Summary
CTCF is a conserved transcriptional regulator with binding sites in DNA insulators identified in vertebrates and invertebrates. The Drosophila Abdominal-B locus contains CTCF binding sites in the Fab-8 DNA insulator. Previous reports have shown that Fab-8 has enhancer blocking activity in Drosophila transgenic assays. We now confirm the enhancer blocking capability of the Fab-8 insulator in stably transfected Drosophila S2 cells and show this activity depends on the Fab-8 CTCF binding sites. Furthermore, knockdown of Drosophila CTCF by RNAi in our stable cell lines demonstrates that CTCF itself is critical for Fab-8 enhancer blocking.
Keywords: DNA insulator, Fab-8, CTCF, RNAi, S2 cells
DNA insulators are thought to help partition and/or maintain eukaryotic genomes into transcriptionally active and inactive domains, and prevent the status of one domain from affecting the other domain. Based on this idea, regulatory elements are classified as DNA insulators if they either suppress position effects when flanking a gene, i.e. barrier activity, or if they block an enhancer when placed between the enhancer and gene promoter. The molecular basis for this functional definition has been assigned for several DNA insulators. For instance, the Drosophila DNA insulators scs/scs’ and gypsy require proteins ZW5 and BEAF-32, and SU(HW) and MOD(mdg4), respectively 1–4. Roles for homologs of these trans factors have not been defined at vertebrate insulators. However, a consistent molecular component among vertebrate insulators is the presence of binding sites for CTCF 5. CTCF is a highly conserved, ubiquitously expressed transcription factor that binds different DNA sequences using different combinations of its eleven Zn-fingers 6,7. CTCF was first linked to insulator function when it was shown to interact with the well-characterized insulator from the chicken β-globin locus, cHS4 5. Interestingly, cHS4 can function as an insulator in transgenic Drosophila 8. Because cHS4 can function in an invertebrate, it was predicted that insulator factors would be conserved among different species. Indeed, CTCF sites were identified in the Drosophila insulator Fab-8 that resides at the Abdominal-B locus 9–11. Fab-8’s role at the Abd-B locus is to specify expression of the Abd-B gene in the proper segment of Drosophila embryos by insulating the cis-regulatory element iab-7 from iab-8 10. In Drosophila transgenic assays and mammalian cell lines, the enhancer blocking activity of Fab-8 depends on CTCF binding sites 11. This observation suggests that CTCF could be a core component of a conserved insulator complex.
While CTCF binding sites have been shown to be important for enhancer blocking, depletion of CTCF has been studied in other biological processes. DNA methylation status of the H19 differentially methylated domain is disrupted and developmental potential is decreased in oocytes deficient for CTCF 12. Interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1 is disrupted and expression of Wsb1/Nf1 is decreased when CTCF is knocked-down 13. A recent report also described a failed enhancer blocking phenotype after CTCF knockdown 14. Chen et al. used an elegant dual fluorescent reporter in a transient transfection system to demonstrate that an 800 bp region from the latency-associated transcript (LAT) intron of the Herpes Virus-1 genome, containing multiple CTCF binding sites, can block enhancer activity on a downstream reporter. DsRNA against CTCF abrogates enhancer blocking activity on their transient reporter. However, transient transfection-based assays fail to account for the possible influence of local chromatin structure on insulator activity. We generated Drosophila S2 cells with stably integrated reporter constructs that contained variants of the Fab-8 enhancer blocking fragment. We report the role of CTCF in Fab-8’s ability to block an enhancer on our stably integrated reporter.
To test the enhancer blocking activity of the Fab-8 insulator from the Abdominal-B locus in Drosophila S2 cells, stably transfected polyclonal cell lines were established with the fluorescent reporter constructs diagrammed in Figure 1. The constructs use an EGFP reporter driven by the OpIE2 enhancer (denoted enh), which is commonly employed in insect expression vectors. A 550bp fragment of the Fab-8 insulator (sequence is listed in Supplementary Material) was placed upstream of OpIE2 enhancer and the EGFP gene in the F8enhGFP construct. An additional copy of the same Fab-8 insulator fragment was placed between the OpIE2 enhancer and the EGFP gene in the F8enhF8GFP construct. Finally, in the F8enhmutF8GFP construct, a Fab-8 insulator carrying mutations in the two CTCF binding sites, described by Moon et al. 11, was placed between the OpIE2 enhancer and the EGFP gene. All constructs contained the Fab-8 insulator upstream of the OpIE2 enhancer to prevent potential activation of a neighboring EGFP gene in integrated transgene arrays.
Fig. 1.

Fab-8 insulator blocks the OpIE2 enhancer in Drosophila S2 cells. Diagrams at left depict constructs diagrammed between parentheses. The OpIE2 enhancer and Fab-8 insulator were amplified from plasmid pIZ/V5-EGFP (Invitrogen) and a fly lysis preparation, respectively, using primers listed in supplemental data. The OpIE2 enhancer and Fab-8 insulator were cloned into a pBluescript plasmid upstream of a minimal promoter EGFP SV40 pA fragment, which was amplified from plasmid pIZ/V5-EGFP (Invitrogen) using the primers listed in supplemental data. The black wavy line represents genomic DNA surrounding the site of integration. The “n” signifies that at the site of integration there may be multiple copies of the reporter construct in our stable polyclonal cell lines. Stable lines were made by co-transfecting Drosophila S2 cells, that were cultured at room temperature in Schneider’s medium (Invitrogen) with 10% FBS, 100 units penicillin, and 100 μg streptomycin, in a 6 well dish with 1 μg of reporter plasmid DNA and 250 ng of pCoHygromycin plasmid DNA. Four days after transfection the cells were passaged and hygromycin was added to the cultures at a final concentration of 500 μg/ml. Cells were maintained in selection for 6 weeks. Panels a, b, and c are fluorescent images, taken with the same exposure settings, that show GFP expression in the stably transfected cell lines. Inserts are bright field images of the same field showing similar cell densities.
After selection, fluorescence microscopy was used to analyze EGFP expression in the three stably transfected S2 cell lines. In the cell line lacking the intervening Fab-8 insulator, construct F8enhGFP, we observed robust GFP expression (Figure 1, panel a). Flow cytometry analysis of this cell line revealed that ~30% of the cells are GFP positive. There are at least two possible explanations for why all cells are not GFP positive. First, prior to integration, the F8enhGFP reporter plasmid may have been cleaved in sequences critical for EGFP expression. Second, the surrounding chromatin at some of the integration sites may repress the transgene expression and CTCF sites alone are not sufficient to prevent position effects 15.
In F8enhF8GFP cells that contain the Fab-8 insulator between the OpIE2 enhancer and EGFP gene, the number of GFP positive cells and the fluorescence intensity is markedly decreased (Figure 1, compare panels a and b). Northern analysis confirmed that the decrease in GFP fluorescence is due to decreased GFP message in the F8enhF8GFP cell line compared to F8enhGFP cell line (Supplementary Figure 1). The decrease in the number of GFP positive cells and GFP expression is likely due to Fab-8 blocking the enhancer on a stably integrated reporter construct, rather than a difference in copy number between the F8enhGFP transgene and the F8enhF8GFP transgene (Supplementary Figure 2). The reduction in GFP positive cells and GFP expression in the F8enhF8GFP cell line is consistent with previous reports that describe Fab-8 as an enhancer blocker 9–11, and indicates that S2 cells with a stably integrated reporter construct are a suitable model to investigate the molecular basis for enhancer blocking.
The Fab-8 insulator is an excellent reagent for the investigation of the molecular basis of enhancer blocking because binding sites for CTCF have been shown to be critical for Fab-8’s enhancer blocking capability. Analyzing the effect of CTCF binding site mutations in the Fab-8 insulator is an additional test of our model system. In the F8enhmutF8GFP cell line, containing an intervening Fab-8 insulator with mutated CTCF sites, flow cytometry analysis revealed that the percentage of GFP positive cells was similar to the F8enhGFP cell line. However, while the percentage of fluorescent cells was comparable, the level of GFP fluorescence, as judged by flow cytometry and fluorescence microscopy, was not equivalent to the level observed in the F8enhGFP cell line (Figure 1, compare panels a and c). The decreased fluorescence intensity may suggest that either the OpIE2 enhancer may be sensitive to distance from the minimal promoter, or factors other than CTCF can bind the mutated Fab-8 insulator and block the enhancer. In either case, the EGFP expression from the F8enhmutF8GFP cell line demonstrates that Fab-8’s enhancer blocking activity depends on the binding sites for CTCF.
To confirm that Drosophila CTCF (dCTCF) is indeed responsible for Fab-8’s enhancer blocking activity we knocked down dCTCF in our stable cell lines using RNAi. Results from three experiments indicate that our stable cell lines are competent for RNAi. First, treating our cells with a dsRNA against Thread/Diap1, an anti-apoptosis gene 16, induced dramatic cell death (data not shown). Second, treating our cells with a dsRNA against the EGFP gene results in a significant decrease in the percentage of GFP positive cells and GFP fluorescence levels (Figure 2A compare panels a to e, and b to f; Figure 2B). The level of EGFP knockdown is comparable to previous reports 17. Finally, Northern analysis showed that after treating cells with dsRNA targeted to dCTCF mRNA, dCTCF mRNA was drastically reduced (Figure 2C). The smear in the lower portion of the gel represents the dsRNA. It is present in only two of the four lanes treated with dsRNA against dCTCF because the probe only overlaps with the dsRNA amplicon dCTCF1 and not dCTCF2.
Fig. 2.

Knockdown of Drosophila CTCF reduces enhancer blocking activity of Fab-8. DNA templates for dsRNA were amplified using the primers listed in supplemental data. Two dsRNA templates were amplified from dCTCF cDNA: dCTCF1 spanned nucleotides 755 to 1051 and dCTCF2 spanned nucleotides 1452 to 1954 of the dCTCF cDNA. DsRNA was produced using Promega T7 RiboMax Express RNAi System. RNAi was performed as previously described 19. Briefly, 1 million cells were incubated in 1 ml of serum free medium with 20 μg of dsRNA. After 30 minutes 1 ml of serum containing medium was added. Three or four days later the cells were analyzed by fluorescence microscopy and FACS. Fluorescent images of live cells at similar cell densities were captured with a Leica DML fluorescence microscope and SPOT RT software using auto exposure setting to capture the images of cells with the F8enhGFP construct. This exposure time was used to capture subsequent images of cells with other constructs or dsRNA treatments. The 8-bit grayscale images were pseudo-colored and the dynamic range adjusted to the same levels with SPOT RT software. Single cell suspensions of live stably transfected S2 cell lines were analyzed with a DakoCytomation, Inc. CyAn ADP flow cytometer. Percent of GFP positive cells and GFP fluorescence intensity were determined by analyzing histograms with DakoCytomation Summit software version 4.3. Statistics for the percentage of GFP positive cells and mean level of GFP fluorescence were performed using a paired Student’s t-test. RNA was extracted from stably transfected S2 cell lines using Trizol reagent. 5 μg of total RNA was separated by formaldehyde-agarose gel electrophoresis, transferred to Hybound-N nylon membrane from Amersham, and probed with 32P labeled dCTCF fragment. The dCTCF fragment used as a probe was amplified using the primers listed in supplemental data; it spanned nucleotides 363 to 861 of the dCTCF cDNA. The probed membrane was exposed to a Molecular Dynamics phosphor imager screen and scanned with an Amersham Typhoon variable mode imager. A. Fluorescent images of F8enhGFP and F8enhF8GFP cell lines, taken with the same exposure settings, show GFP expression after treatment with dsRNAs indicated to the left of images. Panels a, c, e, and g are of cell line F8enhGFP following mock dsRNA treatment or treatment with dsRNAs against Drosophila CTCF (dCTCF), EGFP, or mouse Ctcf (mCtcf) mRNAs, respectively. Panels b, d, f, and h are of cell line F8enhF8 GFP following treatment identical to cell line F8enhGFP. B. Flow cytometry analysis used to determine percentage of GFP positive cells and the mean GFP fluorescence is summarized from multiple independent RNAi experiments. Solid bars represent cell line F8enhGFP; open bars, F8enhF8GFP. Values for the mock treated cells were set at one. The number of independent experiments for different dsRNAs are as follows: mock=5, dCTCF1=5, dCTCF2=3, EGFP=4, mCtcf=2. The percentage of GFP positive cells and mean level of GFP fluorescence are significantly different in F8enhF8GFP cell line treated with dsRNA against dCTCF compared to the non-treated cell line with the following p-values: *0.0065, #0.0029, §0.0037, and ‡0.024. C. Northern blot shows dCTCF mRNA (2.87kb) from F8enhGFP and F8enhF8GFP cell lines 4 days after mock dsRNA treatment, treatment with dsRNA dCTCF1, and dsRNA against mouse Ctcf mRNA (mCtcf). The ethidium bromide stained gel, showing the 18S and processed 28S rRNA bands, indicates that total RNA loaded was similar for each sample.
The effect of the loss of dCTCF on Fab-8 function was determined by analyzing GFP levels with fluorescence microscopy and flow cytometry. In the absence of dsRNA, the Fab-8 insulator effectively blocks GFP expression as GFP fluorescence is much weaker in the F8enhF8GFP cell line (Figure 2A, panel b) compared to the F8enhGFP cell line (Figure 2A, panel a). After incubating cells with a dsRNA against dCTCF mRNA, the number of GFP positive cells and the level of GFP fluorescence is clearly increased in the F8enhF8GFP cell line (Figure 2A, panel d). Flow cytometry analysis of 5 separate RNAi experiments confirmed this result. The number of GFP positive cells increased nearly 2-fold and the level of GFP fluorescence increased nearly 3-fold in the F8enhF8GFP cell line following treatment with dsRNA against dCTCF mRNA compared to untreated cells (Figure 2B). Confirming the observed increase in GFP fluorescence, northern analysis revealed that the level GFP mRNA increased following treatment with dsRNA against dCTCF (Supplementary Figure 1). This is consistent with dCTCF acting at the level of transcriptional enhancers.
Interestingly, the F8enhGFP cell line also responded to treatment with dsRNA against dCTCF mRNA in a distinct and reproducible manner. The number of GFP positive cells in the F8enhGFP cell line is unaltered regardless of dsRNA treatment, but after treatment with dsRNA against dCTCF mRNA the level of GFP fluorescence increased nearly 1.5-fold (Figure 2B). The increased GFP fluorescence may be explained by the fact that stable S2 cell lines have multiple copies of the reporter construct tandemly integrated 18. In this transgene array, the absence of dCTCF renders the upstream Fab-8 insulator incapable of blocking the OpIE2 enhancer from activating in a bidirectional manner. Thus, an EGFP gene can be activated by flanking enhancers.
The specificity of the effect of CTCF knockdown is demonstrated by three experiments. First, in cell lines with a reporter construct where the Fab-8 insulator was replaced with the gypsy insulator--which does not contain CTCF binding sites--treatment with a dsRNA against dCTCF did not increase EGFP expression (Supplementary Table 1). Second, treatment with dsRNA designed against a different region of the dCTCF mRNA resulted in decreased dCTCF mRNA and increased GFP fluorescence similar to dsRNA dCTCF1 (Figure 2B and 2C, dCTCF2). Third, cells treated with a dsRNA directed against mouse Ctcf mRNA (mCtcf) had virtually the same GFP fluorescence compared to untreated cells (Figure 2A panel g, h; 2B, mCtcf). The dsRNA amplicon against mCtcf is only 53.3% identical to dCTCF mRNA with no more than 9 continuously identical nucleotides. Northern analysis revealed that the amount of dCTCF mRNA was unaltered in cells treated with dsRNA directed against mCtcf (Figure 2C). In summary, the data presented in Figure 2 on the loss of function analysis of dCTCF demonstrate the requirement of dCTCF for Fab-8’s enhancer blocking activity.
If dCTCF is required for Fab-8 to block an enhancer, we would predict that restored levels of dCTCF would restore the enhancer blocking activity. We tested this by measuring dCTCF mRNA and reanalyzing GFP expression 10 days after treatment with dsRNA dCTCF1 (6 days after analyzing GFP expression). Northern analysis revealed that dCTCF mRNA levels returned to normal 10 days after treatment with dsRNA dCTCF1, when the dsRNA dCTCF1 had declined and was no longer detectable (Figure 3B). This is consistent with a published report that showed knocked down targets begin to recover 6 days after treatment with dsRNA 19. Importantly, 10 days after administering dsRNA dCTCF1, and concomitant with the restored levels of dCTCF mRNA, the level of GFP fluorescence in the F8enhF8GFP cell line declined to levels nearly equivalent to those prior to dsRNA treatment (Figure 3A, panel f). This provides further evidence that dCTCF is required for Fab-8’s insulator function.
Fig. 3.
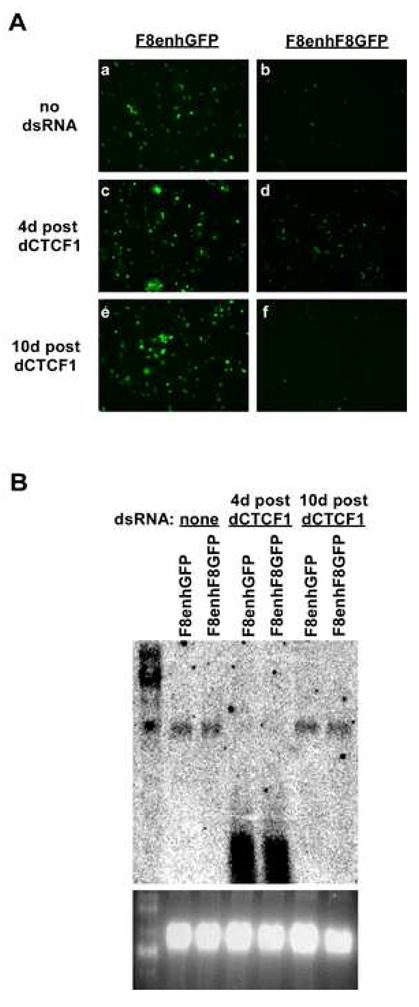
Recovery of dCTCF mRNA restores Fab-8 enhancer blocking. A. Fluorescent images of F8enhGFP and F8enhF8GFP cell lines show GFP expression at two time points after treatment with dsRNA against dCTCF mRNA. Panels a, c, and e are of cell line F8enhGFP 4 days after mock dsRNA treatment, 4 days after treatment with dsRNA dCTCF1, and 10 days after treatment with dsRNA dCTCF1, respectively. Panels b, d, and f are of cell line F8enhF8 GFP following treatment identical to cell line F8enhGFP. All images were taken with the same exposure settings. B. Northern blot shows levels dCTCF mRNA from F8enhGFP and F8enhF8GFP cell lines 4 days after mock dsRNA treatment, 4 days after treatment with dsRNA dCTCF1, and 10 days after treatment with dsRNA dCTCF1. The ethidium bromide stained gel, showing the 18S and processed 28S rRNA bands, indicates that total RNA loaded was similar for each sample.
While our loss of function analysis of dCTCF confirms the data previously reported by Chen et al. 14, using a transiently transfected dual fluorescent reporter, our experimental design is distinct because we used an endogenous Drosophila insulator stably integrated in the Drosophila genome. Both systems are amenable to RNAi screens for other insulator or dCTCF interacting factors. The screens should confirm each other’s results, and the distinctions in the reporter design could potentially yield different targets, as there may be additional or different factors required to block an enhancer in a chromosomal context.
Supplementary Material
Acknowledgments
We would like to thank Eric Wagner for providing plasmid pIZ/V5-EGFP and Robert Duronio for fly DNA. We are grateful to Greg Rogers and members of the Magnuson laboratory for comments and suggestions. This work was supported by NIH grants to T.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Modolell J, Bender W, Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983;80:1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgiev PG, Gerasimova TI. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet. 1989;220:121–126. doi: 10.1007/BF00260865. [DOI] [PubMed] [Google Scholar]
- 3.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 5.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 6.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 7.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development. 1999;126:3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]
- 10.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, et al. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 11.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 13.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Lin L, Smith S, Huang J, Berger S, et al. A CTCF-dependent chromatin boundary element exists between the LAT and ICP0 promoters in the HSV-1 genome. J Virol. 2007 doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 17.Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene. 2000;252:95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- 18.Johansen H, van der Straten A, Sweet R, Otto E, Maroni G, et al. Regulated expression at high copy number allows production of a growth-inhibitory oncogene product in Drosophila Schneider cells. Genes Dev. 1989;3:882–889. doi: 10.1101/gad.3.6.882. [DOI] [PubMed] [Google Scholar]
- 19.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


