less abundant when RNA from muscle is compared with that from ES cells but increased in quantity when compared with the middle region. Error bars indicate standard deviation and asterisks indicate: *P < 0.01; **P < 0.001; ***P < 0.0001; however, the biological significance should be interpreted cautiously due to the small number of independent samples. See Supplementary Material, Table S2 for RT and PCR primer sequences.
In addition, we used strand-specific RT–PCR and identified several sense (relative to DUX4) and anti-sense transcripts throughout D4Z4 in myoblasts and myotubes from both the control and FSHD samples that match the λ42 4qA161 sequence (shown schematically in Fig. 1). We also identified transcripts in similar regions with polymorphisms relative to the 4qA161 sequence, suggesting that multiple different D4Z4 units are transcribed.
Evidence for RNA processing that generates small RNA fragments
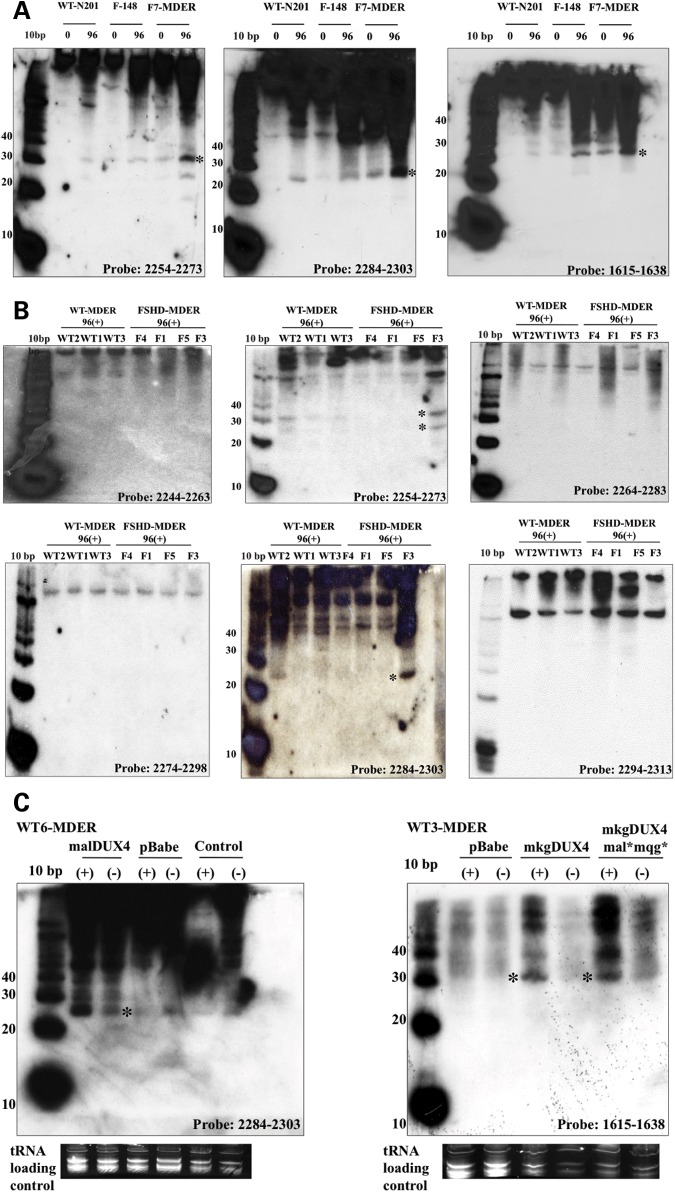
RT–PCR did not detect any splice forms that would remove the central region of the DUX4 transcript and maintain both the 5-prime and 3-prime regions. We therefore sought to determine whether the combined sense and antisense transcripts might generate siRNA fragments, or whether hairpin structures might be processed into miRNA fragments. We took two approaches to determine whether si- or miRNA-sized fragments might be generated from the D4Z4 region. We used RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and Stem-Loop Finder (Combimatrix) to identify regions with predicted RNA stem-loop structures and miR-SCAN (http://genes.mit.edu/mirscan/) to predict potential miRNAs in these regions. We then used probes to those regions on northern blots. This first approach identified two small miRNA-sized fragments generated from a predicted stem-loop-structure present in the middle of the DUX4 ORF (Fig. 4A, probes 2254–2273 and 2284–2303). Shifting the probe 10 nt in either direction eliminated the hybridization to each of the miRNA-sized fragments (Fig. 4B), establishing the location of the miRNA-sized fragment within a region plus or minus several nucleotides.
Figure 4.
Northern detection of miRNA-sized fragments generated from regions of the DUX4 transcript. (A) Micro-RNA northern of RNA from normal muscle cells (N201), FSHD-derived muscle cells (F-148) and FSHD-derived fibroblasts transduced with the MyoD-ER (F7-MDER) with a 21 mer probe to positions 2254–2273, a 20 mer probe to positions 2284–2303 and a 24 mer probe to positions 1615–1638. Each probe detects fragments in the 20–30 nt range; however, multiple larger RNA species are also detected. (B) Micro-RNA northern of wild-type and FSHD-derived fibroblasts with MyoD-ER probed in the region of the predicted miRNA (center panel) and with probes shifted 10 nt in either direction (flanking panels). Fragments in the 20–30 nt range (asterisks) are restricted to the central probes, whereas all probes identify some larger RNA species. All cells contained MDER and were induced to differentiate for 96 h (96+). (C) Micro-RNA northern of wild-type fibroblasts with MyoD-ER probed for the fragments at 2284 and 1615 showing that the fragment abundance is substantially increased in cells transduced with a viral construct expressing the DUX4 RNA and induced to differentiate into muscle (+, indicates the addition of beta-estradiol to activate the MyoD-ER). Control, no viral vector; pBABE, viral vector without insert; pBabe-DUX4, viral vector expressing a DUX4 transcript with nomenclature as indicated in Figure 4A. Sequences of the probes are in Supplementary Material, Table S3.
As a second approach, we searched a database from an unrelated deep-sequencing project designed to identify novel miRNA-sized RNAs in stem cells and cancers (S. Wyman and M. Tewari, personal communication). Several sequences perfectly match the 4qA161 D4Z4 sequence (Table 1) and northern analysis with a probe to one of these sequences identified a small miRNA-sized fragment (Fig. 4A, probe 1615–1638). Together, these two approaches identified three miRNA-sized RNA fragments that might be generated from D4Z4 transcripts. Their locations in D4Z4 are shown schematically in Figure 1 (asterisks) and Supplementary Material, Figure S1. When blasted against the human genome sequence (UCSC-hg18), perfect matches to these sequences are restricted to the D4Z4 repeats on chromosomes 4 and 10, with the exception of one sequence that is also present on chromosome 3 (Table 1). Northern analysis of the three wild-type and three FSHD muscle cell cultures showed that the three miRNA-sized fragments were expressed at detectable levels in all of the cells (Table 2), although there was no clear correlation between the abundance of the signal and FSHD status. A similar northern analysis detected these miRNA-sized fragments in wild-type and FSHD fibroblasts, with some cell cultures showing an increased abundance following conversion to muscle by MyoD (Supplementary Material, Table S1 and Fig. 4).
Table 1.
Sequence of miRNA-sized fragments confirmed by northern
| D4Z4 location | Sequence | Genomic sitesa |
|---|---|---|
| 1615–1634 | UGAAGGGGUGGAGCCUGCCU | Chr 4 and 10 |
| 2254–2273b | caggGUGGCAGGGCGCCCGCGCAGgcaggc | Chr 3, 4 and 10 |
| 2284–2303b | ggccUGUGCAGCGCGGCCCCCGGCggggc | Chr 4 and 10 |
| Additional sequences identified in sequencing projects but not confirmed by northern | ||
| 18–40 | CGCCUACUGCGCACGCGCGGGUU | Chr 4, 10 and 18 |
| 105–92 | GCGGGGGUGGGCUGGUGGAGA | Chr 4 and 10 |
| 1513–1530 | CCACCACCACCACCACCA | Masked |
| 3140–3157 | UGGCUGGCUACGGAGGGG | Chr 4 and 10 |
aSequence blatted to human genome.
bApproximate location.
cFragments identified by hybridization are inferred plus or minus four nucleotides.
Table 2.
Summary of miRNA northerns in myoblasts and muscle cells
| Cell line | Clinical status | EcoRI Frag-kb | D4Z4 repeat no. | Probe |
|||||
|---|---|---|---|---|---|---|---|---|---|
| 2254 |
2284 |
1615 |
|||||||
| 0 | 96 | 0 | 96 | 0 | 96 | ||||
| NR-209 | Control | 87 | 25 | +++ | ++++ | + | + | + | ++ |
| NR-201 | Control | 158 | 46 | + | ++ | + | ++ | + | ++ |
| NR-135 | Control | 65 | 18 | ++++ | ++ | ++ | +++ | ++ | + |
| FSHD-183 | FSHD | 23 | 5 | ++ | + | ++ | + | ++ | + |
| FSHD-073 | FSHD | 31 | 8 | +++ | ++++ | + | ++++ | ++ | ++++ |
| FSHD-148 | FSHD | 16 | 3 | + | ++ | + | ++ | ++ | +++ |
+ indicates relative intensity of the hybridization signal in high serum growth conditions (0) or 96 h of low serum differentiation conditions (96).
It is interesting that the regions generating the small RNA fragments coincide with the regions of transcript discontinuity in the DUX4 ORF in non-ES cell cultures. Therefore, to determine whether the DUX4 transcript can be processed to generate miRNA-sized fragments, we transduced wild-type human fibroblasts with a retroviral construct driving the DUX4 sequence from the viral LTR and performed northern analysis. The miRNA-sized fragments were detected in DUX4-expressing cells and were most abundant after conversion to muscle by MyoD (Fig. 4C). Shifting the probe 10 nt in either direction failed to detect miRNA-sized signal, indicating the specificity of the fragment to the predicted region (Supplementary Material, Fig. S2). Therefore, the 4qA DUX4 transcript can be processed to generate the small miRNA-sized fragments. Similar to the endogenous fragments, the processed fragment from the exogenously expressed DUX4 transcript appears more abundant in cells converted to differentiated muscle, presumably due to differential processing because the LTR is active in both muscle and non-muscle cells.
DUX4 RNA inhibits myogenesis in the absence of the DUX4 protein
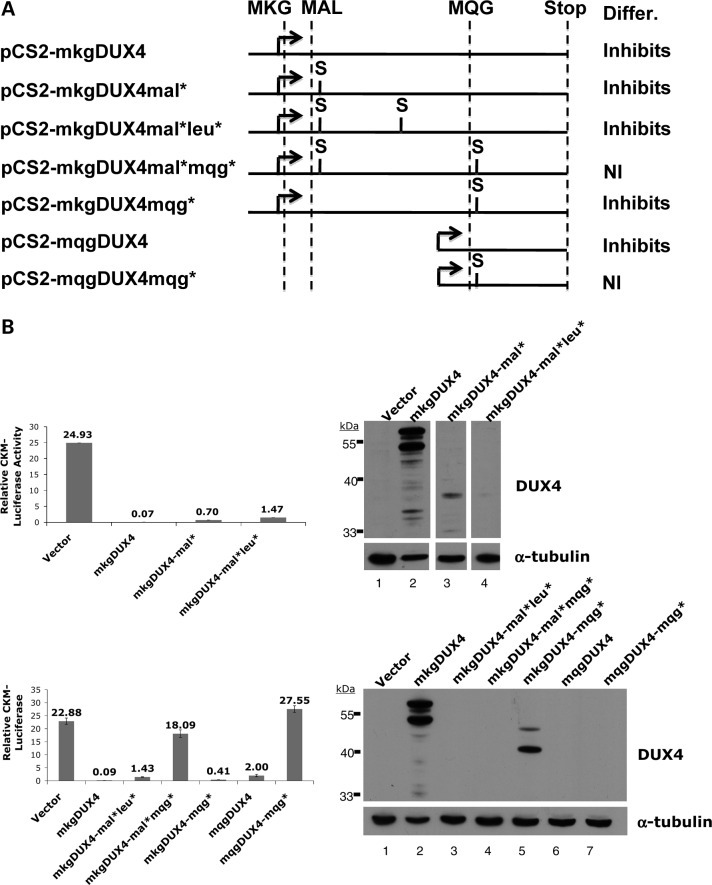
Expressing the DUX4 RNA and protein can be toxic to cells and can inhibit differentiation (6,9). However, these studies have not determined whether the RNA contributes to the toxicity. We transfected C2C12 cells with the pCS2 expression vector expressing a DUX4 transcript initiating just upstream of the DUX4 ORF at the MKG codons (pCS2-mkgDUX4), or the same cDNA with stop codons to prevent RNA translation (shown schematically in Fig. 5A), and assayed differentiation based on expression of a co-transfected luciferase reporter driven by the Ckm regulatory regions (pCkm-Luc). Transfection of pCS2-mkgDUX4 markedly reduced the activity of Ckm-luc (Fig. 5B). Western analysis with the 9A12 monoclonal antibody (7) showed that this construct expressed a full-length DUX4 protein consistent with translation initiation at both the MKG and MAL codons (Fig. 5B western, lane 2). A translation stop codon placed downstream of the MAL codons (pCS2-mkgDUX4mal*) eliminated production of the full-length DUX4 protein (lane 3) and maintained significant suppression of differentiation, as determined by the suppressed level of Ckm-Luc. Similar results were obtained measuring the expression of the endogenous myosin heavy chain, or reporters driven by the desmin promoter (pDes-Luc) or a multimerized E-box (p4R-TK-Luc) (data not shown). Note that all reporter assays were normalized to expression from a co-transfected CMV-beta-galactosidase reporter to control for general cellular toxicity and the assays were performed at a relatively early 24 h time point. At later time points, the full-length DUX4 protein can induce apoptosis, whereas the stop-codon mutations preserve the inhibition of differentiation but largely eliminate the cellular toxicity (data not shown).
Figure 5.
Inhibition of myogenesis in the absence of DUX4 protein. (A) Schematic of the coding regions and stop-codon placements of the expression constructs tested. The methionines in an open reading frame with DUX4 are depicted with the two following amino acids (MKG, MAL, MQG) and the DUX4 translation stop codon indicated by STOP. S shows the regions where we have introduced a new translation stop codon. The column labeled Differ. indicates whether the expression of the indicated vector inhibited C2C12 differentiation (Inhibits) or did not inhibit differentiation (NI). (B) C2C12 cells transiently transfected with pCkm-luc and CMV-beta-galactosidase together with the indicated DUX4 expression. Bar graphs show luciferase activity relative to beta-galactosidase activity 24 h after induction in differentiation medium and western shows abundance of protein containing the epitope recognized by the 9A12 monoclonal antibody to DUX4. The two major bands in the western represent translation initiation at the MKG and the MAL methionines and they migrate at their predicted size. Note that the monoclonal antibody does not recognize in vitro translated mqgDUX4 (data not shown) indicating that the epitope is not contained within this region of the protein.
The size of the small protein produced by the mkgDUX4-mal* (lane 3) indicates that it is produced by translation initiation at one, or both, of two internal leucine residues, consistent with reports that the CUG codon of leucine can be used as a translational-initiation codon (13–15). Placing a second stop codon downstream of these two leucine codons (mkgDUX4-mal*leu*) completely eliminated protein on western (lane 4) yet maintained significant inhibition of myogenesis. Therefore, the DUX4 transcript significantly inhibits myogenesis in the absence of full-length DUX4 protein or any protein recognized by the 9A12 monoclonal antibody.
Internal translation initiation generates a small protein from the DUX4 RNA that inhibits myogenesis
Because the DUX4-mal* construct demonstrated that internal initiation can occur on the DUX4 RNA, we wanted to determine if the inhibition of differentiation from the DUX4mal*leu* was due to RNA or an internally initiated protein not recognized by the 9A12 monoclonal antibody. Although the DUX4-mal*leu* inhibited myogenesis, moving the second stop codon downstream of the MQG codons (mkgDUX4-mal*mqg*) eliminated the myogenic inhibition (Fig. 5B, lower panel), suggesting that a C-terminal peptide of DUX4 can be translated by initiating at the MQG methionine. Translation initiation at the MQG codons would produce a 76 amino acid protein consisting of the highly conserved carboxyterminal region of DUX4 (8). Expressing this isolated 76 amino acid peptide (pCS2-mqgDUX4) was sufficient to inhibit differentiation, and introducing a translation stop codon downstream of the MQG codons (mqgDUX4mqg*) eliminated the myogenic inhibition (Fig. 5B, lower panel). The MQG protein is not recognized by the 9A12 monoclonal antibody (data not shown) and therefore was not identified on the western (lane 6, lower panel). Therefore, the carboxy-terminal portion of DUX4 can inhibit myogenesis. However, the amino-terminal region also inhibited muscle differentiation because constructs that have a stop codon after MQG (mqgDUX4mqg*) and make a truncated form of DUX4 protein (lane 5, lower panel) predicted to contain the full amino-terminal portion without the carboxy-terminal portion also inhibited muscle differentiation.
To determine whether the 76 amino-acid C-terminal protein can inhibit myogenesis in a developmental context, mRNAs encoding this C-terminal peptide or the full-length DUX4 were injected into zebrafish embryos at the two-cell stage, resulting in mainly unilateral distribution of the mRNAs as the embryos develop. The full-length DUX4 (mkgDUX4) was either incompatible with development past gastrulation or caused severe developmental abnormalities in the surviving embryos, such as spina bifida, and broadly suppressed myogenic markers (Fig. 6). The C-terminal protein (mqgDUX4) did not alter normal morphological development, and the embryos had normal expression of MyoD RNA in the somites, but the expression of two markers of muscle differentiation, myogenin (myog) and myosin light chain (mylz2) was significantly diminished. Because MyoD activates the expression of myog and mylz2, we conclude that the C-terminal peptide specifically impedes myogenesis at a step between the transcription of MyoD and the activation of MyoD-regulated genes. A translation stop codon immediately downstream of the MQG codons eliminated the biological activity of the C-terminal protein, indicating that the protein, and not the RNA, was inhibiting myogenic development.
Figure 6.
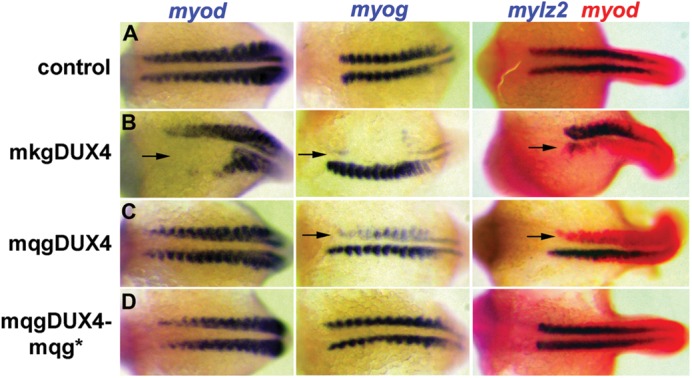
Inhibition of myogenesis in zebrafish embryos by the c-terminal peptide sequence of DUX4. Zebrafish embryos were injected with mRNA encoding the full-length DUX4 (mkgDUX4, see Fig. 4A for nomenclature), the c-terminal fragment of DUX4 (mqgDUX4) or the c-terminal fragment with a stop codon to prevent protein translation (mqgDUX4-mqg*). Full-length DUX4 is highly toxic and broadly interferes with the development on the injected side. The mqgDUX4 injected side shows nearly normal development with normal expression of MyoD RNA, but has a very specific inhibition of muscle gene expression with decreased expression of myogenin (myog) and myosin light chain (mylz2). The stop-codon mutant of mqgDUX4 does not inhibit muscle gene expression, demonstrating that the mqg protein is required for inhibitory activity.
Polyadenylated transcripts contain an IRES-like element and the MQG ORF
Oligo-dT selection of polyadenylated RNA from cultured FSHD myotubes showed enrichment of the MQG ORF in the poly-adenylated (poly-A) fraction (Fig. 7A, 3-prime DUX4). Three-prime RACE demonstrated that the 3-prime region containing the MQG ORF extended into the pLAM region (the sequence telomeric to the D4Z4 units) and had several alternative 3-prime splice forms that matched the previously reported 3-prime UTR of DUX4 (7). A summary of the spliced mRNAs identified is shown in Figure 7B with locations of miRNA-like fragments indicated by asterisks. It is interesting to note that three of the miRNA-sized fragments previously identified (3140–3157, 18–40 and 105–86) the map to the introns in the 3-prime UTR (shown schematically in Fig. 7B). The first intron and second exon are in the last partial repeat and could originate from any of the D4Z4 units; however, because the last exon and the polyadenylation sites are in the pLAM region, it is likely that the polyadenylated transcript containing the MQG C-terminal ORF originates from the last D4Z4 unit. Furthermore, because almost all of the detectable DUX4 3-prime region fractionates with the poly-A RNA, it is likely that the last D4Z4 unit accounts for nearly all of the stable 3-prime DUX4 transcript.
Figure 7.
Five-prime and 3-prime polyadenylated transcripts with an IRES-like element upstream of the MQG ORF. (A) RT–PCR on random primed RNA from FSHD muscle cells using primers to three regions of the DUX4 transcript (5-prime, Central and the 3-prime region of the MQG ORF) on total RNA (T), the poly-adenylated fraction that binds oligo-dT (P), or the unbound fraction that does not bind oligo-dT (U). Primers used were: 5-prime, 1707 and 1906; central, 2307and 2815; 3-prime, 6315 and 7074. In vitro transcribed full-length DUX4 RNA was used as a positive control for the RT reaction (I). 100 bp ladder (L) with 100 bp as lowest band. (B) A schematic of the DUX4 region with representations of the transcripts identified by a combination of 5-prime RACE and 3-prime RACE on the poly-A fraction and location of miRNA-like fragments indicated by asterisks. Top schematic shows locations of potential translation start codons (MKG, MAL and MQG) and stop codon (STOP); asterisks indicate locations of miRNA-like fragments; LR-EX1, cloned sequence matches last repeat-ExonI; LR-EX2, last repeat Exon 2; LR EX3, last repeat Exon 3; IR-EX1, cloned sequence does not match either first or last repeat (Supplementary Material, Fig. S4); IR-EX2, in this case cloned sequence does match LR-EX2 but the tandem repeat indicates it comes from an internal repeat. (C) Sequence of the DUX4 ORF and pLAM region showing the locations of the capped 5-prime ends (Blue, positions 4941–4944 and 4970–4971) and uncapped 5-prime ends (Red, positions 5715 and 5863), representing sites of transcription initiation and RNA cleavage, respectively. The polyadenylation sites are indicated in Orange (postions 7155–7156 and 7166); introns are underlined; miRNA-sized fragments are shown in green (note that the last partial D4Z4 unit is between the last full D4Z4 unit and the pLAM sequence (Fig. 1), and therefore, the last two miRNA-sized fragments are also present in the beginning of the D4Z4 repeat). (D) The construction of the dual cistronic pRF backbone is detailed in (33). The locations of SV40 promoter and chimeric intron are indicated, and Poly-A is the SV40 polyadenylation signal. Inserting test sequences between the EcoRI and NcoI sites of pRF created the constructs pRF + 423DUX4, pRF + 275DUX4. pRF + HCVir was created by inserting the previously characterized IRES element from the Hepatitis C virus as a positive control, and its IRES activity has been previously described (34). An empty pRF plasmid without insert was used as a negative control. Each of the constructs was transfected into mouse myoblast C2C12 cells as two sets of triplicates. Twenty-four hours post-transfection, one triplicate set of cells, designated as ‘growth,’ was harvested and their lysates assayed for FLuc and RLuc activities as described in Materials and Methods. The remaining set was switched to ‘differentiation’ media and assayed for luciferase activities 48 h post-transfection. FLuc activity was normalized to RLuc and plotted as the mean ± SD relative to the empty plasmid pRF.
We performed 5-prime RACE on the poly-A RNA from FSHD muscle cells using protocols to separately detect RNA with a 7-methylguanosine cap and uncapped RNA 5-prime ends, representing polymerase II transcriptional start sites and possible sites of RNA cleavage, respectively. Using a reverse primer for RT near the MQG codons, prominent uncapped 5-prime ends map to position 5715 and 5863 in the last D4Z4 unit (Fig. 7C and Supplementary Material, Fig. S1), whereas we did not detect any capped 5-prime ends. These uncapped 5-prime ends indicate RNA cleavage in the central portion of the DUX4 transcript, which is in the region of the transcript that is difficult to amplify by RT–PCR (corresponding to positions 2417 and 2565 in the first unit).
If a polyadenylated and uncapped C-terminal fragment containing the MQG C-terminal ORF is produced by cleavage of the DUX4 mRNA, could it possibly be translated into a protein? There is precedent for translation of uncapped viral RNAs in mammalian cells through internal ribosomal entry site (IRES) elements and some mammalian transcripts have been shown to contain IRES elements, although the characterization and biological role of these elements remains controversial (16,17). To determine whether the 3-prime uncapped and polyadenylated fragment contains an IRES-like element, we cloned the regions between each of the two putative cleavage sites, as defined by uncapped 5-prime ends, and the sequence ending immediately prior to the methionine of MQG—from 5715 to 6138 (423 nt) and from 5863 to 6138 (275 nt) (see Fig. 7C) into a bicistronic reporter construct between the renilla and the firefly luciferase reporter genes (shown schematically in Fig. 7D). C2C12 cells were transfected with the bicistronic reporter constructs and the expression of each reporter gene was measured under growth (myoblast) and differentiation (myotube) conditions. There was a modest increase in firefly luciferase activity with the longer 423-nt fragment in myoblasts and a robust 15-fold increase in myotubes, suggesting a cell-type-specific IRES-like activity. We conclude that the DUX4 transcript contains a region that facilitates the translation of the C-terminal portion of the protein (the MQG ORF), either through IRES activity, a cryptic splice site or a cryptic promoter element.
Additional poly-adenylated and spliced transcripts encode the double-homeobox domain of DUX4
The 5-prime region of the DUX4 transcript (just downstream of the MAL codons) was also enriched in the poly-A RNA from FSHD myotubes, whereas the central portion and the full-length RNA were not enriched in this fraction (Fig. 7A). Performing 5-prime RACE on this RNA with a reverse RT primer downstream of the MAL codons identified several capped 5-prime ends upstream of MAL, indicating a region of transcription initiation. These capped transcripts initiate at 4941–4944 and 4970–4971 (corresponding to positions 1644–1647 and 1673–1674 in the first repeat), which represents a cluster of transcription start sites upstream of the MAL ORF (Fig. 7C and Supplementary Material, Fig. S1).
It is tempting to conclude from these results that a transcript initiates upstream of the MAL ORF, has spliced introns in the 3-prime UTR and is poly-adenylated in the pLAM sequence, consistent with Dixit et al. (7), and that this mRNA is cleaved to produce the poly-adenylated 3-prime end with the MQG ORF. Our data are, at least partly, inconsistent with this conclusion. This is because the central portion of the DUX4 transcript appears to be under-represented in RT–PCR assays on oligo-dT enriched mRNA, whereas the 5-prime and 3-prime regions are strongly represented. We do not detect a spliced mRNA that joins the 5-prime region containing the MAL ORF with the 3-prime region containing the MQG ORF by 5-prime RACE or by standard RT–PCR (data not shown). Therefore, our data suggest that there might be two different polyadenylated mRNAs, one that includes the region near the MAL codons, and one that includes the region of the MQG ORF.
Indeed, 3-prime RACE on the oligo-dT selected mRNA with primers located near the MAL codons identified two novel splice forms that remove the region of the MQG ORF and splice the 5-prime region directly to the second exon (shown schematically in Fig. 7B). Oddly, one of these two splice forms contains a direct repeat of the second exon. At first, we considered this to be an artifact of the PCR process; however, we note that the sequence of the second exon overlaps the KpnI site (see Supplementary Material, Fig. S1) and is present at the junction of each D4Z4 unit, including the junctions of the internal repeats and the last partial repeat. We conclude, therefore, that a capped transcript originates from an internal repeat, is spliced to an internal exon 2 and then this exon 2 is spliced to the exon 2 at the junction with the last partial repeat, resulting in a direct duplication of exon 2 in the mRNA.
Translation of the ORF in these capped, spliced and polyadenylated transcripts would make a protein containing the paired homeodomains but not the highly conserved C-terminal portion of DUX4, replacing that region with an ORF that extends into the second exon. This protein is likely to inhibit myogenesis, because myogenesis was inhibited when we placed a stop codon immediately downstream of the MQG codons in the DUX4 expression constructs (Fig. 5B, lower panel, mkgDUX4-mqg*). In addition, DUX4C is nearly identical to DUX4 but lacks the conserved C-terminal region, and DUX4C has been shown to inhibit myogenesis (18). It is interesting to note that the splice donor sites in the first and last repeat do not have a consensus donor sequence. Although non-consensus splice donor sites have been described, it is possible that one, or more, internal D4Z4 unit has a polymorphism that creates a consensus splice donor site. Additional sequencing of internal D4Z4 units in the 4qA161 and other haplotypes will be necessary to determine whether this is the case.
Finally, a full-length DUX4 transcript can occasionally be amplified using nested RT–PCR on polyadenylated RNA from muscle cells. The difficulty of amplifying the full-length DUX4 is likely due to very low abundance, which would be consistent with mRNA splicing and/or cleavage.
DISCUSSION
In summary, our data demonstrate capped transcripts initiating upstream of the DUX4 MAL codons that continue into the pLAM region where they are polyadenylated. Although a full-length DUX4 transcript is initially produced, and can be detected at low levels, spliced and cleaved forms are more readily detected, presumably indicating higher abundance of these processed forms of the DUX4 mRNA. The carboxy-terminal portion of DUX4 is either removed by a splice event that creates an mRNA encoding for the double-homeobox region of DUX4, or is isolated as an uncapped and polyadenylated RNA, presumably by cleavage of the full-length DUX4 transcript. The presence of a direct repeat of the second exon in some mRNA strongly indicates that some of the spliced and polyadenylated transcripts initiate from internal D4Z4 units and progress through intervening D4Z4 repeats to terminate at the poly-adenylation site in the pLAM region.
Our study is consistent with a recent publication (7) showing that a transcript from the last D4Z4 unit extends into the pLAM region, has specific splice sites in the 3-prime UTR and is polyadenylated at a specific site. We extend that study by demonstrating (i) several overlapping sense and anti-sense RNA transcripts from the 4q D4Z4 units; (ii) RNA cleavage and processing of the central portion of the DUX4 containing transcript to generate mi/siRNA-sized fragments; (iii) additional miRNA-sized fragments generated from the introns in the 3-prime UTR of the DUX4 transcript; (iv) biological activity of DUX4 transcripts that do not produce a full-length DUX4 protein; (v) translation of the highly conserved C-terminal portion of DUX4, possibly through internal ribosomal initiation, is sufficient to inhibit myogenic differentiation and (vi) novel splice forms of the DUX4 transcript that lack the highly conserved C-terminal region. These findings suggest several new candidate mechanisms for FSHD pathophysiology that deserve rigorous exploration (Supplementary Material, Fig. S3).
Small RNA fragments might have a biological function
The northern analysis demonstrates that restricted regions of the D4Z4 transcripts give rise to the small si or miRNA-sized fragments, because moving the probe 10 nt in either direction fails to detect the small fragments, and multiple other probes to the D4Z4 regions failed to detect small fragments. Some of the fragments map to regions of predicted hairpin RNA and others map to introns in the 3-prime UTR of the DUX4 transcript, both consistent with a miRNA mechanism. However, we are reluctant to conclude that these are si-, mi- or pi-RNAs without further confirmatory study. The northern blots show more hybridization to larger precursor-like RNA fragments than is typically seen for the precursors to the miRNA (pre-miRNA or priRNA) signals and this suggests a fragmentation process distinct from miRNA generation and might be more consistent with the formation of endogenous siRNAs from double-stranded RNA. In either case, however, these small RNA fragments might have a biological role.
The overlapping sense and anti-sense transcripts that apparently span the D4Z4 region, with areas of discontinuity (see Figs 1 and 2), might generate double-stranded RNA that can subsequently be cleaved to generate siRNAs. This is of interest because repeat associated heterochromatin in numerous species has been shown to be mediated by an RNAi mechanism (19). Transcripts from the repetitive regions are converted to siRNA fragments through Dicer-mediated cleavage and the siRNA induce local heterochromatin through recruitment of HP1 and other factors. This creates the paradox that transcription of the repetitive element is necessary for heterochromatic silencing. Therefore, the bidirectional transcripts and small RNA fragments we describe at the D4Z4 repeats are consistent with the emerging model of repeat associated heterochromatic silencing.
In addition to silencing the transcribed locus in cis, endogenous siRNA can also target other DNA loci in trans, or RNA transcripts. A recent example reveals that siRNA generated from pseudogenes can target transcripts from corresponding genes in mammalian ES cells (20,21), either through double-stranded RNA generated from the pseudogene, or a single-stranded pseudogene RNA that hybridizes with the spliced RNA from the cognate gene. Alternatively, the ∼21 nt siRNA fragments, or the ∼25–27 nt piRNA fragments can target retroposons, or potentially other DNA elements to induce heterochromatic silencing in cis (20,21). Therefore, the transcripts and small RNA fragments we identified at the D4Z4 repeats might be associated with local chromatin silencing, chromatin silencing at distant loci or might target RNA from other loci. We should note that a prior publication failed to detect RNA transcripts or PolII association with the D4Z4 repeats (22); however, this might represent differences in cell type or relative sensitivity of the assays compared with our current study.
In contrast to siRNAs, miRNAs are generated from a single RNA strand that forms a double-stranded hairpin structure, which frequently are encoded in introns of transcribed genes. Several of the small RNA fragments identified in this study map to regions of predicted RNA hairpin structure and several also map to introns, both characteristics common to miRNA. It is interesting to note that standard miRNA prediction algorithms identify several RNAs involved in muscle cell differentiation (S.J.T., unpublished data), and it will be important to determine whether these have a role in normal development or FSHD.
Internal translation initiation can produce a C-terminal fragment of DUX4 that blocks myogenesis
Transfection experiments with stop codons introduced in the DUX4 ORF indicate that internal translation initiation can result in protein translation of the C-terminal region of DUX4, and that this small protein (76 amino acids) can block specific steps of myogenic differentiation. Prior studies have demonstrated that transcriptional targets of MyoD are expressed at lower levels in FSHD muscle cells (23,24) and it is interesting that the C-terminal protein beginning at the MQG codons appears to block myogenesis at a step between MyoD RNA transcription and the activation of MyoD target genes. It is important to note that prior studies have not identified the presence of this 76 aa protein in either FSHD or wild-type muscle and our study only shows that it is expressed from transfected RNA, not from the endogenous RNA. However, the epitope recognized by the 9A12 monoclonal antibody we, and others, have used to detect DUX4 is not contained in the 76 aa MQG protein and it will be necessary to generate antibodies to this protein to assess its expression in wild-type and FSHD tissues.
RNA containing this C-terminal MQG ORF fractionates with poly-adenylated mRNA; however, 5-prime race identifies only uncapped 5-prime ends upstream of the MQG codons, suggesting that the MQG ORF containing RNA is generated through cleavage of a longer transcript, possibly initiating upstream of the DUX4 MAL codons. Normally, it would be anticipated that an uncapped RNA would not be translated; however, the transient transfection studies provided definite evidence for internal translation initiation in the region upstream of the MQG codons. Our demonstration of IRES activity in this region of the RNA further indicates that this uncapped RNA fragment can be translated. Uncapped and polyadenylated viral RNA has been shown to be translated in mammalian cells through IRES elements, although as noted above, there remains some disagreement regarding the molecular mechanisms (16,17). Therefore, our suggestion that the MQG ORF might be translated from an uncapped and polyadenylated RNA needs rigorous validation. However, the ability of this 76-aa protein to inhibit myogenesis in C2C12 cells and in zebrafish embryos suggests a possible role in FSHD, particularly since this 76-aa protein inhibits a specific stage of myogenesis—after the expression of MyoD and the before the activation of Myog and Mylz2 (a fast myosin isoform) (Fig. 6)—whereas, DUX4 appears to be broadly toxic to both cells and embryos. It is interesting to note that protein and RNA expression studies on FSHD muscle identified both a decreased expression of MyoD targets and a transition from fast-glycolytic to slow-oxidative fibers in FSHD (23,24). In addition, a prior study demonstrated partial inhibition of C2C12 differentiation when transfected with D4Z4 repeats but did not identify the full-length DUX4 protein (25). Our findings provide a new basis for extending these earlier studies.
Novel splice sites suggest continuous transcripts through the D4Z4 units producing a protein similar to DUX4C
We have also identified poly-adenylated mRNA containing the 5-prime region of the DUX4 transcript. In our studies, these transcripts lack the 3-prime region containing the MQG ORF due to an internal splice donor site that connects with the splice acceptor of the second exon located in the region of the KpnI site that arbitrarily determines the repeat boundaries. It might be quite revealing that the majority of these transcripts contain a direct repeat of the second exon. Our best interpretation at this time is that the duplicated second exon is strong evidence that the polyadenylated transcript originates within an internal D4Z4 unit, continues through one or more additional D4Z4 units until there is a successful splice to an internal second exon splice acceptor site, and then this internal second exon is spliced to the second exon in the last repeat followed by polyadenylation in the pLAM region. This interpretation is consistent with our original observation that sense transcripts appear to span the entire D4Z4 unit with some regions of interruption that are likely secondary to RNA processing. In addition, the 5-prime region of the transcripts containing the duplicated second exon have a polymorphism that does not match the first or last D4Z4 sequence in λ42, again suggesting that this transcript arises from an internal repeat. It will, however, be necessary to accurately identify intra-allelic polymorphisms in the 4qA161 D4Z4 units to validate our interpretations.
If a protein is produced from this spliced transcript, it would contain the double-homeobox region of DUX4 but lack the highly conserved C-terminal region. This would be very similar to the DUX4c transcript and protein, which has been shown to inhibit myogenesis and suppress expression of both MyoD and Myf5 (18). Together with our data, it appears that the amino-terminal portion of DUX4 might suppress MyoD and Myf5 expression, whereas the carboxy-terminal portion can suppress myogenesis at a step following the expression of MyoD. The presence of alternative splice forms and RNAs that potentially differentially regulate the expression of each of these DUX4 regions suggests that each might have a distinct developmental role that needs further exploration.
Finally, it is important to mention that we do find evidence of full-length DUX4 transcripts; however, these appear to be of significantly lower abundance than RNAs containing the 5-prime or 3-prime regions.
Macrosatellite repeats and an emerging model for FSHD
At this time, a biological role for the D4Z4 arrays remains speculative, but recent studies on retrotransposons, chromatin regulation and other macrosatellite repeats reveal striking parallels to our current findings at D4Z4 and suggest a biological role for these repeats. A strong parallel to our work on D4Z4 is the DXZ4 macrosatellite repeat (26). Similar to D4Z4, DXZ4 is a 3 kb GC rich unit repeated 50–100 times on the X-chromosome. On the active X-chromosome, bidirectional transcription of DXZ4 results in small RNA fragments, presumably siRNA generated from dsRNA. The locus also has H3K9 and CpG methylation that are associated with a siRNA-mediated induction of heterochromatin. On the inactive X-chromosome, the insulator factor CTCF binds adjacent to a bidirectional promoter in a region that remains unmethylated at CpG residues, and this is associated with epigenetic marks of euchromatin and longer RNA transcripts, possibly secondary to decreased production and processing of double-stranded RNA. Similar to DXZ4, our study finds bidirectional transcription of D4Z4 associated with small RNAs. In addition, the D4Z4 units have CTCF binding sites and we find enriched CTCF binding at hypomethylated sites on the deleted pathogenic allele, as well as enriched CTCF binding to the D4Z4 units in undifferentiated ES cells (Filippova et al., in preparation).
At least two other macrosatellite repeats contain genes. TSPY is in the DYZ5 repeat on the Y chromosome and is expressed in the placenta, and USP17 encodes a deubiquitinating enzyme in the RS447 repeat (27–29). Similar to the coding region of DUX4, the USP17 gene does not contain introns. Also similar to our findings at DUX4, USP17 is transcribed in both sense and anti-sense directions and the anti-sense transcripts are believed to have a role in regulating USP17 expression.
Many intronless genes and pseudogenes were generated by retrotransposition of a spliced mRNA into the genome. It was initially suggested that DUX4 was generated following a retrotransposition of DUXA (30); however, an elegant study of the evolution of the human DUX4 and the D4Z4 repeat indicates that this region arose from a retrotransposition of the DUXC gene (8). DUXC has apparently been lost in the primate lineages but is still present in dogs, cows and armadillo. Generation and propagation of multiple retrotransposed genes indicates germ-line expression and thereby suggests a potential role for DUXA and DUXC in germ cell or early embryonic stem cell. The coding region for DUX4 has been conserved (8) and it is possible that DUX4 protein expression might substitute for the original DUXC function. The mouse DUX4 ortholog is also transcribed in the sense and anti-sense orientation with partial fragments of the RNA detected more readily than the full length (8), indicating that our findings at human DUX4 are conserved in the murine ortholog.
Although DUX4 is not a pseudogene because of its conserved ORF, it has some similarities to emerging properties of some pseudogenes. Recent studies in Drosophila (31) and mammals (20,21) demonstrate that transcripts from pseudogenes, sometimes occurring in subtelomeric clusters, can suppress transposable elements in the germ-line and regulate RNA stability or translation from the related gene family. One pathway for this regulation is through pi-RNA, but siRNA is also generated through bi-directional transcription of these pseudogenes. In this context, it is very interesting that we have identified bi-directional transcripts through the subtelomeric cluster of D4Z4 units that contain the pseudogene-like DUX4, and also have demonstrated DUX4 RNA expression in ES cells. Although a more thorough analysis is needed, the apparent decreased RNA processing of the DUX4 transcript in ES cells (Fig. 1D) suggests that there is a special function for this RNA in ES cells and possibly for the cleaved RNA in the process of ES cell differentiation. It will be interesting to determine whether the small RNAs generated from DUX4 function to suppress DUX4 expression in the germ-line or regulate DUXC in some species or other DUX paralogs.
Therefore, our studies on D4Z4 are very consistent with the recent findings that bidirectional transcription of pseudogenes and genes in macrosatellite repeats is developmentally regulated and also serves a regulatory function. The contraction of the repeats in FSHD likely alters the efficiency of one of these functions, such as maintaining regional heterochromatin. Because of the apparent restriction of FSHD to D4Z4 deletions of the 4qA161 allele, it is likely that a polymorphism results in the production of an abnormal product by affecting RNA splicing or polyadenylation, CTCF (or other factor) binding or small RNA production or targeting. Our current study has provided a strong foundation for this new model of FSHD and identified several new biological processes associated with D4Z4 that warrant further investigation as candidate mechanisms of disease pathophysiology.
MATERIALS AND METHODS
Cell culture
Myoblasts were obtained through the Fields Center at the University of Rochester (http://www.urmc.rochester.edu/fields%2Dcenter/) and derived from a needle biopsy of the vastus lateralis (http://www.urmc.rochester.edu/fields-center/protocols/needle-muscle-biopsy.cfm) and established as primary cultures through dispase and collagenase dispersion (http://www.urmc.rochester.edu/fields-center/protocols/myoblast-cell-cultures.cfm). The size of the 4q alleles are indicated in Table 2 for each cell line. Primary myoblast cell lines were maintained in F-10 nutrient media (Gibco), 20% FBS (Gibco), 1% Pen/Strep, 10 ng/ml bFGF (Promega) and 1 µm Dexamethasone. To differentiate cells, we switched cells at 100% confluency from maintenance media to F-10 differentiation media (DM) containing 1% heat-inactivated horse serum, 1% pen/strep and 0.1% insulin and 0.1% transferrin.
The embryonic stem cell line H1 (WiCell) was purchased and maintained according to University of California IRB/ESCRO regulations. Colonies were cultured in KO-DMEM/F12, 20% KO Serum, 2 mm glutamine, 0.1 mm NEAA, 0.1 mm b-mercaptoethanol and 4 ng/ml bFGF. For RNA extraction, cells were trypsinized and resuspended in Trizol. Protein extracts were generated by physical disruption of colonies and lysis in RIPA buffer. The mesenchymal stem cell line derived from human bone marrow (hMSC, LonzaBiotech) was maintained in MSCGM medium (Lonza) to prevent adipo-, chondro- or osteogenesis.
Primary fibroblast cell lines from both unaffected individuals and FSHD patients were infected with retrovirus containing the pBABE vector expressing a puromycin-resistance gene and inducible MyoD-ER (12). Control cells were infected with virus containing only vector and selectable marker. MyoD-ER-infected fibroblast cell lines were maintained in growth medium, which consisted of DME containing 1% l-glutamine, 10% bovine calf serum (Hyclone), 10% fetal bovine serum (Hyclone) and 1% pen/strep. Infected cells were selected in 1.2 µg/ml puromycin for 4 days after infection. To induce expression of MyoD-ER, cells were switched to DM, which consisted of DME containing 1% glutamine, 1% heat-inactivated horse serum, 10 µg/ml insulin, 10 ug/ml transferrin and 10 µM β-estradiol.
Semi-quantitative RT–PCR
Cultured cells were harvested by first lifting the cells off each plate with 0.05% trypsin. Cells were spun down at 250g for collection. Total RNA was prepared using Trizol (Invitrogen) extraction, as per the manufacturer's instructions. Trizol purification was followed by acid phenol extraction to reduce DNA contamination. The isolated RNA was then treated with DNase I, amplification grade (Invitrogen) prior to cDNA synthesis. Two microgram of total RNA from MyoD-induced fibroblasts were used for each cDNA synthesis. cDNA was generated with random hexamers and Superscript II reverse transcriptase (Invitrogen). The reverse transcription reaction was incubated first at 75° for 5 min, followed by 45° for 45 min, 50° for 15 min and 55° for 15 min, and 75° for 15 min in a total of 30 µl reactions. One microliter of cDNA generated from reverse transcription was used in subsequent 20 µl PCR reaction. To establish the linear range for each gene-specific and control primer set, aliquots of the PCR reaction were collected every two cycles ranging from 30 to 40 cycles and the relative amounts determined by ethidium bromide staining of an agarose gel. Typical thermal cycling condition was 5 min at 94°, 30 s at 94°, 30 s at annealing temperature, 1:30 min at 68° for extension and 10 min at 68°. Amplification of RT–PCR was done using Platinum Taq (Invitrogen). Each PCR reaction was run on 1% agarose gel in 1X TBE.
Strand-specific RT–PCR
Total RNA was prepared using Trizol (Invitrogen) extraction, as per the manufacturer's instructions. Trizol purification was followed by acid phenol extraction to reduce DNA contamination. The isolated RNA was then treated with DNase I, amplification grade (Invitrogen) prior to cDNA synthesis. Four microgram of total RNA from myoblast cell lines was used for each cDNA synthesis. cDNA was generated using gene-specific primers which had a linker (LK) sequence, LK 5′-CGACTGGAGCACGAGGACACTGA-3′, attached to 5′ end. The following primary/nested primer combinations were used for strand specific RT–PCR: 1646/1670, 2206/2270, 2270/2307, 2808/2922 and 1707/1797. All primer sequences are listed in Supplementary Material, Table S2. Briefly, cDNA was generated with Superscript III (Invitrogen) at 55° for 60 min, 60° for 15 min and 75° for 15 min. PCR amplification of the strand-specific transcripts was performed using PCRx enhancer, the LK sequence alone as primer and gene-specific forward/reverse primer in a 20 µl total reaction. Thermal cycling conditions for strand-specific RT–PCR is as followed: 94° for 30 s, 55° for 30 s, 68° for 1:30 min and 68° for 10 min for a total of 20 cycles. All 20 µl of PCR products were then purified using QIAQuick column (QIAGEN) and eluted with 30 µl of EB buffer. Four microliter of the eluted products were then used as templates for subsequent nested PCR reactions (25 cycles, at 94° for 30 s, 55° for 30 s, 68° for 1:30 min and 68° for 10 min). Amplification of RT–PCR was done using Platinum Taq (Invitrogen). Nested PCR products were cloned into the pCRw4-TOPO vector (Invitrogen) and sequenced using an ABI Prism 3730Xl DNA analyzer (Applied Biosystems).
Northern blot analysis
Cultured cells were harvested by first lifting cells off each 15 cm plate with 0.05% trypsin. Cells were spun down at 250g for collection. Total RNA was prepared using Trizol (Invitrogen) extraction, as per the manufacturer's instructions. Trizol purification was followed by acid phenol extraction to remove DNA contamination. Each RNA sample was validated for integrity using the Bioanalyzer, RNA 6000 Nano Assay (Agilent Technology). Each lane contained 25 µg RNA. Samples were separated electrophoretically in a 20% polyacrylamide/8 M urea/1X TBE gel. RNA was electroblotted onto Nytran SPC nylon membrane in 1X TBE at 250 mA for 45 min, and was fixed to the membrane by UV cross-linking. Blots were hybridized overnight at 35° in Ultrahybe Oligo Buffer (Ambion) with radiolabeled oligonucleotide probes complementary to the predicted microRNA sequence of interest. Blots were washed the next day for 3 h with 2X SSC/0.5% SDS. Images were captured on film, digitized, and if needed, minor linear adjustments in contrast were made using Photoshop software (Adobe). To re-probe, blots were stripped by incubation in 1% SDS at 85° for 2 h. Loading controls were determined by either probing for miR-16 or cutting off the top part of the gel prior to transfer and ethidium bromide staining in 1X TBE.
Real-time PCR
Real-time PCR was performed using TaqMan universal PCR mix reagent according to the manufacture's instruction (Applied Biosystems). For detection of each gene, the primers and probe sequences are listed in Supplementary Material, Table S2. Real-time PCR was performed on a Sequence Detection System instrument (ABI Prism 7900HT; Applied Biosystems), and expression levels were quantified using SDS 2.1 software (Applied Biosystems). Each sample was assayed in triplicate; data represent the mean and SD for the triplicates. The relative expression levels of each gene were normalized to those of Timm17b in the same samples. Standard curves for each amplicon were generated using serial dilutions of known quantities of reverse transcribed products or cDNA generated from in vitro transcription.
5′ and 3′ race on capped and uncapped transcripts
RACE was performed using the Invitrogen GeneRacer kit, closely following the manufacturer's instructions. We used a polyA+ enriched fraction of RNA (Dynabeads mRNA Purification Kit) as our starting material to minimize detection of off-target transcripts. Uncapped transcripts were detected by omitting the de-phosphorylation and de-capping steps prior to ligation of the RNA oligo; the capped transcripts will not be available for ligation under these conditions. cDNA was generated using random hexamers or an oligo dT primer and we used nested PCR primers to increase sensitivity and specificity of the reactions. Gene specific primary/nested primer combinations were as follows: 5′ RACE at MAL, 5163/5147; 5′ RACE at MQG, 6142/6083/5850 and 6142/6113; 3′ RACE at MQG 6236/6315; 3′ RACE near MAL, 5105/5174 and 5339/5504 (See Supplementary Material, Table S2 for primer sequences).
Zebrafish mRNA injections
mRNAs were synthesized in vitro from pCS2 constructs using the mMessage Machine kit (Ambion). We injected approximately 120pg of mqgDUX4, 120pg of mqgDUX4-mqg* or 15pg of mkgDUX4 at the two-cell stage. Increased amounts of mkgDUX4 caused severe embryonic lethality. RNA in situ staining and mRNA probe syntheses were as previously described (32).
IRES element assays
The plasmid pRF was kindly provided by A. Willis (Leicester University, Leicester, UK) and its construction was described in (33). To create the test constructs, amplified DUX4 DNA fragments were ligated into pRF between the EcoRI and NcoI sites. A 423 bp DNA fragment corresponding to the region within DUX4 from position 5715 to 6138 just upstream of the MQG start site was amplified from pCS2-mkgDUX4 using primers 5′-CCGGAATTCCGTGAGCCAGGCAGCGAGGGC-3′ (sense) and 5′-CATGCCATGGCTGCCCCTGCCGCGCGGAG-3′ (antisense; EcoRI and NcoI sites are underlined) and inserted into PRF to make pRF + 423DUX4. A 275 bp DNA fragment that is contained within the previous fragment (corresponding to position 5863 to 6138 in DUX4) was amplified in the same as the previous fragment except the sense primer 5′-CCGGAATTCCCTCGCTGGCCTCCGCACCCG-3′ and inserted into pRF to make PRF + 275DUX4. To make pRF + HCVir, the HCV IRES was amplified from pRL-HL (a gift from R. Lloyd) using primers 5′-CCGGAATTCGCCAGCCCCCGATT-3′ (sense) and 5′-CATGCCATGGGACGTCCTGTGGGCGG-3′ (antisense). All fragments were first digested with EcoRI and NcoI and then ligated into pRF between the same sites; constructs were verified by restriction enzyme analysis and automated DNA sequencing.
Mouse C2C12 myoblast cells were maintained in Dulbecco's Modified Eagle's Medium (Gibco) supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin. Growth in a humidified incubator was at 37°C in 5% CO2, and cells were kept below 70% confluency unless otherwise specified. Transient DNA transfections were performed using SuperFect reagents (Qiagen) according to the manufacturer specifications. Briefly, 8 × 104 cells were seeded per 35 mm plate 24 h prior to transfection. Cells were transfected with DNA (3 µg/plate) and cultured at 37°C. A set of cells, designated ‘growth,’ were harvested 24 h post-transfection by removing the medium and then lysed in Passive Lysis Buffer (PLB; Promega). The remaining cells, which at this point are ∼100% confluent, were switched to DM. This DMEM media contained 0.1% insulin, 0.1% transferrin and 1% pen/strep. ‘Differentiation’ cells were harvested and lysed 48 h post-transfection and assayed for enzyme activity as the previous ‘growth’ set.
Upstream cistron Renilla luciferase (Rluc) and Firefly luciferase (Fluc) activities were quantified using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Typically, 20 µl of post-transfection lysate was combined sequentially with Fluc- then Rluc-specific substrates. Light emission was measured 3 s after addition of each of the substrates and integrated over a 10 s interval using a BioTek Synergy2 luminometer. Luciferase data are given as the averages ± SEM of triplicates.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health AR045203, NS046788 and HD047157 to S.J.T.; the Pacific Northwest Friends of FSH Research to D.G.M. and S.J.T.; the Shaw Family Foundation to G.N.F. and S.M.M. and S.T.W.; the National Center for Research Resources (NCRR) UL1RR024160; the Fields Center for FSHD and Neuromuscular Research to S.M.M. and R.T.; the Netherlands Organization for Scientific Research NWO 917.56.338; Marjorie Bronfman Fellowship grant from the FSH Society to R.J.L.F.L.; Breakthrough Project Grant by the Netherlands Genomics Initiative NWO 93.51.8001 to R.J.L.F.L.; the Muscular Dystrophy Association to G.N.F., S.T.W. and S.M.M.; and the FSH Society to S.T.W.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Wyman and M. Tewari for sharing deep sequencing data of small RNAs from stem cells and cancer cells, A. Belayew's group for providing the 9A12 monoclonal antibody to DUX4 and DUX4 vectors, B. Trask, R. Endicott, E. Linardopoulou and K. Siebenthall for helpful discussions, and T. Bird and the University of Washington Clinical Core for cells.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tawil R., Van Der Maarel S.M. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 3.de Greef J.C., Frants R.R., van der Maarel S.M. Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat. Res. 2008;647:94–102. doi: 10.1016/j.mrfmmm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 5.Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 6.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clapp J., Mitchell L.M., Bolland D.J., Fantes J., Corcoran A.E., Scotting P.J., Armour J.A., Hewitt J.E. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O., et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 10.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol. Genet. 1993;2:2037–4202. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 11.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 12.Hollenberg S.M., Cheng P.F., Weintraub H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl Acad. Sci. USA. 1993;90:8028–8032. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemeth A.L., Medveczky P., Toth J., Siklodi E., Schlett K., Patthy A., Palkovits M., Ovadi J., Tokesi N., Nemeth P., et al. Unconventional translation initiation of human trypsinogen 4 at a CUG codon with an N-terminal leucine. A possible means to regulate gene expression. FEBS J. 2007;274:1610–1620. doi: 10.1111/j.1742-4658.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- 14.Pasumarthi K.B., Doble B.W., Kardami E., Cattini P.A. Over-expression of CUG- or AUG-initiated forms of basic fibroblast growth factor in cardiac myocytes results in similar effects on mitosis and protein synthesis but distinct nuclear morphologies. J. Mol. Cell. Cardiol. 1994;26:1045–1060. doi: 10.1006/jmcc.1994.1125. [DOI] [PubMed] [Google Scholar]
- 15.Schwab S.R., Shugart J.A., Horng T., Malarkannan S., Shastri N. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Eden M.E., Byrd M.P., Sherrill K.W., Lloyd R.E. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosnakovski D., Lamb S., Simsek T., Xu Z., Belayew A., Perlingeiro R., Kyba M. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp. Neurol. 2008 doi: 10.1016/j.expneurol.2008.07.022. Epub. [DOI] [PubMed] [Google Scholar]
- 19.Grewal S.I., Elgin S.C. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 22.Alexiadis V., Ballestas M.E., Sanchez C., Winokur S., Vedanarayanan V., Warren M., Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim. Biophys. Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., van der Maarel S.M., Hayashi Y., Flanigan K.M. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 24.Celegato B., Capitanio D., Pescatori M., Romualdi C., Pacchioni B., Cagnin S., Vigano A., Colantoni L., Begum S., Ricci E., et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 25.Yip D.J., Picketts D.J. Increasing D4Z4 repeat copy number compromises C2C12 myoblast differentiation. FEBS Lett. 2003;537:133–138. doi: 10.1016/s0014-5793(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 26.Chadwick B.P. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manz E., Schnieders F., Brechlin A.M., Schmidtke J. TSPY-related sequences represent a microheterogeneous gene family organized as constitutive elements in DYZ5 tandem repeat units on the human Y chromosome. Genomics. 1993;17:726–731. doi: 10.1006/geno.1993.1393. [DOI] [PubMed] [Google Scholar]
- 28.Okada T., Gondo Y., Goto J., Kanazawa I., Hadano S., Ikeda J.E. Unstable transmission of the RS447 human megasatellite tandem repetitive sequence that contains the USP17 deubiquitinating enzyme gene. Hum. Genet. 2002;110:302–313. doi: 10.1007/s00439-002-0698-2. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh Y., Miyamoto N., Okada T., Gondo Y., Showguchi-Miyata J., Hadano S., Ikeda J.E. The RS447 human megasatellite tandem repetitive sequence encodes a novel deubiquitinating enzyme with a functional promoter. Genomics. 2000;67:291–300. doi: 10.1006/geno.2000.6261. [DOI] [PubMed] [Google Scholar]
- 30.Booth H.A., Holland P.W. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. doi: 10.1016/j.gene.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 32.Maves L., Waskiewicz A.J., Paul B., Cao Y., Tyler A., Moens C.B., Tapscott S.J. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- 33.Stoneley M., Paulin F.E., Le Quesne J.P., Chappell S.A., Willis A.E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 34.Honda M., Kaneko S., Matsushita E., Kobayashi K., Abell G.A., Lemon S.M. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.