Abstract
Actin plays fundamental roles in a wide array of plant functions, including cell division, cytoplasmic streaming, cell morphogenesis and organelle motility. Imaging the actin cytoskeleton in living cells is a powerful methodology for studying these important phenomena. Several useful probes for live imaging of filamentous actin (F-actin) have been developed, but new versatile probes are still needed. Here, we report the application of a new probe called Lifeact for visualizing F-actin in plant cells. Lifeact is a short peptide comprising 17 amino acids that was derived from yeast Abp140p. We used a Lifeact–Venus fusion protein for staining F-actin in Arabidopsis thaliana and were able to observe dynamic rearrangements of the actin meshwork in root hair cells. We also used Lifeact–Venus to visualize the actin cytoskeleton in the liverwort Marchantia polymorpha; this revealed unique and dynamic F-actin motility in liverwort cells. Our results suggest that Lifeact could be a useful tool for studying the actin cytoskeleton in a wide range of plant lineages.
Keywords: Actin, Arabidopsis thaliana, Lifeact, Liverwort, Marchantia polymorpha
Introduction
Actin microfilaments are a major component of the cytoskeleton and play crucial roles in a wide variety of cellular activities in eukaryotic cells. In animal cells, actin plays essential roles in cell migration and cytokinesis. In plants, actin is utilized differently, probably reflecting the immobile nature of the cells due to the rigid cell wall. Many cellular functions in plants, including cytoplasmic streaming, cell division, cell elongation, cell morphogenesis and organelle movement, require filamentous actin (F-actin) (Ketelaar and Emons 2001, Hasezawa and Kumagai 2002, Hussey et al. 2006).
The establishment of fluorescent probes, particularly green fluorescent protein (GFP), to visualize F-actin in living cells has dramatically facilitated the understanding of actin dynamics. Initially, actin–GFP fusion proteins were used successfully to visualize F-actin. However, direct labeling of actin impaired its function; thus, the actin–GFP fusion protein had to be mixed with non-tagged actin (Westphal et al. 1997). In addition, the fluorescence from unpolymerized GFP–actin in the cytosol could impair clear visualization of F-actin. These problems were overcome with the advent of GFP-tagged actin side-binding proteins. These comprised GFP fused to the actin-binding domain (ABD) from mouse talin (mTn) (Kost et al. 1998) or the ABD from fimbrin1 of Arabidopsis thaliana (ABD2) (Sheahan et al. 2004). These two probes are currently the state of the art for visualizing F-actin in plant cells. They allow investigators to identify various structures and dynamics of actin filaments concomitant with various plant functions (for example, Mathur et al. 2003, Cheung and Wu 2004, Holweg et al. 2004, Gu et al. 2005, Voigt et al. 2005, Higaki et al. 2007, Yoneda et al. 2007, Higaki et al. 2008, Staiger et al. 2009). On the other hand, these F-actin probes have also been reported to exert unexpected side effects on actin organization or myosin-dependent motility of organelles (Ketelaar et al. 2004, Sheahan et al. 2004, Higaki et al. 2007, Holweg 2007). This spurred interest in developing a new F-actin marker with different molecular properties.
It was recently demonstrated that a peptide comprising the first 17 amino acids of the yeast protein Abp140p is sufficient for localizing GFP to F-actin. This short peptide, called ‘Lifeact’ (Riedl et al. 2008), did not affect actin polymerization or depolymerization in vitro. The usefulness of Lifeact was also demonstrated in vivo with the expression of the Lifeact–GFP fusion protein. In mammalian cells, Lifeact–GFP did not influence actin-dependent processes, including neuronal polarization, lamellipodia flow and leukocyte chemo-taxis. These features are probably due to the following facts: (i) the low binding affinity of Lifeact for F-actin presents little competition against major endogenous actin-binding proteins; and (ii) the absence of homologous sequences in higher eukaryotes obviates the titration of endogenous proteins (Riedl et al. 2008). Furthermore, the small size of Lifeact allows construction of chimeric markers with a single round of PCR amplification.
We hypothesized that these properties would also be advantageous in plants. In this study, we used Lifeact to visualize F-actin in Arabidopsis thaliana and an emerging bryophyte model, Marchantia polymorpha (Bowman et al. 2007, Ishizaki et al. 2008). Our study demonstrated that Lifeact could be a useful tool for the live imaging of F-actin in broad lineages of plants, including angiosperms and bryophytes.
Results and Discussion
Lifeact–Venus-labeled actin microfilaments in A. thaliana
We constructed a chimeric gene that contained the cauliflower mosaic virus (CaMV) 35S promoter, the Lifeact actin-binding sequence and the Venus protein sequence (modified yellow fluorescent protein). The resulting Lifeact–Venus fusion protein comprised the two peptides, separated by a four amino acid linker (Gly–Gly–Ser–Gly) (Fig. 1). To verify the availability of Lifeact in plant cells, we introduced Lifeact–Venus into A. thaliana. We observed actin structures and dynamics under a spinning-disk confocal laser scanning microscope in homozygous or hemizygous plants for the transgene in which a marked deleterious effect of Lifeact–Venus on plant growth was not observed (Supplementary Fig. S1).
Fig. 1.
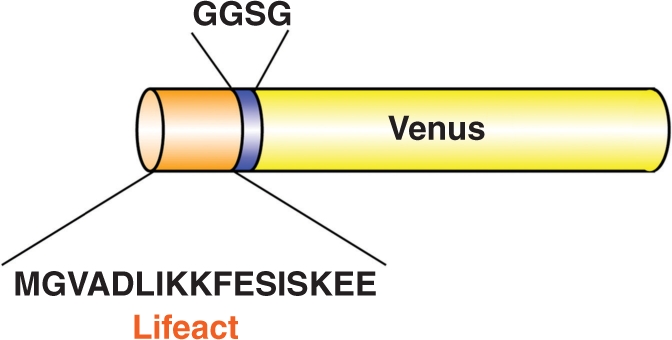
Schematic structure of Lifeact–Venus. Lifeact, comprised of 17 amino acids, is fused to the N-terminus of Venus by a four amino acid linker.
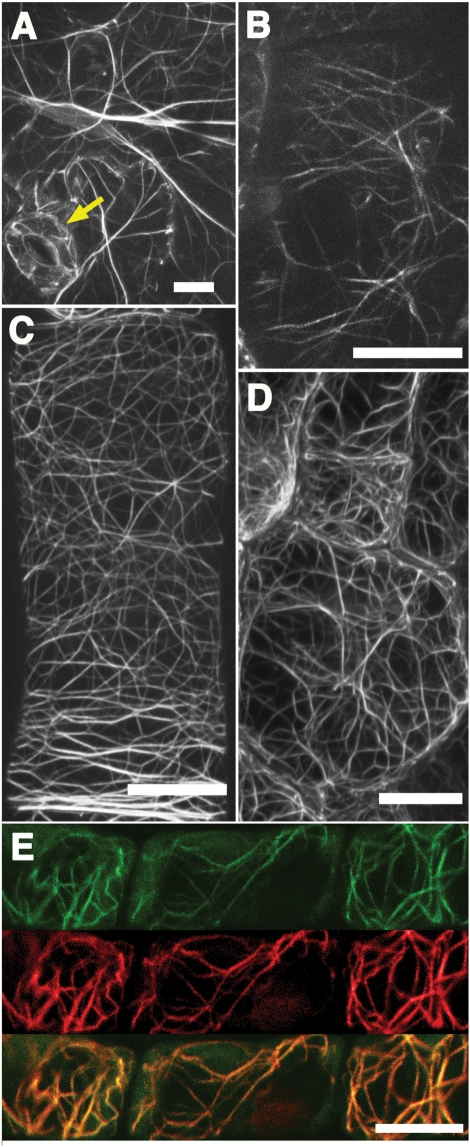
Lifeact–Venus permitted the visualization of filamentous networks in various types of A. thaliana cells, including cotyledon epidermal cells (Fig. 2A), guard cells (Fig. 2A, arrow), hypocotyl cells (Fig. 2B), root cells (Fig. 2C) and true leaf epidermal cells (Fig. 2D). The observed networks of fine and dense filaments were quite similar to the actin networks observed with GFP–ABD2 (Sheahan et al. 2004, Higaki et al. 2007), suggesting that Lifeact had been successfully integrated into Arabidopsis cells. To confirm that Lifeact was bound to F-actin, we used rhodamine-conjugated phalloidin to stain F-actin in root epidermal cells that expressed Lifeact–Venus. As shown in Fig. 2E, the signal from rhodamine entirely overlapped the fluorescence from Venus, indicating that Lifeact–Venus does successfully visualize actin filaments by binding to F-actin.
Fig. 2.
Lifeact–Venus was successfully used to visualize actin filaments in A. thaliana. (A) Maximum intensity projection images of actin filaments in epidermal cells of the cotyledon, (B) hypocotyl, (C) root and (D) young true leaf. (E) Actin filaments in root epidermal cells expressing Lifeact–Venus (green) were stained with rhodamine–phalloidin (red). Lifeact–Venus and phalloidin co-localized (yellow, merge). Scale bars = 10 μm.
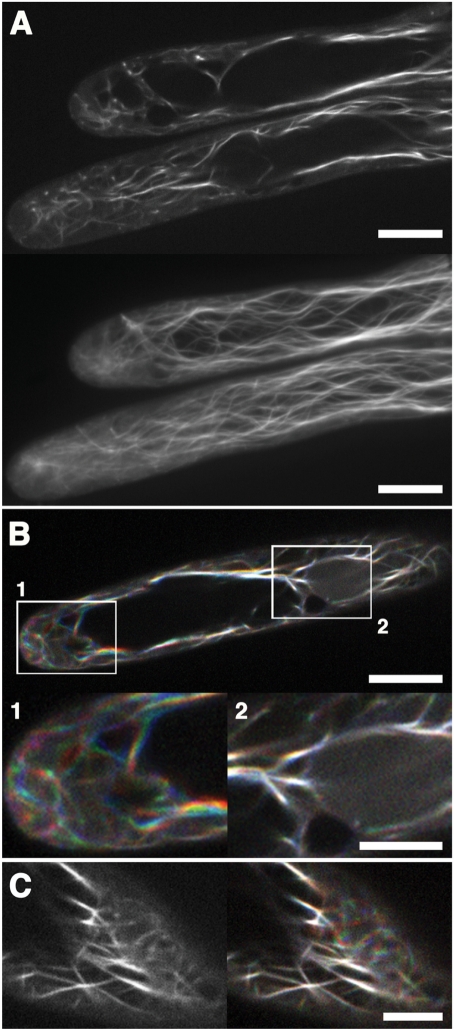
We observed F-actin buckling in several tissues (data not shown), consistent with observations in previous reports when F-actin was visualized with other probes (Sheahan et al. 2004, Higaki et al. 2007, Staiger et al. 2009). Among the observed A. thaliana tissues presented in Figs. 2 and 3, the most dynamic behavior of F-actin was observed in growing root hair cells. Dense actin cables were aligned along the long axis of root hair cells and seemed to branch into a fine meshwork at the apex (Fig. 3A). This pattern was similar to the distribution of endogenous F-actin visualized with immunostaining (Miller et al. 1999), further supporting the reliability of this new probe. Time-sequential observations of actin filaments in root hair cells revealed that the fine actin filaments in the apical region were highly dynamic (Fig. 3B, Supplementary Movie 1). In contrast, the longitudinally oriented dense actin cables were more static, but they did exhibit lateral waving movements. Recently, GFP–ABD2 expression was used to observe dynamic behavior of fine actin filaments that was dominated by rapid growth and severing activity in hypocotyl cells of A. thaliana (Staiger et al. 2009). We found that fine actin filaments at the cortical region of the cytoplasm in root hair cells, between the plasma membrane and the static dense actin cables, were also dynamically rearranged (Fig. 3C, Supplementary Movie 2). These results suggest that hypocotyl and root hair cells may share a common mechanism for actin filament dynamics. Thus, these results demonstrated that Lifeact could be a new and useful probe for visualizing F-actin in A. thaliana.
Fig. 3.
Structure and dynamics of actin filaments in root hair cells of A. thaliana, visualized with Lifeact–Venus. (A) A confocal section (upper panel) and a maximum intensity projection image (lower panel) of actin filaments in root hair cells are presented. Scale bars = 10 μm. (B) Three images of actin filaments in a root hair cell, taken every 3 s; sequential images are colored red (0 s), green (3 s) and blue (6 s), and projected in a single frame. Magnified images of boxed areas are shown in the lower panels. White pixels indicate static actin filaments. Note that the fine actin filaments in the apical region are highly motile, while dense cables in the proximal region are more static. Scale bars = 10 μm in the upper panel and 5 μm in the lower panel. (C) Fine motile actin filaments were observed beneath the plasma membrane in root hair cells. A confocal section (left) and a projection of three images taken every 3 s (right); sequential images are colored red (0 s), green (3 s) and blue (6 s). Scale bar = 5 μm. Note the high motility of fine actin filaments located between the plasma membrane and the static, dense actin cables.
Actin filaments in M. polymorpha are highly dynamic
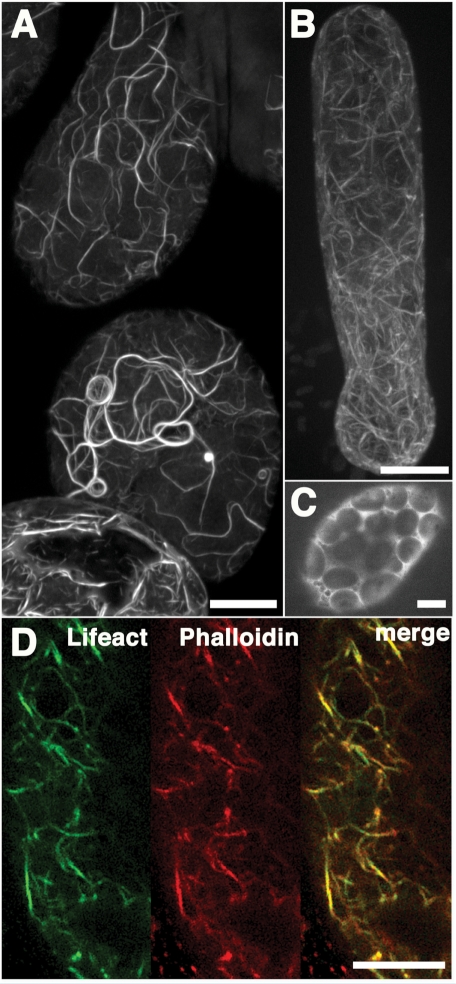
Given its importance in the evolutionary chain, M. polymorpha is expected to be a new model bryophyte (Bowman et al. 2007). The F-actin dynamics in M. polymorpha have not been well characterized; thus, we introduced Lifeact–Venus into this emerging model plant for the characterization of F-actin organization. We transformed M. polymorpha by co-cultivating immature thalli with Agrobacterium tumefaciens that harbored a binary vector containing Lifeact–Venus (Ishizaki et al. 2008). This probe allowed the visualization of filamentous networks and fragmented filaments in thalli and growing rhizoids (Fig. 4A, B). The Lifeact fluorescence pattern completely overlapped with the signal from Alexa Fluor 568-conjugated phalloidin (Fig. 4D); thus Lifeact– Venus also efficiently bound to the actin filaments in M. polymorpha cells. The identity of the filamentous structures was also confirmed by the observation that the fluorescence was dispersed throughout the cytosol upon treatment with the actin-depolymerizing drug, bistheonellide A (BA) at a concentration of 4 μM for 2 h (Fig. 4C). These results indicate that Lifeact is applicable to studies on a wide range of plant taxa, ranging from bryophytes to angio-sperms.
Fig. 4.
Actin filaments in M. polymorpha visualized by Lifeact–Venus. (A) Maximum intensity projection images of actin filaments in epidermal cells of thallus and (B) young rhizoid obtained using a spinning-disk confocal microscope. (C) Thallus cells expressing Lifeact–Venus were treated with bistheonellide A. (D) Actin filaments in thallus cells expressing Lifeact–Venus were stained with phalloidin conjugated to Alexa Fluor 568. Lifeact–Venus and phalloidin co-localized. Scale bars = 10 μm.
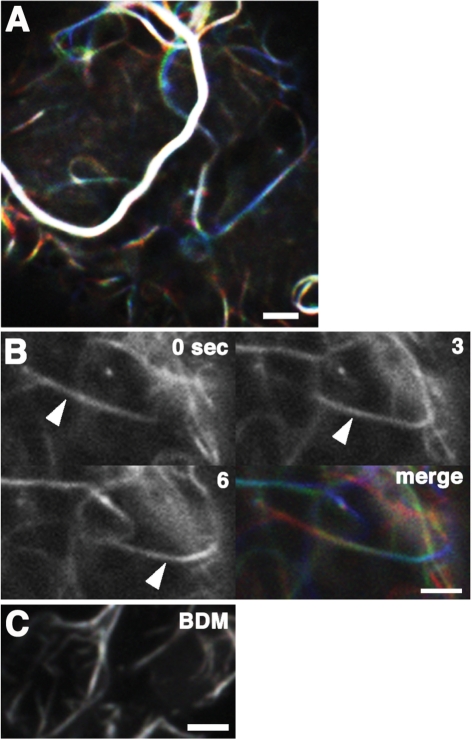
It has been reported that actin filaments can actively swing or move laterally in plant cells, depending on the myosin activity (Sheahan et al. 2004, Higaki et al. 2006). To examine whether actin filaments also have dynamic behavior in M. polymorpha cells, we performed time-sequential imaging of the actin filaments in epidermal cells of developing M. polymorpha thalli. We found that the actin filaments of M. polymorpha cells were highly dynamic. In most cells that we observed, actin networks were rapidly moving with motilities that depended on their thickness; fine actin filaments were highly motile, while dense actin cables appeared only slightly motile (Fig. 5A, Supplementary Movie 3). Stabilization of actin filaments by thickening was also reported in vitro and in A. thaliana hypocotyl cells (Michelot et al. 2007, Staiger et al. 2009). In addition to lateral movement, we observed actin cables that could slide directionally (Fig. 5B, Supplementary Movie 4). The sliding movement was reminiscent of the gliding assay performed with F-actin on coverslips coated with myosin. Therefore, we examined whether myosin was involved in the F-actin sliding in M. polymorpha cells. We treated young thalli expressing Lifeact–Venus with 30 mM 2,3-butanedione-2-monoxime (BDM), an inhibitor of myosin ATPase (Tominaga et al. 2000, Funaki et al. 2004). The F-actin movement completely ceased in the presence of this drug (Fig. 5C, Supplementary Movie 5); this suggested that the motility of F-actin was dependent on myosin in M. polymorpha. However, this result did not rule out the possible involvement of actin assembly proteins (e.g. formins) or actin-nucleating proteins (e.g. the Arp2/3 complex) in the actin dynamics of M. polymorpha.
Fig. 5.
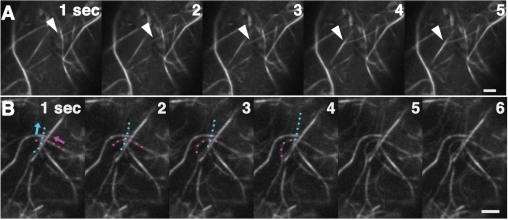
Actin filaments are highly dynamic in M. polymorpha cells. (A) Images of actin filaments in a thallus cell were taken every 3 s, colored red (0 s), green (3 s) and blue (6 s), and projected together. Note that the thick actin cables are more static than the fine filaments. (B) Sliding movement of an actin bundle. Three serial images taken every 3 s (0, 3 and 6 s) and their projections in which each image was colored red, green or blue (merge) are presented. The arrowhead indicates a sliding actin bundle. (C) Projection of three images of a BDM-treated cell taken every 3 s and colored red, green and blue, respectively. Treatment with BDM terminated actin filament movement. Scale bars = 2 μm.
There appeared to be a track that defined the path of actin cable movements, because actin cables were frequently observed sliding along the same routes in cells. This notion was supported by the observation that sliding actin cables sometimes merged and bundled into a dense cable (Fig. 6A, Supplementary Movie 6). Intriguingly, in one observation, two actin cables moving in opposite directions met on the same track (Fig. 6B, Supplementary Movie 7). This observation may reflect the flexibility of the motor proteins that enables movement in opposite directions in M. polymorpha cells. It is also an interesting possibility that an actin bundle functions as a track for the bundle oriented in the opposite direction.
Fig. 6.
Time-lapse montage of actin filament bundling. (A) An actin bundle (arrowhead) sliding toward another bundle to be merged into a single thick bundle. Images were taken every second. (B) Two actin bundles (marked by pink and light blue dots) sliding in distinct directions (arrows) appear to merge into a single actin bundle. Images were taken every second. Scale bars = 2 μm.
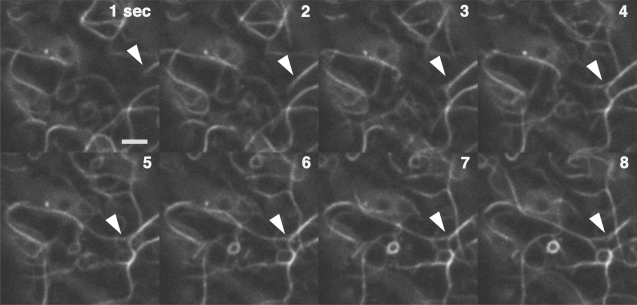
Another interesting behavior of F-actin in M. polymorpha was the branching of actin bundles. As shown in Fig. 7 and Supplementary Movie 8, bundled actin filaments were observed to branch into two or more sub-bundles. The branching of actin bundles has been observed in vivo in animal cells (Schaefer et al. 2002), but has not yet been clearly reported in plant cells, although some time-lapse micrographs in Staiger et al. (2009) imply the existence of a similar phenomenon in A. thaliana. It would be interesting in a future study to investigate the molecular mechanisms underlying the actin filament dynamics.
Fig. 7.
Time-lapse montage of a branching actin bundle. An actin bundle (arrowhead) divides into two sub-bundles. Images were taken every second. Scale bar = 2 μm.
Intriguingly, we did not observe definitive cytoplasmic streaming, even in cells with highly dynamic actin filaments. This was surprising, because actin–myosin-dependent cytoplasmic streaming is generally observed in other plants, from charophytes to angiosperms (Shimmen and Yokota 1994). From this observation, we can speculate that the actin–myosin system might have distinctive functions in M. polymorpha. Further analysis of actin involvement in various physiological events, including organelle motility, cell morphogenesis and cell division, might reveal novel functional roles for actin in M. polymorpha. In addition, future investigations could compare the dynamics and function of actin among different plant lineages. It might be particularly insightful to compare the previous model bryophyte, Physcomitrella patens, with M. polymorpha. The unique characteristics of Lifeact, particularly its conveniently small size, will facilitate future studies.
Materials and Methods
Plant materials and growth conditions
Seeds of A. thaliana (accession Col-0) were surface-sterilized and plated on Murashige and Skoog medium [× 1 Murashige–Skoog salt mixture, 1% (w/v) sucrose, 0.01% (w/v) myoino-sitol and 0.5% (w/v) gellangum, pH 5.8], incubated for 4 d at 4°C in the dark, and grown in a growth chamber under continuous light at 23°C. Seedlings were transferred to soil and grown in a temperature-controlled (23°C) growth room under continuous light.
Marchantia polymorpha was asexually maintained and propagated through gemma growth as previously described (Okada et al. 2000, Takenaka et al. 2000).
Construction and transformation of plants
The chimeric gene encoding Lifeact–Venus was generated by PCR using the following primers: 5′-CACCATGGGTG TCGCAGATTTGATCAAGAAATTCGAAAGCATCT CAAAGGAAGAAGGCGGCAGCGGCATGGTGAG CAAGGGCGAGGA-3′ and 5′-TTACTTGTACAGCTCGTC CA-3′. The forward primer contained the whole sequence for Lifeact and linker peptides. The PCR product was cloned into the pENTR vector (Invitrogen, Carlsbad, CA, USA), and then transferred by recombination into the pGWB2 binary vector containing the CaMV 35S promoter (a kind gift from T. Nakagawa). The transformation of A. thaliana plants was performed by floral dipping using A. tumefaciens (strain GV3101::pMP90) (Clough and Bent 1998). Transgenic plants with the T-DNA in one locus were selected, and homozygous or hemizygous T2 plants were observed. The transformation of M. polymorpha was performed by the co-cultivation of immature thalli with A. tumefaciens (strain GV3101 : : pMP90) carrying the binary vector as described previously (Ishizaki et al. 2008).
Drug treatments
Young thalli approximately 5 mm at the longest diameter were treated for 2 h at room temperature with 30 mM BDM (Wako Pure Chemical Industries, Osaka, Japan) or 4 μM BA (Sigma) dissolved in water.
Microscopy
For the observation of actin filaments visualized by Lifeact–Venus in A. thaliana and M. polymorpha, samples were observed under a fluorescence microscope (model BX51; Olympus, Tokyo, Japan) equipped with a confocal scanner unit (model CSU10, Yokogawa Electric, Tokyo, Japan) and cooled CCD camera (model ORCA-AG or ORCA-ER, Hamamatsu Photonics, Hamamatsu, Japan). Serial confocal images were obtained every 0.5 μm. Images were processed with the IPLab (BD Biosciences, Rockville, MD, USA) or iVision Mac (BioVision Technologies, Exton, PA, USA) software, and projection images were constructed using VoxBlast (VayTek, Fairfield, IA, USA) or ImageJ software (http://rsb.info.nih.gov/ij/).
For the double labeling of actin filaments with Lifeact–Venus and rhodamine–phalloidin in A. thaliana, 4-day-old A. thaliana plants expressing Lifeact–Venus were fixed with 4% paraformaldehyde in MTSB (50 mM PIPES, 5 mM MgSO4, 5 mM EGTA, pH 7.0) for 1 h at room temperature. Fixed plants were rinsed with wash buffer (0.1% Triton X-100 in MTSB) three times and then with distilled water three times. The samples were then treated with 2% driserase (Sigma) in MTSB for 40 min at room temperature, rinsed with wash buffer three times and treated with MTSB plus 10% dimethylsulfoxide (DMSO) and 3% NP-40 for 1 h. After rinsing with wash buffer, samples were stained with 100 nM rhodamine–phalloidin (Molecular Probes, Eugene, OR, USA) in MTSB for 20 min. After rinsing with wash buffer, samples were mounted on a glass slide in MTSB and observed under a confocal microscope (model LSM510; Carl Zeiss, Tokyo, Japan). Actin filaments in M. polymorpha expressing Lifeact–Venus were double-stained by phalloidin conjugated to Alexa Fluor 568. Young thalli were fixed with 2% glycerol PMEG [50 mM PIPES, 1 mM MgSO4, 5 mM EGTA and 2% (v/v) glycerol, pH 6.8] for 1 h, washed three times in phosphate-buffered saline (PBS), stained with 165 nM Alexa Fluor 568–phalloidin in PBS, and then observed using a confocal microscope (model LSM710, Carl Zeiss, Tokyo, Japan).
Supplementary data
Supplementary data are available at PCP online.
Funding
The Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid for Scientific Research and the Targeted Proteins Research Program (TPRP) (to A.N. and T.U.); Japan Society for the Promotion of Science research fellowship for young scientists (No. 195010 to K.E.).
Acknowledgments
We thank Dr. Tsuyoshi Nakagawa (Shimane University) for providing the pGWB2 vector.
Glossary
Abbreviations:
- ABD
actin-binding domain
- BA
bistheon-ellide A
- BDM
2,3-butanedione-2-monoxime
- CaMV
cauliflower mosaic virus
- F-actin
filamentous actin
- GFP
green fluorescent protein
- PBS
phosphate-buffered saline
References
- Bowman JL, Floyd SK, Sakakibara K. Green genes—comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell. 2004;16:257–269. doi: 10.1105/tpc.016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Funaki K, Nagata A, Akimoto Y, Shimada K, Ito K, Yamamoto K. The motility of Chara corallina myosin was inhibited reversibly by 2,3-butanedione monoxime (BDM) Plant Cell Physiol. 2004;45:1342–1345. doi: 10.1093/pcp/pch154. [DOI] [PubMed] [Google Scholar]
- Gu Y, Fu Y, Dowd P, Li S, Vernoud V, Gilroy S, et al. A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 2005;169:127–138. doi: 10.1083/jcb.200409140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasezawa S, Kumagai F. Dynamic changes and the role of the cytoskeleton during the cell cycle in higher plant cells. Int. Rev. Cytol. 2002;214:161–191. doi: 10.1016/s0074-7696(02)14005-8. [DOI] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Okubo E, Sano T, Hasezawa S. Actin microfilaments regulate vacuolar structures and dynamics: dual observation of actin microfilaments and vacuolar membrane in living tobacco BY-2 cells. Plant Cell Physiol. 2006;47:839–852. doi: 10.1093/pcp/pcj056. [DOI] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Hasezawa S. Quantitative analysis of changes in actin microfilament contribution to cell plate development in plant cytokinesis. BMC Plant Biol. 2008;8:80.. doi: 10.1186/1471-2229-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Sano T, Hasezawa S. Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 2007;10:549–556. doi: 10.1016/j.pbi.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Holweg CL. Living markers for actin block myosin-dependent motility of plant organelles and auxin. Cell Motil. Cytoskel. 2007;64:69–81. doi: 10.1002/cm.20164. [DOI] [PubMed] [Google Scholar]
- Holweg C, Susslin C, Nick P. Capturing in vivo dynamics of the actin cytoskeleton stimulated by auxin or light. Plant Cell Physiol. 2004;45:855–863. doi: 10.1093/pcp/pch102. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006;57:109–125. doi: 10.1146/annurev.arplant.57.032905.105206. [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 2008;49:1084–1091. doi: 10.1093/pcp/pcn085. [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Anthony RG, Hussey PJ. Green fluorescent protein–mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor’s actin depolymerizing activity in vitro. Plant Physiol. 2004;136:3990–3998. doi: 10.1104/pp.104.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Emons MC. The cytoskeleton in plant cell growth: lessons from root hairs. New Phytol. 2001;152:409–418. doi: 10.1046/j.0028-646X.2001.00278.x. [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP–mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003;15:1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Miller DD, de Ruijter NCA, Bisseling T, Emons AMC. The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 1999;17:141–154. [Google Scholar]
- Okada S, Fujisawa M, Sone T, Nakayama S, Nishiyama R, Takenaka M, et al. Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J. 2000;24:421–428. doi: 10.1046/j.1365-313x.2000.00882.x. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 2004;136:3968–3978. doi: 10.1104/pp.104.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmenm T, Yokota E. Physiological and biochemical aspects of cytoplasmic streaming. Int. Rev. Cytol. 1994;155:97–139. [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L. Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 2009;184:269–280. doi: 10.1083/jcb.200806185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Yamaoka S, Hanajiri T, Shimizu-Ueda Y, Yamato KT, Fukuzawa H, et al. Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L. Transgenic Res. 2000;9:179–185. doi: 10.1023/a:1008963410465. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Yokota E, Sonobe S, Shimmen T. Mechanism of inhibition of cytoplasmic streaming by a myosin inhibitor, 2,3-butanedione monoxime. Protoplasma. 2000;213:46–54. [Google Scholar]
- Voigt B, Timmers AC, Samaj J, Muller J, Baluska F, Menzel D. GFP–FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur. J. Cell Biol. 2005;84:595–608. doi: 10.1016/j.ejcb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, et al. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP–actin fusion protein. Curr. Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Kutsuna N, Higaki T, Oda Y, Sano T, Hasezawa S. Recent progress in living cell imaging of plant cytoskeleton and vacuole using fluorescent-protein transgenic lines and three-dimensional imaging. Protoplasma. 2007;230:129–139. doi: 10.1007/s00709-006-0237-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.