Abstract
Proper limb development requires concerted communication between cells within the developing limb bud. Several molecules have been identified which contribute to the formation of a circuitry loop consisting in large part of secreted proteins. The intracellular actin nucleator, Formin 1 (Fmn1), has previously been implicated in limb development, but questions remain after the identification of a Gremlin transcriptional enhancer within the 3′ end of the Fmn 1 locus. To resolve this issue, a knockout mouse devoid of Fmn1 protein was created and characterized. The mice exhibit a reduction of digit number to four, a deformed posterior metatarsal, phalangeal soft tissue fusion as well as the absence of a fibula to 100% penetrance in the FVB genetic background. Importantly, this mutant allele does not genetically disrupt the characterized Gremlin enhancer, and indeed Gremlin RNA expression is upregulated at the 35 somite stage of development. Our data reveal increased Bone Morphogenetic Protein (Bmp) activity in mice which carry a disruption in Fmn1, as evidenced by upregulation of Msx1 and a decrease in Fgf4 within the apical ectodermal ridge. Additionally, these studies show enhanced activity downstream of the Bmp receptor in cells where Fmn1 is perturbed, suggesting a role for Fmn1 in repression of Bmp signaling.
INTRODUCTION
Vertebrate limb development starts as an outgrowth of mesoderm surrounded by ectoderm from the lateral flanks of embryos. Key molecular signals contributing to limb development have been extensively studied and characterized in limb outgrowth and patterning. At the distal margin of limb buds lies a specialized region of ectoderm, the apical ectodermal ridge (AER), which coordinates the proliferation of cells within the underlying mesenchyme. Members of the Fgf family are secreted from the AER, and are thought to mediate a large part of AER activity. Genetic analysis of Fgf function in limb development has revealed a reduction in digit number for mice lacking Fgf8 limb expression (1,2), but not in mice with conditional inactivation of Fgf4 within limb buds (3,4). When both Fgf4 and Fgf8 were inactivated, a more severe phenotype was observed, resulting in a virtual absence of mature limbs (5,6). Limb patterning along the anterior–posterior axis requires communication between the AER and the zone of polarizing activity (ZPA). The ZPA is positioned within the distal posterior mesenchyme and produces the morphogen, Sonic hedgehog (Shh), which directs anterior–posterior patterning and digit identity. The maintenance of the AER is dependent on the Shh signal, thus creating a regulatory loop with the Fgf proteins (7,8). This was evidenced by the eventual loss of expression of Fgf4 and Fgf8 in Shh−/− embryos (9,10). The pathway leading from Shh back to Fgf is indirect and involves the regulation of Bmps, secreted proteins that repress expression of Fgfs in the AER. Shh can activate Bmp expression (7,11,12), as well as maintain expression of an antagonist of Bmp, Gremlin (9), thus leading to a dynamic control of Fgfs in the AER.
Bmps are multifunctional secreted factors that are expressed in the AER as well as in the mesenchyme of limb buds (reviewed in 13). Bmps bind the heterodimeric Bmp receptors (BmpR) to initiate phosphorylation of Smad1, 5 and 8, which subsequently form a complex with Smad4 and translocate into the nucleus leading to the transcriptional activation of its target genes (reviewed in 14). With the predominant expression of Bmp2, Bmp4 and Bmp7 within the limb bud, the dissection of their individual roles has been challenging. Experiments in which the Bmp inhibitor, Noggin, was misexpressed, led to the observation that Bmps negatively regulated Fgf expression and AER function (15). These results were extended by studies which selectively disrupted BmpR1a and established the requirement of Bmp signaling to specific dorsal-ventral patterning in the ectoderm (16). Conditional inactivation of Bmp4 within the mesoderm leads to an expansion of the AER and both Fgf4 and Fgf8 expression, resulting in an upregulation of Shh signal and a polydactyly phenotype (17). Bmp2 and Bmp4 double null mutant limbs are severely malformed, presumably due to a failure in bone condensation (18). Moreover, the double knock-out of BmpR1a and BmpR1b in cartilage showed an essential role for Bmp signaling in chondrogenesis (19).
The limb deformity (ld) locus was originally identified with the genetic characterization of several ld alleles which either spontaneously arose, or were generated by transgenic insertion. These mice display fusion of the zeugopod in both the forelimb and hindlimb, and syndactyly of the digits and metacarpal bones (20). Two such ld alleles, ldTgHd (created by the insertion of a transgene) and ldIn2 (created by a translocation/inversion), were shown to result in the alteration of formin-encoding transcripts (21), which led to the identification of Fmn1, and its multiple RNA transcripts (22). An additional ld allele, ldTgBri, was identified arising from an intronic disruption within the Fmn1 locus (23). The two trangene-induced alleles, ldTgHd and ldTgBri, produce similar 110 aa carboxy-terminal truncated Fmn1 proteins, while the ldIn2allele produces a less extensive carboxy-terminal truncation of 42 aa (24).
The formins consist of a large family of proteins that are expressed throughout eukaryotes (reviewed in 25–27). In mammalians, members of the formin family are differentially expressed in various tissues and cell types. A universal feature of the formin proteins is the presence of a formin homology 2 (FH2) domain. The FH2 domain of the yeast Formin, Bni1 (28,29), was originally shown to be sufficient for the nucleation of actin filaments at its barbed end, and subsequently FH2 domains derived from other formins have been shown to function similarly (30–32). In addition to the FH2 domain, the majority of formins possess a formin homology 1 (FH1) domain that is highly proline-rich and functions in recruiting profilin to the emerging actin filament (33–35).
Fmn1 is a large gene consisting of 24 exons and spanning approximately 400 kb within chromosome 2. Fmn1 transcripts have been detected in the developing kidney, and found within the AER and the mesenchyme of developing limb buds (36). Moreover, Fmn1 is expressed in tissues of human lung and kidney as well as various tumors, suggesting the possibility of involvement in human disorders (37). The ld phenotype was attributed to a disruption in signaling between the ZPA and the AER, and it was believed that the absence of Fmn1 contributed to these observations. The identification of Gremlin in this relay circuitry was demonstrated by its exogenous addition within the developing ld limb bud which resulted in the restoration of the Fgf/Shh loop (9). Gremlin acts as a secreted protein which can bind Bmp2 and Bmp4 to block its function (38). This led to a proposed pathway where Gremlin antagonizes Bmps, which act as repressors of AER structure and function. Interestingly, the genomic position of Gremlin is directly adjacent to Fmn1 in a tail-to-tail orientation (39). Generation of mutant alleles of Gremlin displayed a disruption of the Fgf/Shh feedback loop as well as the AER (40,41), which placed Gremlin downstream of Shh and upstream of Fgf. The further identification of Gremlin-affecting, cis-regulatory elements within Fmn1 highlighted the existence of a long-range transcriptional control region within the 3′ end of the Fmn1 (39). This was confirmed in a spontaneously arising deletion of the Fmn1 coding region (42).
In this report, we examined the role of Fmn1 in limb development by analyzing mice that fail to produce detectable Fmn1 proteins or any of the full-length splice variants of Fmn1. We show that Fmn1Pro/Pro mice display an oligodactyly phenotype. Further, our mutation led to a downregulation of Fgf4 and Shh, and upregulation of Gremlin expression at the E10.5 stage of development. These expression patterns are distinct from the previously characterized ld alleles which display downregulated expression of Fgf4, Shh and Gremlin. Further analysis suggests a model where in Fmn1 acts to repress Bmp signaling just downstream of the BmpR and, thus, affects the proper patterning of digit formation.
RESULTS
Disruption of the Fmn1 gene results in both kidney aplasia and limb development defects
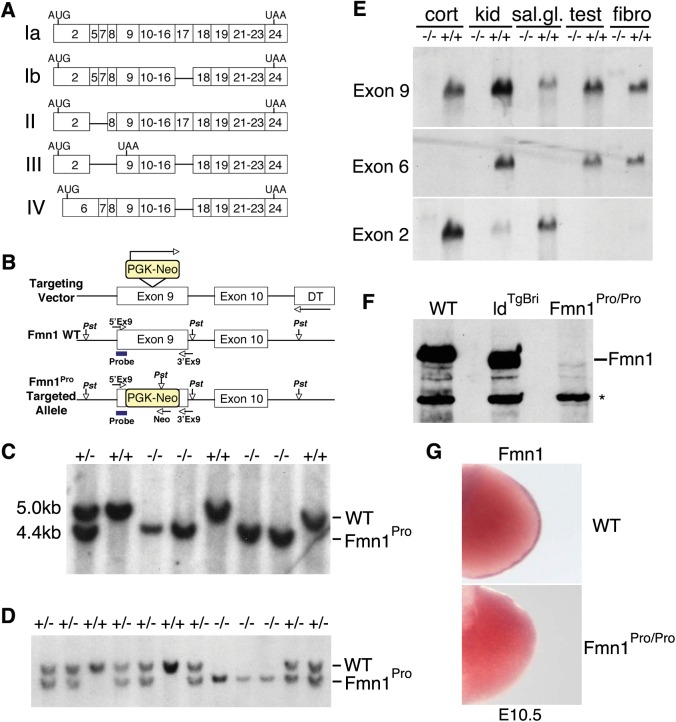
The Fmn1Pro allele, which represents a disruption at the proline-rich FH1 domain, was generated to assess the role for Fmn1 in development. The Fmn1 coding region leads to the expression of several splice variants (Fig. 1A). In this study, we sought to generate an allele of Fmn1 that disrupts all of its gene products by targeting exon 9, which is universally expressed among the various RNA splice products and encodes the FH1 domain (Fig. 1B). Correctly targeted ES-cell clones were used to generate Fmn1Pro mice. Mice were subsequently screened for the PGK-Neo targeted allele by Southern blot (Fig. 1C) and confirmed by PCR analysis to distinguish wild-type, heterozygous and mutant animals (Fig. 1D). Fmn1Pro/Pro mice were viable, fertile and the allele displayed Mendelian segregation. Analysis of tissue expression patterns was performed on polyadenylated RNA and subjected to northern blotting (Fig. 1E). Using a probe to exon 9 revealed broad tissue expression of the Fmn1 RNA isoforms derived from wild-type mice, while Fmn1Pro/Pro mice failed to express full-length transcripts in the tissues examined. Using probes to exons 6 and 2 further showed no detectable full-length mRNA in tissues of Fmn1Pro/Pro mice, suggesting that the disruption of the allele at exon 9 leads to the destabilization of full-length Fmn1 mRNAs generally. The RNA isoform containing sequences corresponding to exon 6 were enriched in kidney, testis and embryo fibroblasts. Exon 2 encoding transcripts by contrast were enriched in the cerebral cortex and salivary gland, with trace amounts in the kidney, and none detected in testis and embryo fibroblasts. Protein expression was analyzed by western blot with rabbit antiserum against Fmn1, and revealed no detectable full-length protein in embryo fibroblasts derived from Fmn1Pro/Promice (Fig. 1F). Interestingly, when embryo fibroblast protein extracts derived from ldTgBri were examined for Fmn1 expression (24,43), a band of near full-length size was detected at similar abundance to cells from wild-type mice, suggesting that Fmn1 is indeed expressed in these mice. An antibody cross-reacting background band is present in all samples, and has been detected in cell extracts known not to express Fmn1. Since the ldTgBri allele led to a disruption of the gremlin regulatory region of the ld locus, it is thought that perturbation of Gremlin expression is responsible for the limb phenotype (39). The data presented in this study show that ldTgBri embryo fibroblasts still express the Fmn1 proteins (Fig. 1F). Moreover, embryo fibroblasts derived from other ld alleles, such as ldTgHd and ldIn2 also express Fmn1 proteins, albeit slightly truncated versions (43). Examination of Fmn1 transcripts within the developing limb from Fmn1Pro/Pro mice displays disruption of its expression (Fig. 1G). The generation of the Fmn1Pro/Promice by insertion of a PGK-Neo cassette approximately 200 kb away from the characterized gremlin regulatory region allowed us to probe the role of the Fmn1 proteins in development.
Figure 1.
Generation and expression pattern characterization of Fmn1Pro/Pro mice. (A) Depiction of the major splice variants expressed from the Fmn1 allele are marked as Ia, Ib, II, III and IV. Boxes filled with numbers running from 2 to 24 indicate the exons expressed within each of the splice variants. AUG and UAA correspond to positions of the translational start and stop sites, respectively (schematic diagram adapted from 24). (B) Shown is the targeting scheme for disruption of exon 9 of Fmn1, by insertion of a PGK-Neo cassette (yellow box). WT and Fmn1Pro targeted alleles are shown. The positions of the PstI restriction enzyme sites are shown. The probe used for Southern blotting is positioned (blue line) at the 5′ end of exon 9. PCR primers used for genotype analysis are shown as 5′Ex9, 3′Ex9 and Neo with an arrow indicating the orientation of the primers. (C) Representative Southern blot analysis of tail DNA derived from Fmn1Pro allelic disruption mice, digested with PstI and hybridized to a probe derived from the 5′ end of exon 9. (D) Representative 3-primer PCR genotyping from tail DNA derived from Fmn1Pro allelic disruption mice. (E) Northern blots of polyadenylated-positive RNA derived from tissues of wild-type, +/+, or Fmn1Pro/Pro, −/−, mice. Northern blots were probed for expression of transcripts containing sequences derived from exons 9, 6 and 2, as indicted to the left of the blots. Abbreviations used: cerebral cortex, cort; kidney, kid; salivary gland, sal.gl.; testis, test; embryo fibroblasts, fibro. (F) Western blot of mouse embryo fibroblast protein extracts derived from wild-type, ldTgBri, and Fmn1Pro/Pro mice, probed with anti-Fmn1 rabbit antiserum. The full-length Fmn1 protein is indicated, and an unrelated cross-reactive band is labeled with *. (G) Expression pattern of Fmn1 in limb bud. Embryonic stage 10.5 wild-type and Fmn1Pro/Pro limb buds probed for RNA expression by in situ hybridization with a probe to Fmn1.
Examination of Fmn1Pro/Pro mice demonstrated defects in kidney development in the form of unilateral renal aplasia. Kidney phenotypes were observed to varying degrees of penetrance dependent on the genetic background of the mice, with Fmn1Pro/Pro mice on the 129/SvEv background displaying a 4.4% penetrance, whereas the FVB background gave a 50% penetrance (Table 1). The Fmn1Pro/Pro mice additionally exhibited reduced digit number to four in both the forelimb and hindlimb, with a differing degree of penetration dependent on the genetic background. The 129/SvEv Fmn1Pro/Pro mice displayed 66% penetrance, whereas the FVB background gave a 100% penetrance. Of the animals affected, all four limbs displayed a limb defect. The different degrees of genetic background-dependent penetrance suggest that additional genetic factors contribute to these phenotypes.
Table 1.
Genetic background of mice contributes to the phenotypic penetrance of the Fmn1Pro/Pro allele
| Genotype | Limb phenotype | No. of mutants/total | Kidney phenotype | No. of mutants/total |
|---|---|---|---|---|
| Fmn1Pro/+ | Normal | 0/88 (0%) | Normal | 0/88 (0%) |
| Fmn1Pro/Pro (129/SvEv) | Four digits | 90/136 (66%) | Renal aplasia | 6/136 (4.4%) |
| Fmn1Pro/Pro (FVB) | Four digits | 32/32 (100%) | Renal aplasia | 16/32 (50%) |
The table represents the number of animals examined that displayed a mutant phenotype as a percentage of the total, no. of mutants/total. Mice were examined for both a limb and kidney phenotype. The genotypes of the animals were heterozygous for Fmn1, Fmn1Pro/+, homozygous on the 129Sv/Ev background, Fmn1Pro/Pro (129Sv/Ev) or homozygous on the FVB background, Fmn1Pro/Pro (FVB).
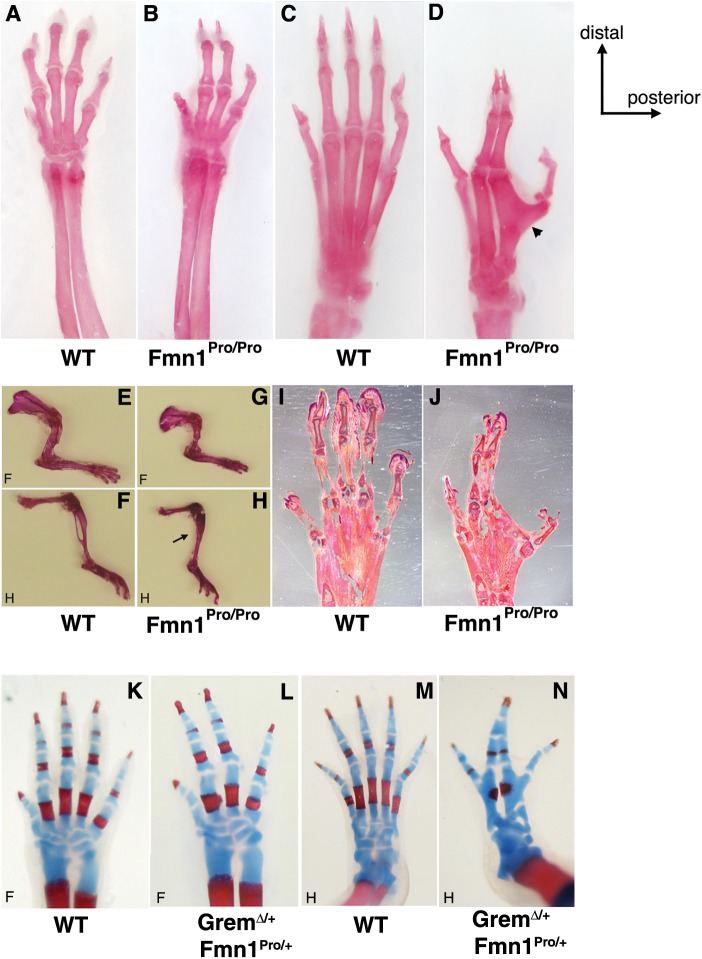
Examination of the skeletal structures of limbs showed differentiation of the radius and ulna in the forelimb (Fig. 2B), whereas the hindlimb zeugopod consisted of only one element in the Fmn1Pro/Pro mice, with the fibula absent (Fig. 2H). The autopods displayed defects with the loss of a digit (Fig. 2B and D) and soft tissue fusion of the two middle digits (Fig. 2J). Hindlimb autopods presented with a metatarsal fusion of the two posterior digits (Fig. 2D). This limb phenotype can be distinguished from that of the Gremlin-null mice which display extreme shortening of all limb elements, fusion of the two bones of the zeugopod for both the forelimb and hindlimb, and general reduction in digit size and number (40,41).
Figure 2.
Limb phenotype of Fmn1Pro/Pro mice. Alizarin red stained adult wild-type and Fmn1Pro/Pro mouse fore and hindlimbs. (A) Wild-type forelimb, (B) Fmn1Pro/Pro forelimb, (C) wild-type hindlimb and (D) Fmn1Pro/Pro hindlimb. Fmn1Pro/Pro limbs possessed only four digits and a defined radius and ulna in the forelimb. The arrowhead indicates the fused posterior metatarsal bones of the Fmn1Pro/Pro hindlimb. Alizarin red stained adult (E and F) wild-type and (G and H) Fmn1Pro/Pro mouse forelimbs and hindlimbs. The arrow indicates the absence of the fibula in the Fmn1Pro/Pro hindlimb. Sectioned hematoxylin and eosin stained hindlimb autopods of (I) wild-type and (J) Fmn1Pro/Pro mice. Alcian blue and alizarin red stained neo-natal wild-type and GremΔ/+; Fmn1Pro/+ mouse fore and hindlimbs. (K) Wild-type forelimb, (L) GremΔ/+; Fmn1Pro/+ forelimb, (M) wild-type hindlimb, (N) GremΔ/+; Fmn1Pro/+ hindlimb. Abbreviations used: forelimb, F; hindlimb, H.
To view possible complementation effects of our Fmn1 mutation on Gremlin, we generated a compound heterozygote harboring the Fmn1Pro mutation on one allele and GremΔ mutation on the other allele (39). Interestingly, although neither the GremΔ /+ heterozygote nor the Fmn1Pro/+ heterozygote display a limb phenotype (data not shown), the compound heterozygote does. Shown are both the forelimbs and hindlimbs from wild-type controls and compound heterozygotes (GremΔ/+; Fmn1Pro/+), which resemble the Fmn1Pro/Pro phenotype (Fig. 2K–N). The limbs did not display a GremΔ/Δ phenotype with extreme reduction in digit size and number of digits to three.
Fmn1 regulates expression of the Fgf/Shh loop in the developing limb bud
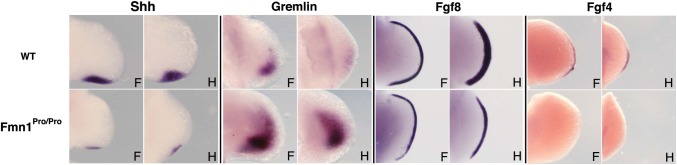
To investigate the possible molecular controls of limb development, we examined the expression of the Fgf/Shh loop components. Given the limb phenotype observed in Fmn1Pro/Pro mice, it might have been anticipated that some or all of the members of the AER loop might be downregulated. In situ hybridization was performed on embryonic stage 10.5 mice for Shh, Gremlin, Fgf4 and Fgf8 (Fig. 3). Reduced Shh expression was witnessed in both the forelimb and hindlimb of Fmn1Pro/Pro mice. The Fmn1Pro/Pro mice possessed an intact AER as evidenced by the pattern of expression of Fgf8, which contrasts with the disruption of the AER structure in Gremlin null mice (40,41), Fgf4 on the other hand, was not detectable in the AER of Fmn1Pro/Pro mice, which is consistent with the notion that Shh secreted from the ZPA reinforces AER activity. Somewhat surprising is the finding that Gremlin RNA was significantly upregulated in the Fmn1Pro/Pro limb buds (Fig. 3). The downregulation of Shh would normally lead to a reduced expression of Gremlin RNA. In contrast, the Fmn1Pro/Pro mice appear to have interfered with this regulation. With the induction of Gremlin, these data further suggest that the Fmn1Pro allele has not negatively disrupted the gremlin regulatory region located within the ld locus.
Figure 3.
Expression pattern of Fgf/Shh loop. Embryonic stage 10.5 wild-type and Fmn1Pro/Pro limb buds probed for RNA expression by in situ hybridization. Forelimbs and hindlimbs were probed for sonic hedgehog, Shh, Gremlin, Fgf8 and Fgf4 as indicated. Abbreviations used: forelimb, F; hindlimb, H.
Upregulation of Bmp activity in Fmn1Pro/Pro limb buds, with modest change in RNA expression
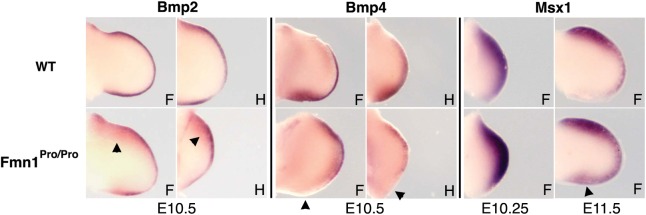
Molecular components that have been shown to upregulate Gremlin expression include the Bmps (12). Bmp expression in the AER is involved in limb outgrowth and patterning by antagonizing Fgf signaling (15,44). We examined the levels of Bmp2 and Bmp4 expression in the context of Fmn1Pro/Pro mice, both of which displayed comparable levels of RNA signal within the AER with evidence of increased levels of Bmp2 expression in the underlying mesoderm (Fig. 4; as indicated by the arrowheads). Of interest were the reduced levels of Bmp4 expression within the posterior mesenchyme (Fig. 4; as indicated by the arrowheads). It remains unclear why the levels of Bmp4 within the AER remain normal, whereas the signal within the ZPA is reduced. This may be due to the reduction in Shh and its inability to induce Bmp locally (12). To further examine the extent of Bmp activity, the levels of Msx1 were probed by in situ hybridization (Fig. 4). Consistent with the upregulation of Gremlin, an additional target of Bmp activity, Msx1 was also upregulated in the Fmn1Pro/Pro limb buds, suggesting increased Bmp protein activity.
Figure 4.
Expression patterns of Bmps and Msx1. Wild-type and Fmn1Pro/Pro limb buds probed for RNA expression of Bmp2, Bmp4 and Msx1 by in situ hybridization. Arrowheads indicate increased expression of Bmp2 in the underlying mesoderm of the AER, the absence of expression of Bmp4 within the posterior mesenchyme and the expanded expression of Msx1 in the posterior mesenchyme. Embryonic stages are indicated below image panels. Abbreviations used: forelimb, F; hindlimb, H.
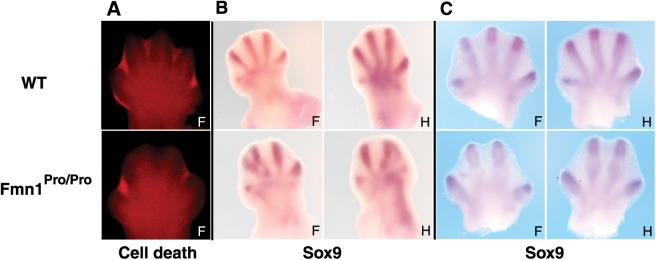
Bmp has pleiotrophic effects on limb development including the repression of AER expansion (15,17) and the promotion of osteogenesis (18). The phenotype observed in the Fmn1Pro/Pro mice is presented as a reduction in digit number to four (Fig. 2B and D), as well as a soft tissue fusion between the middle digits (Fig. 2J). Interestingly, the middle two digits are aligned in parallel and not arrayed outward. To address whether cell death played a role in this phenotype, we assessed the extent of Lysotracker Red staining in E13.5 limb buds (Fig. 5A). Interdigital cell death was evident between all of the digits in the wild-type. The Fmn1Pro/Pro limb buds also displayed interdigital Lysotracker Red staining, however not between the two middle digits, which may have contributed to the soft tissue fusion of the middle digits.
Figure 5.
Patterning defects in limbs of Fmn1Pro/Pro mice. (A) Analysis of cell death by Lysotracker Red staining in the developing limb of wild-type and Fmn1Pro/Pro mice at embryonic stage 13.5. Analysis of chondrogenesis was performed by probing embryonic stage (B) 12.5, and (C) 13.5 limb buds with the cartilage marker, Sox9. Abbreviations used: forelimb, F; hindlimb, H.
To understand the reduction in digit number, we investigated the placement of cartilage within the hand and foot plates in the Fmn1Pro/Pro limbs. This was addressed by probing for the RNA expression of Sox9, a marker for cartilage, at the E12.5 and E13.5 stages. A clear alteration in patterning of condrogenesis was evident in limbs lacking Fmn1 (Fig. 5B and C). Sox9 detected primordia of possibly five digits in the forelimb, with digits 2 and 3 seeming to fuse. Placement of five digits in the hindlimb was not evident, with more prominent defects in the posterior digits. Additionally, at E12.5 cartilage placement proximal to the digits was evident in patterns not witnessed in the wild-type limbs, with noted intense expression in the posterior region of the hindlimb (Fig. 5B). Examination of Sox9 expression patterns at E13.5 demonstrated patterning of four digits only for both of forelimb and hindlimb of the Fmn1Pro/Pro mouse, with an absence of additional proximal cartilage staining.
Fmn1 repression of Smad phosphorylation through the Bmp receptor
The reduction in Fgf RNAs within the AER and the increase in both Gremlin and Msx1 suggested that Bmp activity had increased within the context of Fmn1Pro/Pro limb buds. Since the Bmp signals were comparable within the AER, it was of interest to test whether signaling downstream of the BmpR was altered. As in situ hybridization assesses the levels and localization of RNA, an alternative approach was employed to examine the levels of Smad phosphylation, a direct downstream effect of induction of Bmp signaling from the BmpR (14). Embryo fibroblasts derived from wild-type and Fmn1Pro/Pro mice were cultured overnight under serum starvation conditions followed by a 10 min exposure to increasing levels of Bmp4 (Fig. 6A; 0, 1.5, 6.25, 25 and 100 ng/ml; denoted by the solid inclining triangle). A significant increase in the phosphorylation of Smad levels was observed in the Fmn1Pro/Pro cells, as determined by the immunoreactivity of protein extracts with an antibody specific to phospho-Smad1, 5 and 8. To confirm this finding, 293 cells were transfected with a dominant negative construct encoding the coiled-coil domain and part of the FH1 domain of Fmn1, similar to that previously used (45). In these experiments, transfected 293 cells were serum-starved and induced with Bmp4 in a similar manner as performed with mouse embryo fibroblasts. In cells that received the Fmn1 fragment, Smad phosphorylation was significantly induced compared with the vector only transfected cells (Fig. 6B). For these Bmp induction experiments, unphophorylated Smad levels were additionally assessed. These experiments bypass the potential regulatory influences of Gremlin to antagonize Bmps by not allowing its accumulation within the tissue culture media. Several intracellular regulators of Smad signaling have been identified which guide processes such as receptor interaction, regulation of phosphorylation, ubiquitin ligation and nucleocytoplasmic shuttling (14,46). Unlike the members of the Fgf/Shh loop, which are secreted proteins, Fmn1 is found within the cytoplasm of cells to regulate the cytoskeleton (47,48). This suggests that Fmn1 may be acting as a repressor of Smad phosphorylation through its role in coordination of the cytoskeleton.
Figure 6.
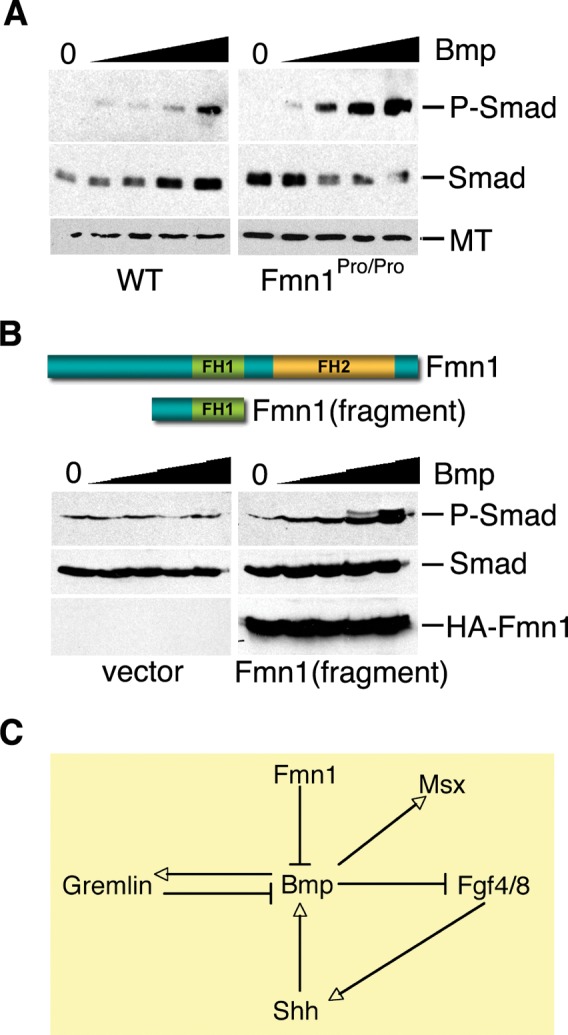
Fmn1 represses Smad phosphorylation through Bmp signaling. (A) Embryo fibroblasts derived from wild-type and Fmn1Pro/Pro mice were induced with increasing concentrations of Bmp4, indicated by solid inclining triangle. Smad1, 5, 8 phosphorylation, P-Smad as a response to Bmp signaling, was assessed by western blot analysis. Cell extracts were additionally blotted with antibodies to unphosphorylated Smad and α-tubulin (MT), lower panels. (B) Two hundred and ninety-three cells transfected with an HA-tagged fragment of Fmn1 or an empty vector were induced with increasing concentrations of Bmp4. P-Smad, Smad and HA-Fmn1 were analyzed by western blot. (C) Depiction of the role of Fmn1 in regulation of Bmp. This model indicated that Fmn1 acts as a repressor of Bmp signaling. This in turn leads to an increase in Msx expression, a decrease in Fgf4/8 and consequently, a decrease in Shh. Additionally, this explains the upregulation of Gremlin that was observed in the developing limb bud. Gremlin acts as an antagonist of Bmp which might lead to subsequent downregulation of Bmp activity.
DISCUSSION
Using a genetic approach to remove all of the potential Fmn1 activity resulting from the various full-length splice forms of Fmn1, we find that Fmn1 is important for proper limb patterning. Fmn1, as an intracellular protein that regulates the cytoskeleton, is shown to affect the expression patterns of the Fgf/Shh loop. We showed that RNAs of Gremlin were upregulated and Fgf4 were downregulated in the context of the Fmn1Pro/Pro mice. Since the Bmps have been shown to both induce Gremlin expression and act as a repressor of Fgf4, we decided to examine the role of Fmn1 on Bmp regulation. Our data further revealed an upregulation of the Bmp signaling target, Msx1, which is indicative of increased activity of Bmp. More directly, we provide evidence that Fmn1 is acting downstream of the BmpR to repress Smad signaling.
Gremlin and Fmn1 relationship
Both Gremlin and Fmn1 have previously been implicated in contributing to limb patterning, but recent evidence has raised some questions regarding the role of Fmn1 (39). These authors identified a cis-regulatory region that contributed to Gremlin expression and that overlapped with several ld alleles (ldTgBri, ldTgHd and ldIn2). All three of these ld alleles were found to be genetic disruptions within the 3′ end of the Fmn1 locus. This argues the ldTgBri, ldTgHd and ldIn2alleles led to disruptions in a Gremlin cis-regulatory region. However, it was noted that ldTgBri, ldTgHd and ldIn2 mice do not lead to general disruption in Fmn1 expression in tissues derived from whole embryos (43). Therefore, these ld mice could not address the functional role of Fmn1. The disruption of a single isoform IV of Fmn1, ldGKO, has been described and failed to produce a limb defect (49). Mice with deletions of Fmn1 exons 4 or 5, and exon 10 similarly failed to produce a limb defect, possibly due to an incomplete disruption of all Fmn1 isoforms (39,50). In contrast, the rather large deletion of exons 10 through 24 of Fmn1 produced a limb phenotype similar to that observed in mice carrying the ldTgBri, ldTgHd and ldIn2alleles, and could be attributed to a disruption in Gremlin expression (39). The relationship between the mutation generated for this study, Fmn1Pro, and the GremΔ allele was addressed through complementation experiments by generating compound heterozygous mice (Fig. 2K–N). These mice carry the Fmn1Pro mutation on one allele and the GremΔ mutation on the other, and display a similar phenotype to the Fmn1Pro/Pro mice. A plausible explanation leading to this limb phenotype could involve the Bmp pathway. We have shown that Fmn1 represses the phosphorylation of Smad, a downstream target of the Bmp-R (Fig. 6). Further, it has been shown that Gremlin directly binds Bmp2 and Bmp4 to inhibit its function (38). Both Fmn1 and Gremlin biochemically converge on the Bmp pathway, thus the combined effect in the compound heterozygote could lead to the limb phenotype observed. Although a likely explanation, others are possible. For example, the Fmn1Pro mutation, which is the result of a PGK-Neo insertion, may have disrupted a distant regulatory element within Fmn1, effecting expression of genes within this locus. Although a PGK promoter directed transcript of the 3′ end of Fmn1 could possibly be generated, no specific Fmn1-related protein product was detected (Fig. 1F). The Fmn1Pro/Pro mouse generated in this study fails to produce any full-length Fmn1 splice variants or functional protein, thus allowing us to address the involvement of Fmn1 in limb development.
The genomic position of Gremin and Fmn1 on chromosome 2, exist as neighboring genes in a tail-to-tail orientation that is evolutionarily conserved. A similar orientation was observed for Gremlin2 and Fmn2 on chromosome 1 (51), which suggests genomic duplication of this cassette and raises the notion of an interrelationship between these genes. In addition to the cis-regulatory region in the ld locus which affects the expression of Gremlin, both gene products act as negative regulators of Bmp.
Fmn1 and regulation of the Fgf/Shh loop
Expression of Fmn1 begins early in limb bud development and its disruption influences gene expression of Fgf/Shh loop components. Mice harboring Fgf8 mutant alleles result in a delayed expression of Shh (1), and normal levels of Shh at E10.5 with slightly diminished expression by E11.5 (2). Genetic analysis of Fgf4 mouse mutants failed to display limb development defects (3,4). However, the disruption of both Fgf4 and Fgf8 in the AER produced virtual no expression of Shh (6). In the Fmn1Pro/Pro mice, Fgf4 is not detected and the expression of Fgf8 is normal with only slight reduction in the hindlimb. Although this may contribute to a reduction in Shh, the degree to which it is downregulated in the Fmn1Pro/Pro mice cannot be fully explained through Fgf4 and Fgf8 expression. Nevertheless, the decreased Shh in the Fmn1Pro/Pro mice most likely accounts for some of the disrupted patterning of digit formation.
One of the more surprising findings of this study is the significant upregulation of Gremlin in the Fmn1Pro/Pro mice. With a reduction in Fgf4 and Shh, it would be anticipated that Gremlin expression should be lower. Shh-null limb buds receiving exogenous grafts producing Shh respond with the induction of Gremlin expression (11). However, in mice double null for Fgf4 and Fgf8, Gremlin expression is still detected even though these limb buds are dramatically reduced in size (5). Moreover, Gremlin expression appears to be upregulated in the presence of the FGF signaling inhibitor, SU5402, while Shh expression is undetected (39). Collectively, these studies suggest a more complex control mechanism from Shh to the expression of Gremlin. A recent report using Bmp2 soaked beads showed a Bmp concentration-dependent regulation of Gremlin (12). In fact, this study showed that Bmp2 is sufficient to activate Gremlin in the context of the chick ozd mutant limb, suggesting that this regulation may take place in a Shh-independent manner. This raises the possibility that alteration of the control mechanisms for Bmp activity in the Fmn1Pro/Pro mice led to the Shh-independent regulation of Gremlin expression levels (Fig. 6C). The increase in Gremlin levels could subsequently antagonize Bmp activity. In a recent study, the feedback loop between Bmp4 and the transcriptional activation of Gremlin was shown to be relatively fast-acting and provided of an interlinking regulatory mechanism for the Fgf/Shh loop (52).
Fmn1 repression of Bmp signaling
It has been shown that inhibition of Bmp activity from within the AER expands the Fgf8 expression patterning (53), and that genetic disruption of Bmp4 in the underlying mesoderm upregulates and expands Fgf4 levels within the AER (17). Therefore, regulation of Bmp activity within and adjacent to the AER can contribute to the expression of Fgf. The decreased expression of Fgf4 observed in E10.5 Fmn1Pro/Pro limb buds suggested an increased activity of Bmp. This observation was extended with the upregulation of Msx1 and expansion of its presence within the posterior region of the limb bud at E11.5. Furthermore, disrupted placement of limb autopod cartilage and reduction in digit formation suggest altered Bmp regulation. Based on these results, we could infer that Bmp proteins are either more active, or that the cells expressing the BmpR are more sensitive to the presence of Bmp. As the formins are intracellular proteins that regulate cytoskeleton and contribute to cell polarization events, they would not be directly involved in the regulation of gene expression (27). Rather Fmn1 might influence the localization of factors that could contribute to cell signaling and consequently affect gene expression. Therefore, we favor the notion that Fmn1Pro/Pro cells are more sensitive to the presence of Bmp. The upregulation of Gremlin could temporally affect downstream targets such as Bmp activity, which would setup a dynamic regulation of Bmp activity (Fig. 6C). Within the context of the limb bud, Gremlin as a secreted protein has the potential to affect cells away from the mesenchyme where it is produced. By contrast, Fmn1 must act intracellularly. The dual Bmp repressive actions of Fmn1 and Gremlin could establish respective spatial control of Bmp activity.
Our cell-based studies to assess the sensitivity of Bmp mediated signaling suggest a repressive role for Fmn1 downstream of the BmpR. The intriguing observation that upon cell stimulation, Smad proteins move away from their intracellular association with microtubules suggests a level of Smad regulation involving cytoskeleton (54). The Fmn1 proteins, which function as actin nucleators, also have an association with microtubules (47,48). Based on evidence from this study, Fmn1 may limit Smad phosphorylation by sequestering it in association with cytoskeleton components, thus providing a framework for understanding the mechanistic role of Fmn1 in limb development. It was previously shown that Gremlin also negatively regulates the BMP pathway (38). Thus, the combined effect of Gremlin and Fmn1 mutations on the BMP pathway in the compound heterozygote could lead to the observed limb phenotype. This could come about because the Fmn1 locus affects limb development in two ways, first, it harbors a control element that regulates the transcription of the secreted protein Gremlin (39), and second, because it codes for the cytoplasmic protein Fmn1, which plays a role in the repression of Bmp signaling.
MATERIALS AND METHODS
Generation of Fmn1proallelic mice
A targeting vector was constructed by inserting a PGK-Neo cassette within exon 9 of Fmn1. Homologous recombination events at the endogenous Fmn1 locus were confirmed by Southern blotting analysis using PstI digestion of genomic DNA. Hybridization with an internal probe gave a 5 kb band for WT and a 4.4 kb band for the Fmn1Pro allele. ES clone no. 2–81 was microinjected into blastocytes of C57BL/6J to generate chimeric mice. Germline offspring was identified by Southern blot analysis after breeding chimeric mice with C57BL/6J or 129/SvEv wild-type mice. Genotypes of the offspring were confirmed by Southern blot and PCR analysis. PCR was performed using standard methods with primers for exon 9 of Fmn1: 5′ GCTCAGTTTGAACTCAAGAC and 5′ AAAGGTCCAAGTCCAGGTG, and a primer for PGK-Neo: 5′ TTCGTCCAGATCATCCTGATC. Poly-A enriched RNA derived from various mouse tissues was analyzed by northern blot using probes directed against exons 9, 6 or 2. Protein expression was analysed by western blot of embryo fibroblast extracts derived from wild-type, ldTgBri, and Fmn1Pro/Pro, and reacted with an anti-Fmn1 rabbit anti-serum (43).
Western blot
Embryo fibroblasts were prepared from mice as previously described (47). Embryo fibroblasts and 293 cells were maintained at 37°C in Dulbecco’s Modified Eagle’s Medium (Gibco-BRL), supplemented with 10% fetal calf serum (FBS) and penicillin–streptomycin. Transfections were performed using Fugene6 according to the manufacturer’s protocol (Roche). Embryo fibroblasts and 293 cells were grown in the presence of 0.5% FBS overnight followed by a 10 min incubation with Bmp4 (R&D Systems) at 0, 1.5, 6.25, 25 and 100 ng/ml. Total cell extracts were collected in ice cold PBS and analyzed by western blot with antibodies against Smad5 (Abcam), phosphorylated Smad1, 5, 8 (Cell Signaling), α-tubulin (Abcam) and HA (Covance).
Skeletal analysis
Adult (6 weeks) and neo-natal mice were euthanized by carbon dioxide. Viscera and skin were removed and mice were placed in 90% ethanol for 5 days, followed by 100% acetone for 2 days. Animals were stained with 0.015% Alcian Blue (Sigma) and 0.005% Alizarin Red S (Sigma) for 3 days at 37°C in a mixture of 70% ethanol/5% glacial acetic acid. Adult animals were stained in the absence of Alcian Blue. After staining, skeletons were rehydrated by sequential immersion in 70% ethanol, 40% ethanol, 5% ethanol and 100% H2O, each for 2 h. Destaining was performed by sequential immersion in 1% KOH solutions containing 20% glycerol, 50% glycerol, 80% glycerol, followed by storage in 100% glycerol. Hematoxylin and eosin stain was performed using standard protocols on sectioned limb autopods.
Lysotracker red staining of limb buds
Cell death was detected in whole limb buds by staining live embryos with 250 nM Lysotracker Red (Invitrogen) in Dulbecco’s Modified Eagle’s Medium (Gibco-BRL) at 37°C for 30 min. Embryos were destained in three 5 min washes with PBS, and fixed in 4% parformaldehyde overnight. Following fixation, embryos were methanol dehydrated and limb buds were imaged by microscopy (55).
In situ hybridization of limb buds
In situ hybridization was performed on 4% parformaldehyde fixed and methanol dehydrated whole-mount E10.25–E13.5 embryos as described (36). Staging of embryos was further verified by counting somites (E10.25–E10.5 staged at 30–35 pairs of somites; E11.0–E11.5 staged at 40–45 pairs of somites). All AER staining in this study was done using detergent permeabilization, while Protease K (5 mg/ml) for 10 min was used for staining limb mesoderm. Embryos were hybridized with probes at 70°C overnight, followed by three washes in W1 (50% formamide, 5 × SSC pH 4.5, 1% SDS) at 70°C for 30 min each, three washes in W2 (50% formamide, 5 × SSC pH 4.5) at 65°C for 30 min each and three washes in TBST (0.05 M Tris/HCl, 0.30 M NaCl, 0.1% Tween 20, pH 7.6). Embryos were blocked in 10% sheep serum/TBST at RT for 60 min. Hybridization signal was determined by reaction of bound probes with alkaline phophatase-conjugated digoxigenin antibodies (1:1000; Roche) in 1% blocking buffer and incubated at 4°C overnight. Colorimetric development was performed with NBT/BCIP (Roche). Probes to Fgf4, Fgf8, Shh, Bmp2, Bmp4 and Msx1 were gifts from the Clifford Tabin lab. The probe directed against Gremlin was generated by PCR amplification of a 1 kb fragment of cDNA from mouse tissues. The probe directed against Fmn1 was previously described (36). All probes were verified by sequencing. All in situ hybridization experiments presented were performed at least three times, with the Gremlin results verified in five separate experiments.
FUNDING
This work was supported in part by funding from the Howard Hughes Medical Institute (F.Z., P.L. and M.D.); Swiss National Science Foundation grant (3100A0-112607) and the University of both cantons Basel to A.Z.
ACKNOWLEDGEMENTS
We would like to thank Cliff Tabin for helpful discussions, and Aya Leder, Richard Maas, Volney Sheen and Rolf Zeller for critically reading the manuscript. We are grateful to Cliff Tabin and members of the Tabin lab for generously sharing probes for the in situ hybridization experiments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lewandoski M., Sun X., Martin G.R. Fgf8 signaling from the AER is essential for normal limb development. Nat. Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 2.Moon A.M., Capecchi M.R. Fgf8 is required for outgrowth and patterning of the limbs. Nat. Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moon A.M., Boulet A.M., Capecchi M.R. Normal limb development in conditional mutants of Fgf4. Development. 2000;127:989–996. doi: 10.1242/dev.127.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X., Lewandoski M., Meyers E.N., Liu Y.-H., Maxson R.E., Martin G.R. Conditional inactivation of Fgf4 reveals complexity of signaling during limb bud development. Nat. Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- 5.Sun X., Mariani F.V., Martin G.R. Functions of FGF signaling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- 6.Boulet A.M., Moon A.M., Arenkiel B.R., Capecchi M.R. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev. Biol. 2004;273:361–372. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Laufer E., Nelson C.E., Johnson R.L., Morgan B.A., Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 8.Niswander L., Jeffrey S., Martin G.R., Tickle C. A positive feed back loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- 9.Zuniga A., Haramis A.-P.G., McMahon A., Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 10.Kraus P., Fraidenraich D., Loomis C.A. Some distal limb structures develop in mice lacking sonic hedgehog signaling. Mech. Dev. 2001;100:45–58. doi: 10.1016/s0925-4773(00)00492-5. [DOI] [PubMed] [Google Scholar]
- 11.Panman L., Galli A., Lagarde N., Michos O., Soete G., Zuniga A., Zeller R. Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signaling. Development. 2006;13:3419–3428. doi: 10.1242/dev.02529. [DOI] [PubMed] [Google Scholar]
- 12.Nissim S., Hasso S.M., Fallon J.F., Tabin C.J. Regulation of Gremlin expression in the posterior limb bud. Dev. Biol. 2006;299:12–21. doi: 10.1016/j.ydbio.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Robert B. Bone morphogenetic protein signaling in limb outgrowth and patterning. Dev. Growth Differ. 2007;49:455–468. doi: 10.1111/j.1440-169X.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- 14.Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 15.Pizette S., Niswander L. BMPs negatively regulate structure and function of the limb apical ectodermal ridge. Development. 1999;126:883–894. doi: 10.1242/dev.126.5.883. [DOI] [PubMed] [Google Scholar]
- 16.Ahn K., Mishina Y., Hanks M.C., Behringer R.R., Crenshaw E.B., 3rd BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- 17.Selever J., Liu W., Lu M.-F., Behringer R.R., Martin J.F. Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev. Biol. 2004;276:268–279. doi: 10.1016/j.ydbio.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4 and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:2116–2130. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon B.S., Ovchinnikov D.A., Yoshii I., Mishina Y., Behringer R.R., Lyons K.M. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl Acad. Sci. USA. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeller R., Haramis A.G., Zuniga A., McGuigan C., Dono R., Davidson G., Chabanis S., Gibson T. Formin defines a large family of morphoregulatory genes and functions in establishment of the polarizing region. Cell Tissue Res. 1999;296:85–93. doi: 10.1007/s004410051269. [DOI] [PubMed] [Google Scholar]
- 21.Maas R.L., Zeller R., Woychik R.P., Vogt T.F., Leder P. Disruption of formin encoding transcripts in two mutant limb deformity alleles. Nature. 1990;346:853–855. doi: 10.1038/346853a0. [DOI] [PubMed] [Google Scholar]
- 22.Woychik R.P., Maas R.L., Zeller R., Vogt T.F., Leder P. ‘Formins’: proteins deduced from the alternative transcripts of the limb deformity gene. Nature. 1990;346:850–853. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- 23.Vogt T.F., Jackson-Grusby L., Wynshaw-Boris A.J., Chan D.C., Leder P. The same genomic region is disrupted in two transgene-induced limb deformity alleles. Mamm. Genome. 1992;3:431–437. doi: 10.1007/BF00356152. [DOI] [PubMed] [Google Scholar]
- 24.Wang C.C., Chan D.C., Leder P. The mouse formin (Fmn) gene: genomic structure, novel exons, and genetic mapping. Genomics. 1997;39:303–311. doi: 10.1006/geno.1996.4519. [DOI] [PubMed] [Google Scholar]
- 25.Wallar B.J., Alberts A.S. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 26.Zigmond S.H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Faix J., Grosse R. Staying in shape with Formins. Dev. Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. Role of Formins in Actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 29.Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 30.Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F., Higgs H.N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 32.Moseley J.B., Sagot I., Manning A.L., Xu Y., Eck M.J., Pellman D., Goode B.L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier F. Formin is a processive motor that requires Profilin to accelerate Actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 34.Kovar D.R., Pollard T.D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl Acad. Sci. USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovar D.R., Harris E.E., Mahaffy R., Higgs H.N., Pollard T.D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 36.Chan D.C., Wynshaw-Boris A., Leder P. Formin isoforms are differentially expressed in the mouse embryo and are required for normal expression of fgf-4 and shh in the limb bud. Development. 1995;121:3151–3162. doi: 10.1242/dev.121.10.3151. [DOI] [PubMed] [Google Scholar]
- 37.Maas R.L., Jepeal L.I., Elfering S.L., Holcombe R.F., Morton C.C., Eddy R.L., Byers M.G., Shows T.B., Leder P. A human gene homologous to the formin gene residing at the murine limb deformity locus: chromosomal location and RFLPs. Am. J. Hum. Genet. 1991;48:687–695. [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu D.R., Economides A.N., Wang X., Eimon P.M., Harland R.M. The Xenopus dorsalizing factor gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell. 1998;1:673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 39.Zuniga A., Michos O., Spitz F., Haramis A.-P.G., Panman L., Galli A., Vintersten K., Klasen C., Mansfield W., Kuc S., et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Gene Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khokha M.K., Hsu D., Brunet L.J., Dionne M.S., Harland R.M. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- 41.Michos O., Panman L., Vintersten K., Beier K., Zeller R., Zuniga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- 42.Pavel E., Zhao W., Powell K.A., Weinstein M., Kirschner L.S. Analysis of a new allele of limb deformity (ld) reveals tissue- and age-specific transcriptional effects of the Ld Global Control Region. Int. J. Dev. Biol. 2007;51:273–281. doi: 10.1387/ijdb.062249ep. [DOI] [PubMed] [Google Scholar]
- 43.Chan D.C., Leder P. Genetic evidence that formins function within the nucleus. J. Biol. Chem. 1996;271:23472–23477. doi: 10.1074/jbc.271.38.23472. [DOI] [PubMed] [Google Scholar]
- 44.Yoon B.S., Pogue R., Ovchinnikov D.A., Yoshii I., Mishina Y., Behringer R.R., Lyons K.M. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133:4667–4678. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 45.Kobielak A., Pasolli H.A., Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canalis E., Economides A.N., Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr. Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 47.Zhou F., Leder P., Martin S.S. Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp. Cell Res. 2006;312:1119–1126. doi: 10.1016/j.yexcr.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 48.Dettenhofer M., Zhou F., Leder P. Formin1-isoform IV deficient cells exhibit defects in cell spreading and focal adhesion formation. PLoS ONE. 2008;3:e2497. doi: 10.1371/journal.pone.0002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wynshaw-Boris A., Ryan G., Deng C.-X., Chan D.C., Jackson-Grusby L., Larson D., Dunmore J.H., Leder P. The role of a single Formin isoform in the limb and renal phenotypes of Limb Deformity. Mol. Med. 1997;3:372–384. [PMC free article] [PubMed] [Google Scholar]
- 50.Chao C.W., Chan D.C., Kuo A., Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 51.Zeller R., Zuniga A. Shh and Gremlin1 chromosomal landscapes in development and disease. Curr. Opin. Genet. Dev. 2007;17:428–434. doi: 10.1016/j.gde.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Benazet J.-D., Bischofberger M., Tiecke E., Goncalves A., Martin J.F., Zuniga A., Naef F., Zeller R. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- 53.Wang C.-K.L., Omi M., Ferrari D., Cheng H.-C., Lizarraga G., Chin H.-J., Upholt W.B., Dealy C.N., Kosher R.A. Function of BMPs in the apical ectoderm of the developing mouse limb. Dev. Biol. 2004;269:109–122. doi: 10.1016/j.ydbio.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Dong C., Li Z., Alvarez R., Jr, Fen X.-H., Goldschmidt-Clermont P.J. Microtubule binding to Smads may regulate TGFβ activity. Mol. Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 55.Pajni-Underwood S., Wilson C.P., Elder C., Mishina Y., Lewandoski M. BMP signals control limb bud interdigital programmed cell death by regulating FGF signaling. Development. 2007;134:2359–2368. doi: 10.1242/dev.001677. [DOI] [PubMed] [Google Scholar]