Abstract
Several ribosomal proteins including L11 have been shown to activate p53 by inhibiting oncoprotein MDM2, leading to inhibition of cell cycle progression. Our recent study showed that L11 also inhibits oncoprotein c-Myc. Overexpression of L11 inhibits c-Myc-induced transcription and cell proliferation, while reduction of endogenous L11 increases these c-Myc activities. Interestingly, L11 is a transcriptional target of c-Myc, thus forming a negative feedback loop. We further showed that L11 competes with co-activator TRRAP for binding to c-Myc through the Myc box II (MB II) and reduces histone H4 acetylation at c-Myc target gene promoters. In addition, L11 appears to regulate c-Myc levels. Knocking down L11 markedly increases the mRNA and protein levels of endogenous c-Myc. These results suggest that L11 also inhibits cell cycle progression by regulating the c-Myc pathway. Here we further discuss the implications of this regulation and questions that this finding raises.
Keywords: ribosomal protein, L11, c-Myc, transcription, TRRAP, cell cycle
INTRODUCTION
Ribosomal biogenesis, a complex process for making the ribosome, is of fundamental importance for normal cell growth and proliferation. Interfering with the production of ribosomes severely retards cell growth and animal development. For example, deletion of ribosomal protein L16 in yeast results in a lethal phenotype due to deficiency of the 60 S subunit of the ribosome.1 A class of dominant mutants called minutes in drosophila harbor mutations in genes encoding ribosomal proteins. These minutes display similar phenotypes that are characterized by delayed larval development, short thin bristles, recessive lethality, as well as some variable phenotypes including small body size, female sterility, and malformation of wings and eyes resulting from reduced number of ribosomes and protein synthesis.2,3 In mammals, naturally occurring mutations are found in genes encoding ribosomal proteins S19 and L24. Specifically, heterozygous null mutations in the human S19 gene are present in about 25% of patients with Diamond-Blackfan anemia (DBA), a syndrome characterized by anemia and an increased susceptibility to hematopoietic malignancies.4 Additionally, a spontaneously occurring semindominant and homozygous lethal mutant called Belly spot and tail (Bst) in mice was found to harbor a short deletion mutation of the L24 gene.5 Complete loss of S19 or L24 is embryonically lethal, reinforcing the profound effect of ribosomal biogenesis on cell growth and development.5,6
In addition, genetically manipulated inactivation of individual ribosomal proteins L22, L29, and S6 has recently been reported in mice. Conditional homozygous deletion of the S6 gene in mouse liver resulted in the failure of liver cell proliferation following partial hepatectomy.7 Heterozygous deletion of S6 led to p53-dependent cell cycle arrest in somatic T lymphocytes8 and in embryos during gastrulation.9 These studies suggest that S6-haploinsufficiency triggers the activation of a p53-dependent cell cycle checkpoint.10 In contrast, L29 null mice are viable but display low birth weight, reduced postnatal viability, and a global skeleton growth defect. L29 null MEFs display decreased cell proliferation and protein synthesis.11 L22 null mice are also viable and develop normally, but harbor a selective defect in the development of αβT lymphocytes due to activation of a p53-dependent checkpoint,12 suggesting that certain ribosomal proteins may perform cell-type specific or stage-specific functions. Altogether, genetic studies firmly support that ribosomal biogenesis is essential for cell growth and proliferation as well as animal development.
On the other hand, aberrant over-production of ribosomes and increased translational activity contribute to cell transformation and tumorigenesis.13 For example, overexpression of the ribosomal protein S3a induces transformation of NIH 3T3 cells and tumor formation in nude mice by inhibiting apoptosis.14 Individual overexpression of human translation initiation factor eIF3 subunits and eIF-4E enhances cell proliferation and induces cellular transformation.15–17 Other individual ribosomal proteins, such as S8, L12, L23a, L27 and L30, were up-regulated in various tumors.18,19 Although it is still not clear how the overexpression of individual ribosomal proteins contributes to tumorigenesis and whether increased translation on its own can contribute to tumorigenesis, the above studies point to a clear role for deregulation of ribosomal biogenesis in tumorigeneis. Thus, ribosomal biogenesis must be under tight control in order to constantly coordinate with cell growth and proliferation.
REGULATION OF RIBOSOMAL BIOGENESIS BY TUMOR SUPPRESSORS AND ONCOGENES
Consistent with the need to coordinate ribosomal biogenesis with cell growth and proliferation, the tumor suppressor proteins p53, RB, ARF, and PTEN have all been shown to inhibit ribosomal biogenesis (Fig. 1). Specifically, p53, RB, and RB family member p130 prevent the promoter recruitment of TFIIIB, a RNA Polymerase III (Pol III)-specific transcription factor, leading to repression of Pol III-mediated transcription of tRNA and 5S rRNA required for ribosome function and assembly.20–28 Transcription of rDNA to generate the rRNA components of the ribosome is dependent on basal Pol I-specific transcription factors UBF and TBP-containing SL1/TIF-IB complex. Both p53 and RB interfere with the assembly of the UBF-SL1-Pol I initiation complex on the rDNA promoter, leading to repression of Pol I-mediated transcription of rRNAs.29–32 PTEN also represses Pol I-mediated transcription of rRNA by disrupting the SL1/TIF-IB complex and reducing the occupancy of the SL1 subunits on the rDNA gene promoter.33 Finally, ARF has been shown to inhibit rRNA processing possibly through enhancing proteasome-mediated degradation of nucleophosmin (also called B23), an important nucleolar endoribonuclease required for rRNA processing.34,35 In addition, ARF specifically interacts with the rDNA gene promoter and may play a direct function in rRNA transcription.36 ARF also suppresses Pol III-mediated tRNA synthesis independently of p53.37 In parallel, ARF activates p53 by blocking its negative regulator MDM2, perhaps enhancing the suppression of the synthesis of rRNAs imposed by p53. In summary, all of the above tumor suppressors efficiently suppress ribosomal biogenesis presumably to coordinate with their negative regulation of the cell cycle.
Figure 1.
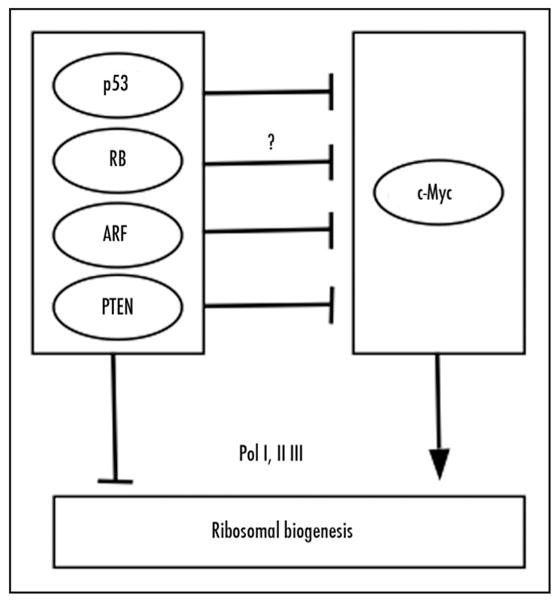
Regulation of ribosomal biogenesis by tumor suppressors and c-Myc. The tumor suppressors p53, RB, PTEN, and ARF reduce the ribosomal biogenesis by either inhibiting Pol I and III-mediated synthesis of rRNAs or rRNA processing, whereas c-Myc enhances ribosomal biogenesis though up-regulation of transcription mediated by all three RNA polymerases. Bars indicate inhibition; arrows denote activation.
On the other hand, the tumor-promoting activity of the c-Myc oncoprotein is associated with its role in enhancing ribosomal biogenesis (Fig. 1). c-Myc has been shown to regulate transcription by all three RNA polymerases.38,39 Specifically, c-Myc enhances Pol I-catalyzed rRNA synthesis by binding to TBP and TBP-associated factors (TAFs), thereby facilitating the recruitment of Pol I to the rDNA promoter.40–42 c-Myc also enhances Pol III-mediated 5S and tRNA transcription by directly interacting with and activating TFIIIB.43 In addition, Pol II-mediated transcription of genes encoding ribosomal proteins, ribosome assembly proteins, and translation initiation and elongation factors is stimulated by c-Myc.44–47 These studies imply that many critical tumor suppressors and oncoproteins exert their function by regulating the dynamics of ribosome biogenesis.
REGULATION OF THE P53-MDM2 FEEDBACK LOOP BY RIBOSOMAL PROTEINS
Nature has evolved many elegant feedback surveillance mechanisms for important cellular processes. Thus an interesting question arises as to whether ribosomal biogenesis components, such as individual ribosomal proteins, in turn regulate the activity of tumor suppressors and oncogenes. This question has recently been investigated by a number of groups, including our own. It is now clear that several ribosomal proteins, including L5, L11, L23, S7, and L26, can regulate p53 activity in response to different stresses by distinct mechanisms.48–55 We found that ribosomal proteins L5, L11, and L23 appear to be the major steady-state components of an MDM2-associated protein complex.49,51 These L proteins as well as S7, a small subunit component, were shown to bind to MDM2 and inhibit MDM2-mediated p53 ubiquitination and degradation, leading to p53 activation.48–53,55 These ribosomal proteins play a crucial role in p53 responses to perturbation of ribosomal biogenesis (ribosomal stress, also called nucleolar stress), such as those induced by the treatment with actinomycin D or 5-FU,51,56,57 serum starvation,58 or genetic disruption of the transcription initiation factor TIF-IA.59 Based upon these studies, it is predicted that expression of dominant negative mutant Bop1,60 or genetic disruption of ribosomal protein S68 may also activate p53 by utilizing these ribosomal proteins. Furthermore, L26 has been shown to enhance p53 translation in response to DNA damage.54 Thus, while p53 inhibits ribosome biogenesis, a group of individual ribosomal proteins, in turn, activates p53, ensuring that ribosome homeostasis is preserved. Since c-Myc is closely linked to ribosome biogenesis, we asked the question of whether individual ribosomal proteins also regulate c-Myc activity. Indeed, our recent study has demonstrated that L11 inhibits c-Myc activity through a negative feedback mechanism.61
FEEDBACK INHIBITION OF c-Myc ACTIVITY BY L11
The c-Myc oncoprotein is a basic helix-loop-helix leucine-zipper (bHLH/LZ) transcriptional factor. It forms a heterodimer with its partner protein Max and binds to cognate DNA sequence elements called E-box (CACGTG).39 The conserved Myc box (MB) II in the N-terminal transcriptional activation domain (TAD) recruits several critical co-activators for c-Myc-mediated transcription, including TRRAP, a core component of the TIP60 and GCN5 containing histone acetyltransferase (HAT) complexes,62 TIP48/TIP49 ATPases, components of chromatin remodeling complexes,63 and Skp2, a component of the SCFskp2 E3 ligase complex.64,65 In addition, the C-terminus of c-Myc recruits co-activators p300/CBP histone acetyltransferase.39,66 By acetlyating histones and remodeling chromatin structure to a transcriptionally active state, these co-activators mediate c-Myc-driven transcription of its target genes that are implicated in cell growth, proliferation, differentiation, apoptosis, metabolism, and neoplastic transformation.39,67,68 Although c-Myc is essential for normal cell growth and animal development,69 deregulated expression of c-Myc due to chromosomal translocations, gene amplification or viral insertions at the c-myc locus is linked to many types of human cancers.39,70 Constitutive or inducible expression of a c-myc transgene leads to neoplastic pre-malignant and malignant phenotypes in mice.70–72 Thus, precise regulation of c-Myc expression and transcriptional activity is critical for normal cellular function. Consistent with this notion, c-Myc expression is regulated at multiple levels, including transcription, mRNA stability, translation, and post-translation protein stability.73,74
In order to test whether individual ribosomal proteins could regulate c-Myc activity, we first employed transfection-luciferase assays. We found that overexpression of L11 significantly inhibited c-Myc-mediated transcription of a luciferase reporter driven by a c-Myc responsive E box-containing E2F2 promoter. Moreover, overexpression of L11 inhibited c-Myc-driven cell proliferation and expression of several endogenous c-Myc target genes. In agreement with these observations, reduction of L11 by siRNA increased these c-Myc activities. Because many ribosomal proteins have been shown as potential transcriptional targets of c-Myc,44–47 we have verified that L11 is indeed a down stream target of c-Myc. Taken together, these results demonstrate that L11 is a negative feedback regulator of c-Myc.61
What is the mechanism underlying the inhibition of c-Myc activity by L11? The finding that L11 actually binds to c-Myc at the MB II region, which is critical for all c-Myc activity and essential for c-Myc to recruit a number of its co-activators, led us to test whether this L11-c-Myc binding would interfere with the recruitment of these co-activators. Indeed, we found that both ectopic and endogenous L11 specifically associated with c-Myc at c-Myc target gene promoters and overexpression of L11 significantly reduced the binding of TRRAP to c-Myc, and thereby histone acetylation at these promoters. In line with these results, L11- and TRRAP-bindings to a c-Myc target promoter displayed reverse profiles in response to growth signals mediated by serum starvation and re-stimulation regimes. These results suggest that L11 attenuates c-Myc-mediated gene transcription via interfering with the recruitment of the TRRAP co-activator to c-Myc target gene promoters,61 further emphasizing the central importance of the MB II region in c-Myc-mediated transactivation and its regulation by c-Myc regulators.39,75
Interestingly, the inhibitory effect of L11 on c-Myc activity resembles the regulation of c-Myc by ARF.76–78 Similarly, ARF binds to the MB II79 and suppresses c-Myc activity, although it has not been tested if ARF may do so by interfering with the TRRAP recruitment and subsequent histone acetylation. Intriguingly, the inhibitory effect of ARF on c-Myc is selective to its transactivational, but not repression, activity.78 It is still unclear whether this selectivity would also be true to L11. Also, it is unknown whether L11 and/or ARF could interfere with the recruitment of other co-factors, such as TIP48/TIP49 or Skp2, by c-Myc to c-Myc target promoters. These are certainly interesting and important questions for future investigation.
L11 REGULATES c-Myc LEVELS
In addition to directly inhibiting c-Myc-dependent transcription by competing with TRRAP for promoter binding, we have also shown that L11 expression is well correlated with the change in c-Myc levels.61 Initially, we found that reduction of L11 by siRNA drastically induced the protein level of c-Myc in human osteosarcoma U2OS cells. This protein induction was at least partly due to increased levels of c-myc mRNA (Fig. 2). The increase in c-myc mRNA might be caused by an increase in either gene transcription or mRNA stability. It has been shown that c-Myc can auto-inhibit its own transcription, though by an unknown mechanism.80,81 It is possible that knockdown of L11 may derepress this autoregulatory inhibition imposed by c-Myc itself. Toward this end, it would be worth investigating whether L11 associates with c-Myc at the promoter region of the c-myc gene. Another possibility is that L11 may regulate c-myc gene transcription independently of its binding to the c-Myc protein. For example, L11 might directly interfere with transcriptional machineries or with the remodeling of chromatin structure in the promoter region of the c-myc gene by binding to histones, since several drosophila ribosomal proteins have recently been shown to associate with linker histone H1 and suppress transcription of a set of genes.9 Correlated with this possibility, we have recently purified an L11-associated complex that also contains the linker histone H1 (data not shown).
Figure 2.
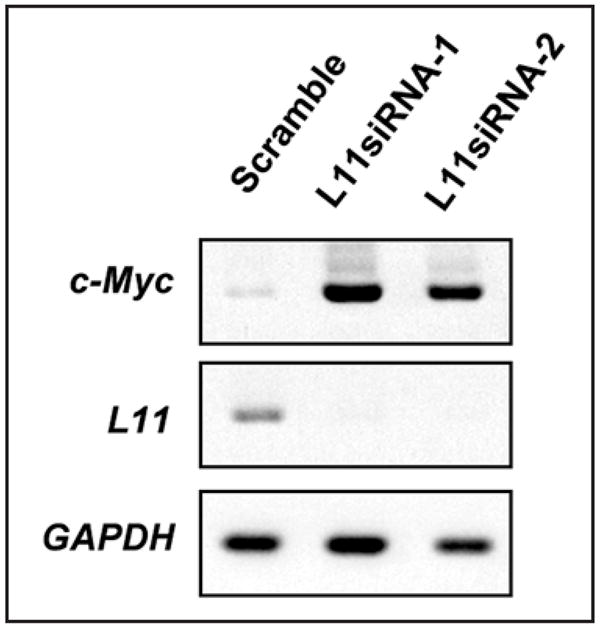
L11 regulates c-myc mRNA levels. U2OS cells were transfected with scrambled or L11 siRNAs against two different sequences followed by semi-quantitative RT-PCR assays.
L11 could also influence c-myc mRNA stability. c-myc mRNA has a short half-life of 15–30 minutes. Two cis-acting sequence elements have been shown to regulate c-myc mRNA turnover: an AU-rich element (ARE) in the 3′-untranslated region (3′-UTR) and a ~250 nucleotide region called coding region instability determinant (CRD). Several ARE binding proteins, including AUF182 and HuR,83 have been found to bind to the c-myc ARE and act as c-myc mRNA destabilizing factors. Furthermore, CRD-binding protein (CRD-BP) binds to the CRD in the c-myc mRNA and protects it from endoribonuclease cleavage within the CRD, leading to stabilization of c-myc mRNA.84–86 The latter regulation has been implicated in the stabilization of c-myc mRNA in response to β-catenin signaling.87 Thus, one interesting and pertinent question for future study would be if L11 regulates c-myc mRNA levels through interaction with these components.
Since knockdown of L11 leads to an increase in c-Myc mRNA and protein expression, one would predict that overexpression of L11 might reduce c-Myc levels. However, we surprisingly observed an increase in the total level of ectopically expressed c-Myc when L11 was overexpressed. This increase was accompanied by a concurrent decrease in the population of NP-40-extractable and soluble c-Myc. One possible explanation is that L11 might facilitate the subcellular localization of c-Myc to insoluble chromatin-bound material or to nucleolar compartments, resulting in the decrease of soluble c-Myc. This indeed was the case, as overexpression of L11 relocalized a fraction of ectopic c-Myc into the nucleolus (Fig. 3). This sequestration required the interaction of c-Myc with L11, as a c-Myc-binding defective mutant of L11 did not re-localize c-Myc into the nucleolus (data not shown). c-Myc was shown to be degraded in the nucleolus by the proteasome,88 however its level was elevated in this subcellular compartment in the presence of overexpressed L11 (Fig. 3), suggesting that L11 may simply block c-Myc degradation in the nucleolus. Also, in our preliminary study using chromatin immunoprecipitation (ChIP) assays, we found that overexpression of L11 increased the residence of c-Myc at its target gene promoters (data not shown), suggesting that L11 may block c-Myc turnover at the promoter as well. Others have shown that Skp2-mediated c-Myc turnover at the promoter is essential for proper c-Myc function and Spk2 also binds to the MB II.64,65 Hence, it is tempting to speculate that L11 may compete with Skp2 for binding to c-Myc at its target gene promoters, leading to accumulation of inactive c-Myc at these promoters.
Figure 3.
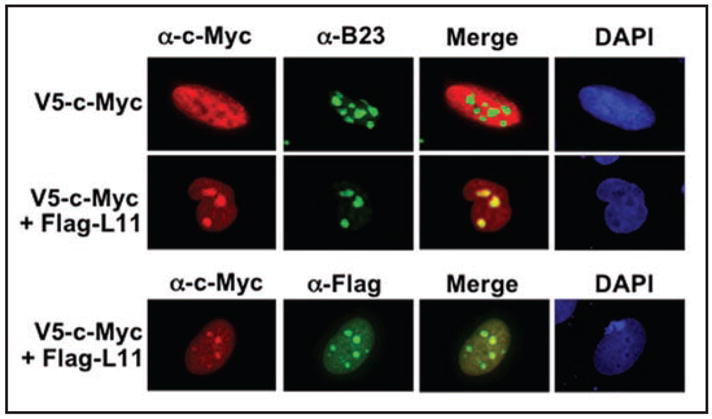
L11 re-localizes ectopic c-Myc into the nucleolus. H1299 cells transfected with V5-c-Myc alone or together with Flag-L11 were immunostained with anti-c-Myc, anti-Flag, or anti-B23 antibodies as indicated.
Taken together, the evidence as described here suggests that multiple mechanisms may account for the inhibitory effect of L11 on c-Myc activity: directly blocking the transactivational activity of c-Myc at c-Myc target gene promoters, controlling c-myc mRNA levels, and sequestering excess c-Myc protein. However, further studies are necessary to verify and dissect the latter two mechanisms. This complex regulation also highlights the important role for L11 in regulating c-Myc and may be a key feedback regulation during ribosomal biogenesis.
IS THE INHIBITION OF c-Myc SPECIFIC TO L11?
In addition to L11, there are other ribosomal proteins that have been shown to play a role in regulating p53 activities.48–55 Thus, an obvious question is whether the effect of L11 on c-Myc is specific to this ribosomal protein. In our recently reported work,61 we have attempted to address this question by testing several other ribosomal proteins for their binding capacity to c-Myc. We found that L29, L30, and S12 do not bind to c-Myc. In addition, neither overexpression of L29 nor knockdown of L29 affected c-Myc activity.61 Thus clearly, the inhibition of c-Myc activity is not a general effect for all ribosomal proteins.
However, L11 is not the only ribosomal protein that binds to c-Myc either. Among our tested ribosomal proteins, L5, L23, and S7, which have all been shown to bind to MDM2,48,49,51,52 were found to bind to c-Myc as well, though with different binding affinity (data not shown). Although further studies are necessary to clarify whether these ribosomal proteins act similarly to L11, these results suggest that a group of individual ribosomal proteins may target both c-Myc and the MDM2-p53 pathways. In support of this hypothesis, our preliminary work shows that L29, L30, and S12, which do not bind to c-Myc, also do not bind to MDM2 (data not shown). The use of a common subset of ribosomal proteins to control both MDM2 and c-Myc activity may ensure the coordination of ribosomal biogenesis with cell cycle progress. It is also possible that these ribosomal proteins may work in concert or synergistically to reach an optimal and efficient effect on c-Myc activity. Or, they may work independently in response to different ribosomal biogenesis stresses. More experiments are necessary to define the individual role for each of these ribosomal proteins in regulating both c-Myc activity and c-myc mRNA levels.
BIOLOGY OF THE L11-C-MYC FEEDBACK LOOP
Our finding that L11 regulates c-Myc level and activity raises additional questions: what is the physiological significance of the L11-c-Myc inhibitory feedback regulation? Under what circumstance is this feedback loop activated? Is it responsive to ribosomal stress signals? Presumably, the excess molecules of L11 that can target c-Myc may be generated from either a net increase in L11 synthesis or release of L11 from the intact ribosome into ribosome-free pools. Deregulated and high levels of c-Myc apparently enhance ribosomal biogenesis and L11 production. Our finding suggests that this increased L11 could then target c-Myc and turn it off. A future project would be to confirm this feedback loop in vivo by employing genetically manipulated mouse models.
The balance of ribosome-bound and ribosome-free L11 molecules may serve as an important signal for activation of the L11-c-Myc feedback loop. Under nucleolar stress, L11 may be released from the nucleolus as the result of the stalled process of ribosomal biogenesis. In such a case, ribosome-free L11 may be in excess and then execute the inhibitory effect on c-Myc activity, as it does to the MDM2-p53 pathway.58 Our study also indicates that L11 regulates c-Myc activity dependent on growth signals.61 In response to serum starvation, the association of L11 with c-Myc target gene promoters increased and thus L11 competed with TRRAP for binding to these promoters, leading to inhibition of c-Myc-dependent transcription. By doing so, L11 may play a role in maintaining a silent or inhibited status of ribosomal biogenesis when growth conditions are unfavorable for cell growth or proliferation. Conversely, In response to serum re-stimulation, c-Myc levels rapidly increased while L11 binding to c-Myc target gene promoters inversely decreased, indicating that the L11 repression of c-Myc activity is de-repressed at a stage when c-Myc activity is required for cells to proliferate.61 It seems that the acutely increased L11 molecules in the early serum re-stimulation stage do not target c-Myc instantly. Instead, these L11 molecules might be used to limit c-Myc activity at the later stage of serum stimulation in order to prevent aberrant cell growth. The increased L11 could just be simply incorporated into ribosomes for protein synthesis. This delayed targeting of c-Myc by L11 may suggest that additional factors, possibly posttranslational modifications, could regulate L11-c-Myc interaction. Nevertheless, our studies suggest that the L11-c-Myc feedback loop is highly regulated in cells and functionally responsive to growth or stress signals. One untested question is whether L11 may regulate c-Myc activity or level in response to other ribosomal stresses, such as those induced by 5-FU, actinomycin D, or UV damage. It has been demonstrated by others that aberrant proliferation signals, such as overexpression of c-Myc, induce ARF through unknown mechanisms. ARF in turn binds to c-Myc and inhibits its activity, similar to L11.24,76–78 An additional, if not the last, question would be whether L11 also represses c-Myc activity in response to oncogenic stress.
CONCLUSION
Recent studies including ours as described here have demonstrated an important role for L11 in cell cycle control by regulating the MDM2-p53 feedback loop as well as the c-Myc pathway.53,55,58,73 Remarkably, L11 resembles ARF in these regulations, but also works independently of ARF. In light of currently available evidence, we propose a model (Fig. 4) for the action of L11 and ARF. In this model, L11 may primarily acts as a sensor of aberrant ribosomal biogenesis, whereas ARF primarily acts as a sensor of oncogenic stress.35 The remaining questions would be: whether these two stress signaling pathways crosstalk with each other, whether L11 itself possesses tumor suppressor function, and whether L11-c-Myc and L11-MDM2 interactions have implications in tumor treatment?
Figure 4.
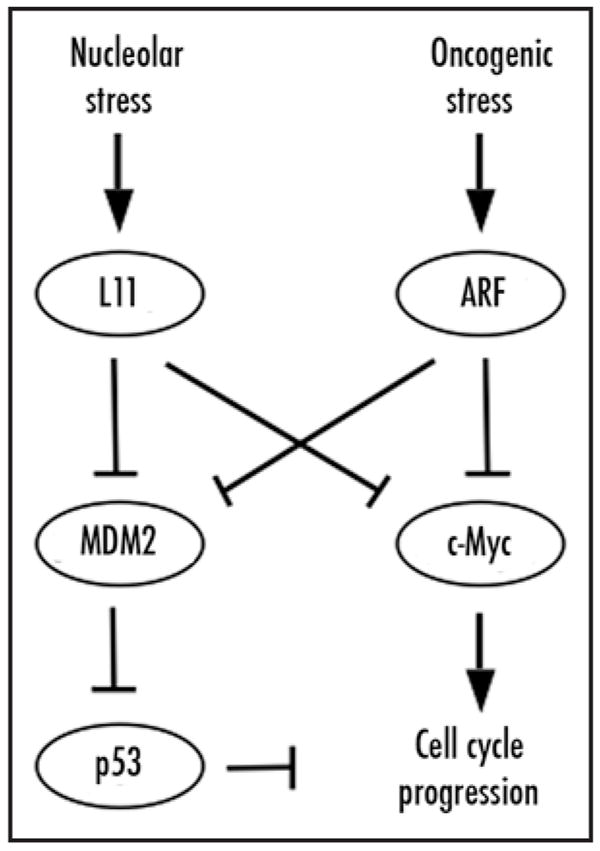
A schematic model illustrating the dual effects of L11 and ARF on cell cycle arrest. L11, in response to nucleolar stress, and, ARF in response to oncogenic stress, bind to MDM2 and suppress MDM2-mediated p53 inhibition, leading to p53 activation. They also bind to c-Myc and inhibit its transactivation activity. Bars indicate inhibition, whereas arrows denote activation.
Acknowledgments
This work was supported in part by NCI grants CA93614, CA095441 and CA 079721 to H.L. and NCI grant K990CA127134 to M.-S.D. R.S. was supported by NIH/NCI grants R01-CA100855 and K01-CA086957.
References
- 1.Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes and development. 1988;2:160–72. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 2.Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- 3.Saeboe-Larssen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: A dose-response relationship. Genetics. 1998;148:1215–24. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 5.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–20. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, Forsberg E, Karlsson S, Dahl N. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Molecular and Cellular Biology. 2004;24:4032–7. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–7. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 8.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes and Development. 2005;19:3070–82. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusic K, Volarevic S. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880–91. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panic L, Montagne J, Cokaric M, Volarevic S. S6-haploinsufficiency activates the p53 tumor suppressor. Cell Cycle. 2007;6:20–4. doi: 10.4161/cc.6.1.3666. [DOI] [PubMed] [Google Scholar]
- 11.Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn. 2007;236:447–60. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- 12.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–72. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 14.Naora H, Takai I, Adachi M. Altered cellular responses by varying expression of a ribosomal protein gene: Sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–53. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 16.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. The Journal of Biological Chemistry. 2007;282:5790–800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 18.Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 2001;21:2429–33. [PubMed] [Google Scholar]
- 19.Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol. 1999;77:197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- 20.Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. The EMBO Journal. 1998;17:3112–23. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chesnokov I, Chu WM, Botchan MR, Schmid CW. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Molecular and Cellular Biology. 1996;16:7084–8. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu WM, Wang Z, Roeder RG, Schmid CW. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. The Journal of Biological Chemistry. 1997;272:14755–61. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 23.Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. The EMBO Journal. 2003;22:2810–20. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–4. [PubMed] [Google Scholar]
- 25.Larminie CG, Cairns CA, Mital R, Martin K, Kouzarides T, Jackson SP, White RJ. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. The EMBO Journal. 1997;16:2061–71. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutcliffe JE, Brown TR, Allison SJ, Scott PH, White RJ. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Molecular and Cellular Biology. 2000;20:9192–202. doi: 10.1128/mcb.20.24.9192-9202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutcliffe JE, Cairns CA, McLees A, Allison SJ, Tosh K, White RJ. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Molecular and Cellular Biology. 1999;19:4255–61. doi: 10.1128/mcb.19.6.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White RJ, Trouche D, Martin K, Jackson SP, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 29.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–24. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 30.Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–80. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 31.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Molecular and Cellular Biology. 1997;17:4230–7. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Molecular and Cellular Biology. 2000;20:5930–8. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Comai L, Johnson DL. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Molecular and Cellular Biology. 2005;25:6899–911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Molecular Cell. 2003;12:1151–64. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell. 2003;11:415–24. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 36.Ayrault O, Andrique L, Larsen CJ, Seite P. Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene. 2004;23:8097–104. doi: 10.1038/sj.onc.1207968. [DOI] [PubMed] [Google Scholar]
- 37.Morton JP, Kantidakis T, White RJ. RNA polymerase III transcription is repressed in response to the tumour suppressor ARF. Nucleic Acids Research. 2007;35:3046–52. doi: 10.1093/nar/gkm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oskarsson T, Trumpp A. The Myc trilogy: Lord of RNA polymerases. Nat Cell Biol. 2005;7:215–7. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- 39.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 40.Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nature Cell Biology. 2005;7:303–10. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 41.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biology. 2005;7:311–8. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 42.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nature Cell Biology. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 44.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–9. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–5. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. Embo J. 2001;20:1383–93. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–8. [PubMed] [Google Scholar]
- 48.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: Binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–37. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 49.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of Biological Chemistry. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 50.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. The Journal of Biological Chemistry. 2006;281:24304–13. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and Cellular Biology. 2004;24:7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Molecular and Cellular Biology. 2004;24:7669–80. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 54.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and Cellular Biology. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. The EMBO Journal. 2006;25:5614–25. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. The Journal of Biological Chemistry. 2007;282:8052–9. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 58.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. Embo J. 2004;23:2402–12. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Molecular Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 60.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G1/S transition. Mol Cell Biol. 2001;21:4246–55. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. The EMBO Journal. 2007;26:3332–45. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 63.Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Molecular Cell. 2000;5:321–30. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 64.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–88. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 65.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 66.Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Luscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–90. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen DE, Prochownik EV. A functional hierarchy for c-Myc target genes? Lessons from MT-MC1 Cell Cycle. 2006;5:392–3. doi: 10.4161/cc.5.4.2484. [DOI] [PubMed] [Google Scholar]
- 68.Cowling VH, Cole MD. HATs off to capping: A new mechanism for Myc. Cell Cycle. 2007;6:907–9. doi: 10.4161/cc.6.8.4123. [DOI] [PubMed] [Google Scholar]
- 69.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes and Development. 1993;7:671–82. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 70.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–34. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 71.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 72.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 73.Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: Ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006;8:630–44. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sears RC. The life cycle of C-myc: From synthesis to degradation. Cell Cycle. 2004;3:1133–7. [PubMed] [Google Scholar]
- 75.Zhang XY, DeSalle LM, McMahon SB. Identification of novel targets of MYC whose transcription requires the essential MbII domain. Cell Cycle. 2006;5:238–41. doi: 10.4161/cc.5.3.2409. [DOI] [PubMed] [Google Scholar]
- 76.Datta A, Nag A, Pan W, Hay N, Gartel AL, Colamonici O, Mori Y, Raychaudhuri P. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. The Journal of Biological Chemistry. 2004;279:36698–707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- 77.Gregory MA, Qi Y, Hann SR. The ARF tumor suppressor: Keeping Myc on a leash. Cell Cycle. 2005;4:249–52. [PubMed] [Google Scholar]
- 78.Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–7. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 79.Amente S, Gargano B, Varrone F, Ruggiero L, Dominguez-Sola D, Lania L, Majello B. p14ARF directly interacts with Myc through the Myc BoxII domain. Cancer Biol Ther. 2006;5:287–91. doi: 10.4161/cbt.5.3.2389. [DOI] [PubMed] [Google Scholar]
- 80.Grignani F, Lombardi L, Inghirami G, Sternas L, Cechova K, Dalla-Favera R. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. The EMBO Journal. 1990;9:3913–22. doi: 10.1002/j.1460-2075.1990.tb07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. The EMBO Journal. 1990;9:1113–21. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Molecular and Cellular Biology. 1993;13:7652–65. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. The Journal of Biological Chemistry. 1996;271:8144–51. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 84.Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. The c-myc coding region determinant-binding protein: A member of a family of KH domain RNA-binding proteins. Nucleic Acids Research. 1998;26:5036–44. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee CH, Leeds P, Ross J. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. The Journal of Biological Chemistry. 1998;273:25261–71. doi: 10.1074/jbc.273.39.25261. [DOI] [PubMed] [Google Scholar]
- 86.Yeilding NM, Lee WM. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Molecular and Cellular Biology. 1997;17:2698–707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, Minamoto T, Ross J, Fuchs SY, Spiegelman VS. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 88.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–7. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]


