Abstract
The tumor suppressor p53 regulates genes involved in progression through the cell cycle, DNA repair, senescence or apoptosis in response to cell stress. Dysregulation of p53 can result in uncontrolled cellular proliferation. Invertebrate homologues to human p53 (Hsp53) have been identified, including a putative p53 gene (Map53) from the soft-shell clam (Mya arenaria). Predicted sequences for human and clam p53 proteins exhibit conservation in key domains. In light of this similarity, and the apparent dysregulation of Map53 under morphologically aberrant/pathologic conditions, we tested the hypothesis that the two proteins function in a similar manner. Plasmids expressing either Hsp53 or Map53 were introduced by transient transfection into the p53-null H1299 cell line. Functionality was assessed by monitoring the p53/mdm2 feedback loop and expression of p53-mediated downstream markers of growth arrest and apoptosis under non-stressed conditions. Hsp53 spontaneously induced markers of growth arrest, while Map53 expression induced neither cell arrest nor apoptosis. The difference in downstream activation is not likely the result of cytosolic sequestration since Map53, like Hsp53, localized almost exclusively to the nucleus. Functional similarity was observed in regulation by human MDM2, suggesting that the clam may have an mdm2 homologue. Protein modeling identified an apparent MDM2 binding site in Map53, supporting the observation of a potential Map53/MDM2 interaction. Significant amino acid differences present in the Map53 tetramerization domain may potentially affect p53 protein/protein interactions. Taken together, these data suggest that the Map53 shares some functional similarity with human p53 as well as with other invertebrates, positioning the mollusk at a critical juncture in evolution of this gene family.
Keywords: Mdm2, apoptosis, cell cycle arrest, bivalve, cancer, post-translational modification
1. Introduction
The mammalian tumor suppressor p53 is one of the most intensely investigated genes due to its dysregulation in the majority of human cancers (Vousden and Lane, 2007). P53 protein is a sequence-specific transcription factor best known for its role in maintaining genetic integrity following cellular stress, particularly in response to DNA damage. Under such conditions, p53 is stabilized by post-translational modifications (PTM), leading to nuclear accumulation and altered transcriptional activity. P53 binds to the p53-responsive elements of many target genes, thereby initiating cellular responses that include growth arrest and DNA repair, senescence or apoptosis. It remains unclear what actions or precursors trigger growth arrest versus apoptosis: tissue specificity, type and extent of DNA damage, post-translational modifications and other physiological conditions are hypothesized to play a role (Vousden, 2002; Sykes et al., 2006). Cells may remain viable following p53-initiated cell cycle arrest and DNA damage repair at the G1/S phase checkpoint (Venot et al., 1998). In this scenario, p53 acts as a transcription factor for p21(WAF1/CIP1), a cyclin-dependent kinase inhibitor capable of blocking cell cycle progression (El-Deiry et al., 1993). In the event DNA damage is too extensive for repair, cells may enter p53-mediated apoptosis, which involves activation of pro-apoptotic genes including members of the caspase family (Agarwal et al., 1998; Shengkan and Levine, 2001). This mechanism of action is crucial for resistance to neoplasia in humans.
In the absence of cellular stress, p53 protein is maintained at low levels by the E3 protein-ubiquitin ligase, MDM2 (Alarcon-Vargas and Ronai, 2002; Oren, 2003). This effector protein modulates p53 levels through a negative feedback loop by both physically blocking the p53 transactivation domain and by targeting it for ubiquitin-mediated degradation (Moll and Petrenko, 2003). Constitutive expression of mdm2 in cell culture also appears to exhibit this feedback mechanism. In vivo, overexpression of mdm2 can inhibit the ability of p53 to arrest cells at the G1/S checkpoint in response to ionizing radiation. Mdm2 overexpression has also been observed in human tumors that lack inactivating mutations in p53 (Perry, 2004).
A cDNA sequence with significant similarity to human p53 (Hsp53) in key functional domains was previously described from the soft-shell clam, Mya arenaria (Map53; Kelley et al., 2001). Conservation of functional domains, altered expression and sub-cellular localization of this purported p53 in neoplastic clam tissues (Kelley et al., 2001; Butler et al., 2004) suggest functional homology to mammalian p53. At least two types of neoplasia have been identified in M. arenaria: a germinoma (gonadal tumor; Barry and Yevich, 1975) and an unrelated proliferative hemocytic disease/disseminated neoplasia (reviewed in Barber, 2004). Previous studies in this laboratory have shown a significant decrease in Map53 expression in neoplastic clam gonadal tissue compared to non-neoplastic tissue (Butler et al., 2004). In addition, clams exposed to the herbicide 2,4-dichlorophenoxyacetic acid exhibited a significant decrease in p53 protein expression concomitant with an altered tissue morphology in reproductive tissue (unpublished). An apparent dysregulation of Map53 also occurs in the clam disseminated neoplasia, where neoplastic hemocytes exhibit aberrant cytosolic localization of p53 (Kelley et al., 2001; Walker et al., 2006, 2008). This was recently confirmed by Böttger et al. (2008).
In light of the sequence similarities between Hsp53 and Map53, and the apparent dysregulation of Map53 under morphologically aberrant/pathologic conditions, we hypothesized that the two proteins function in a similar manner. This hypothesis was tested by transient transfection of a p53-null, human non-small-cell-lung-carcinoma cell line (H1299) with plasmids expressing either human or clam p53. Function was assessed by monitoring the p53/Mdm2 feedback loop and expression of p53-mediated downstream markers of growth arrest and apoptosis.
2. Materials and methods
2.1 Expression Vector Construction
Wild-type Hsp53 (GenBank accession_no. AF192534), kindly provided by Drs. B. Vogelstein and K.W. Kinzler, and Map53 (GenBank accession no. AF253323; Kelley et al., 2001) were cloned into pcDNA3.1/His (Invitrogen) for constitutive expression of N-terminally tagged (Xpress™) fusion proteins. All nucleotide sequences were confirmed (University of Maine DNA Sequencing Facility).
2.2 Transient transfection of H1299 Cells
H1299 cells (ATCC; passages 49 and 50) were grown at 37°C, 5% CO2 in RPMI-1640 medium with 2 mM L-glutamine (Gibco/Invitrogen) and 10% fetal bovine serum (Hyclone). Six-well plates were seeded with 3.5 × 105 cells per well and transfected 24 hours later (80% confluency) at an empirically optimized ratio of 2 μg DNA: 7 μL Lipofectamine 2000 (Invitrogen). Controls were H1299 cells treated with media alone (untreated cells) or transfected with the pcDNA3.1/His vector containing no insert (vector alone).
2.3 Western blot analyses
Cells were harvested following standard trypsinization procedures. Cells were homogenized by three freeze-thaw cycles as described (Kelley et al., 2001) with the addition of Complete Protease Inhibitor Cocktail (Roche). Cytosolic and nuclear fractions were collected for p53 sub-cellular localization as per manufacturer’s directions (Active Motif). Protein concentrations were determined by Bradford assay (BioRad).
Proteins were separated by SDS-PAGE (10% or 12% Bis-Tris gels, 1x MES running buffer; Invitrogen) then transferred to a PVDF membrane (Millipore) and incubated with one of the following primary antibodies: p53 (1:5000; α-Xpress; Invitrogen), p21WAF1/CIP1 (1:333; Calbiochem), pRb (1:1000; Santa Cruz), Caspase-3 (1:1000; Cell Signaling Technologies), human Mdm2 (1:600; Santa Cruz), or Hsp90 (1:1000; Cell Signaling Technologies). Appropriate horseradish peroxidase-conjugated secondary antibodies (1:3000; Cell Signaling Technologies) were used, followed by chemiluminescent detection (ECL Western Blotting Substrate or SuperSignal ELISA Pico chemiluminescent substrate; Pierce). Protein band densities captured on Hyperfilm MP (GE Health Sciences) were analyzed using either UNSCAN-IT™ (Silk Scientific Corporation) or Image J (NIH).
2.4 Flow cytometry
H1299 cells were harvested at given times post-transfection and fixed in ice-cold 70% ethanol. Cells were counted (Beckman-Coulter Automated Cell Viability Counter), washed, treated with RNAse A, then stained with propidium iodide. Samples were analyzed using a Beckman Dickinson Fluorescence-Activated Cell Sorter (FACS Calibur) with Cell Quest software (Cellular and Molecular Imaging Core Facility, University of Southern Maine). Cell cycle distribution was determined using WinMDI Version 2.8 software (Joseph Trotter, Scripps Research Institute, San Diego, CA).
2.5 RNAi knockdown of Mdm2
The role of Mdm2 in regulation of p53 protein levels was investigated using human Mdm2 RNAi (hdm2; Ambion). H1299 cells were transfected with either Map53 or Hsp53 as previously described. Mdm2 RNAi was introduced 4 hours later using Transmessenger Reagent (Qiagen) per manufacturer’s protocol, at a ratio of 2 μg RNAi:10 μL reagent. Cells were harvested at 24, 36, and 48 hours post-transfection and examined for expression of p53 and Mdm2 mRNA and protein. cDNA was generated using random hexamer and oligo d(T15) primers followed by sequence-specific amplification using p53, Mdm2 or β-actin primers. Densitometry was performed on digital images of ethidium bromide-stained agarose gels using Image J (NIH). Relative sequence-specific mRNA expression was based upon p53 or Mdm2 band intensity relative to β-actin. Protein expression was determined by western blotting as described above.
2.6 Molecular modeling
The tetramerization domain of clam Map53 was modeled with Insight II® software based on the reported homologous structure of the Hsp53 tetramerization domain (PDB, 1SAK; Clore et al., 1995). Side chains were altered to conform to the clam predicted protein sequence while maintaining the backbone structure of the human protein. No further optimizations were done. Comparisons of the putative Map53 MDM2 binding site were made utilizing the Map53 aligned sequence and the coordinates of the Hsp53 interaction with the human MDM2 protein (PDB 1YCR; Kussie et al., 1996). Figures were generated utilizing either RasMol (Sayle and Milner-White, 1995) or MolScript (Kraulis, 1991) modeling programs.
2.7 Statistical analyses
Analysis of variance (ANOVA) was performed on all data sets (SYSTAT 11 Software, Inc., 2004). Transformations were performed on the normalized p53 and p21WAF1/CIP1 western blot expression data (4th root and square root, respectively), and on cells in the sub-G1 state as determined by flow cytometry (log transformation). Treatment represents the transfected plasmid or type of control (i.e., Hsp53, Map53, vector alone or untreated cells). Statistically significant (p< 0.05) time, treatment, or interaction led to a Tukey HSD post hoc analysis (p< 0.05).
3. Results
3.1 Significant differences in expression were observed between Hsp53 and Map53 in H1299 cells
Doubling time for H1299 cells was 30.0 ± 2.2 hours, comparable to that reported by Suzuki et al. (2004). H1299 cells that expressed Hsp53 showed little growth over the time course of the study (doubling time > 96 hours). Map53-expressing and vector-alone control cells exhibited similar growth dynamics compared with H1299 cells up to 36 hours post-transfection, but thereafter slowed to doubling times of 46.4± 4.1 and 46.3 ± 4.0 hours, respectively (Fig 1). The effect of the vector alone on cell growth was unexpected and remains unexplained. Lipofectamine treatment alone had no measurable effect on either cell growth or p53 expression.
Fig. 1.
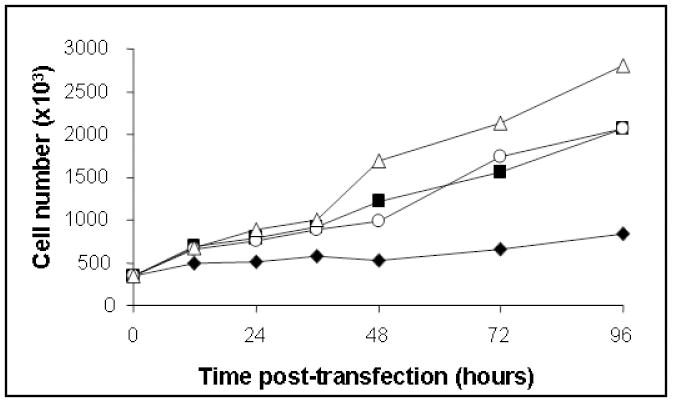
Representative growth curves show that Hsp53 expression significantly increases doubling time of H1299 cells (estimated at 108.1 hr). Untreated H1299, (-Δ-); vector alone-H1299, (-○-); Map53-H1299, (-■-); and Hsp53-H1299 cells, (-◆-). Values are expressed as the mean ± standard error; N = 3.
Significant differences in relative expression of Hsp53 and Map53 proteins were observed at all time points following transient transfection (p< 0.05; Fig 2A, B). Typically, Map53 exhibited maximal expression 24-48 hours post-transfection as compared with 12-36 hours post-transfection for Hsp53. Despite differences in the level of p53 protein expression, Map53 and Hsp53 mRNA levels were not significantly different (Fig 2C). Comparable levels of clam and human p53 proteins were achieved by decreasing the amount of Hsp53 expression plasmid transfected into the H1299 cells. Regardless of the amount of Hsp53 protein expressed, however, the same magnitude of a downstream marker, p21WAF1/CIP1 (see section 3.3), was elicited (Fig. 3). Therefore, we suggest that the low level of Map53 protein expressed should have been sufficient to elicit the p21-mediated growth arrest pathway
Fig. 2.
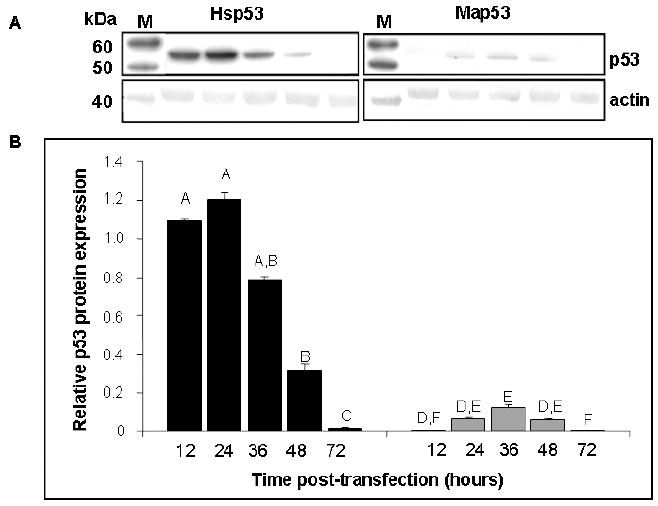
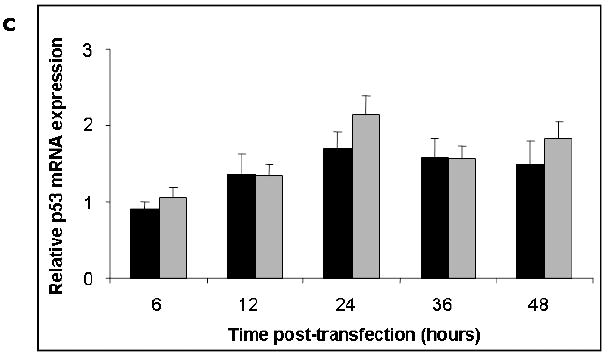
Hsp53 and Map53 protein expression in H1299 cells differ in time and magnitude although mRNA levels do not. (A) Representative western blots illustrate the dramatic difference in levels of Hsp53 and Map53 expression in H1299 cells; M, marker lane. (B) Densitometry analysis shows that at 12 to 48 hours post-transfection, average p53 protein expression is approximately 10-fold higher in Hsp53-H1299 cells (black bars) relative to Map53-H1299 cells (gray bars). Within a treatment, same letters denote no significant difference. (C) There are no significant differences in p53 mRNA levels over time, or between Hsp53-H1299 cells (black bars) or Map53-H1299 cells (gray bars). Values are expressed as the mean ± standard error; N=3.
Fig. 3.
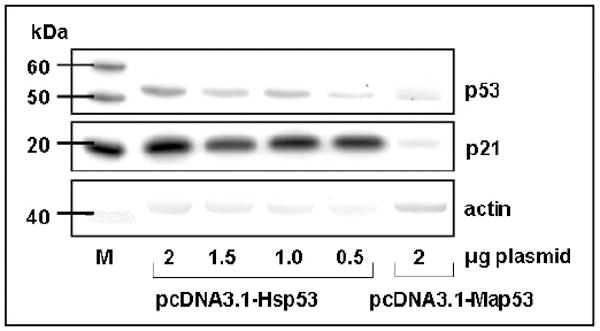
Expression of p21 protein is not altered in response to a range of Hsp53 expression levels. Western blot analysis illustrates a pcDNA3.1-Hsp53 titration series to obtain a level of Hsp53 protein expression comparable to that observed for pcDNA3.1-Map53 (0.5μg and 2μg plasmid, respectively). M, marker lane.
3.2 Map53 functions in the MDM2 feedback loop
The difference in magnitude of Map53 and Hsp53 protein expression in H1299 cells is striking; however, the reason remains unclear. One hypothesis is that turnover of Map53 is accelerated within this heterologous system. The E3 ubiquitin ligase, MDM2, is critical for maintaining low p53 levels under non-stress conditions through an autoregulatory negative feedback loop (Moll and Petrenko, 2003). As mdm2 is a transcriptional target of p53, the introduction of Hsp53 into the p53-/- H1299 cells resulted in an expected up-regulation of MDM2 (HDM2, the human homologue of mouse MDM2) expression: approximately 3-fold increase in mRNA and a 2-fold increase in MDM2 protein (data not shown). Map53 expression induced expression of Mdm2 to approximately the same level as did Hsp53, suggesting Map53 might also participate in a MDM2 feedback loop. To test this hypothesis, we targeted the knock down of mdm2 using RNA interference. MDM2, Hsp53, and Map53 protein levels were monitored 24, 36 and 48 hours following addition of mdm2 RNAi. All cells showed a 60-80% reduction of MDM2 protein at 24 hours post-RNAi transfection relative to cells that did not receive RNAi (Fig. 4). Following a 12-hour lag time, maximal upregulation of Hsp53 and Map53 was observed (Fig. 4). Participation of Map53 in a feedback loop with human MDM2 is supported by structural comparisons. A minimal MDM2-binding site on human p53 has been established by site-directed mutagenesis (Lin et al., 1994) and immunochemical analysis (Picksley et al., 1994). Three critical residues in this site (F19, L22, W23) are conserved in Map53 (Fig. 5). While protein/protein interactions appear to be maintained, other substitutions in this site suggest the possibility of altered regulation of the Map53/MDM2 feedback loop. Specifically, in Map53, glutamic acid and tyrosine are substituted for S20 and D21, respectively, found in Hsp53. Phosphorylation of S20 is a critical modulator of Hsp53 activity, presumably by inhibiting the MDM2/Hsp53 interaction, thus stabilizing the p53 protein (Unger et al., 1999a). Studies have shown that substitution of S20 by alanine or aspartic acid destroys the critical phosphorylation site, thus enhancing MDM2-mediated p53 degradation and reducing the apoptotic response (Unger et al., 1999 a,b; Chehab et al., 1999). The absence of this serine residue may increase Map53 susceptibility to turnover by human MDM2 in a human cell background. It is interesting to note that this specific region of the Map53 MDM2 binding site maintains both a potential phosphorylation site (Y21) and a negatively charged residue (E20), similar to that in the Hsp53 (S20, D21; Fig.5). In the absence of any direct supporting evidence, it is suggested that phosphorylation of this Y21 in Map53 might serve a similar function in the clam as does phosphorylation of Hsp53 S20 in the human.
Fig. 4.
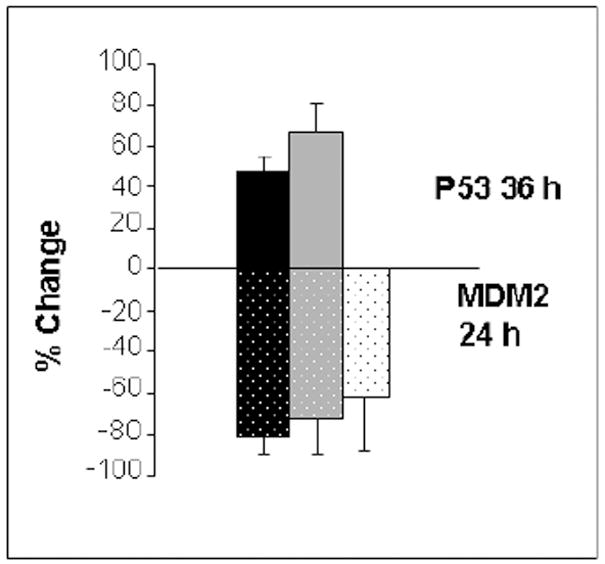
Reduction of MDM2 leads to maximum expression of Hsp53 and Map53 in H1299 cells. Addition of human MDM2 RNAi to Hsp53-H1299 (black bars) or Map53-H1299 cells (gray bars) reduced levels of MDM2 protein (stippled bars) at 24 hours post-transfection relative to cells not transfected with RNAi. Reduction of MDM2 is correlated with an increase in p53 expression (solid bars) at 36 hours post-transfection. White bars are vector alone; values are expressed as the mean ± standard error; N = 3.
Fig. 5.
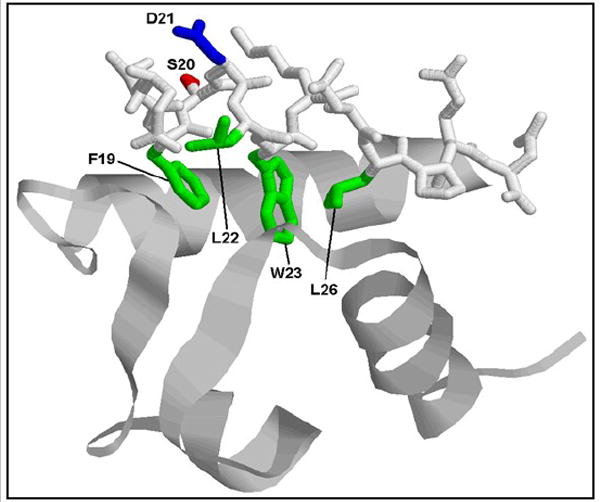
Structural analysis of Map53 shows conservation of a putative MDM2 binding site. Representation of a portion of the human MDM2 protein (ribbon structure) bound to the minimal-binding site of the Hsp53 protein (stick structure). Four residues critical for hydrophobic binding interactions (F19, L22, W23, and L26, shown in green) are conserved between human p53 and clam p53. Two potentially important changes are the substitutions in Map53 of glutamic acid and tyrosine, for S20 (red) and D21 (blue) in Hsp53, respectively. Modified from Kussie et al. (1996) using RasMol software.
Although the data suggest that clam p53 maintains a functional MDM2 binding site, a functional mdm2 homologue has not yet been identified and characterized in invertebrates. Mdm2 is absent from the Drosophila melanogaster and Caenorhabitis elegans genomes, although alternative regulators appear to exist (Lu and Abrams, 2006). Purported invertebrate mdm2 homologues with low similarity to vertebrate mdm2 sequences are predicted for the sea squirts, Ciona intestinalis and C. savignyi, and the limpet, Lottia gigantea (Ensembl, automated annotation analysis; Curwen et al., 2004). A partial sequence with homology to the mdm2 gene has recently been identified in an EST library from the California mussel, Mytilus californianus (GenBank accession no. ES394701).
3.3 Hsp53, but not Map53, elicits downstream markers of growth arrest
The relative level of p21WAF1/CIP1 protein expression and the ratio of hyper- to hypo-phosphorylated pRb were assessed as indicators of growth arrest. Western blot analysis of untreated H1299 cells revealed no significant changes in expression of these protein markers up to 48 hours post-transfection (p>0.05) indicating no cell density effects. Expression of p21WAF1/CIP1 was significantly higher in Hsp53-H1299 compared with Map53-H1299 cells or control groups (Fig. 6A). The elevated p21WAF1/CIP1 in Hsp53-H1299 cells corresponds to the observed increased cell doubling time (Fig. 1), and both are consistent with growth arrest. Growth arrest is also characteristically accompanied by a decrease in the ratio of hyper-: hypo-phosphorylated pRb, as was seen for Hsp53-H1299 (Fig. 6B) and reported by Kannan et al. (2001). Map53-H1299 cells, however, did not elicit markers of growth arrest (Fig. 6A,B). These functional differences also appear to be shared by other invertebrate p53 homologues (Sutcliff et al., 2003).
Fig. 6.
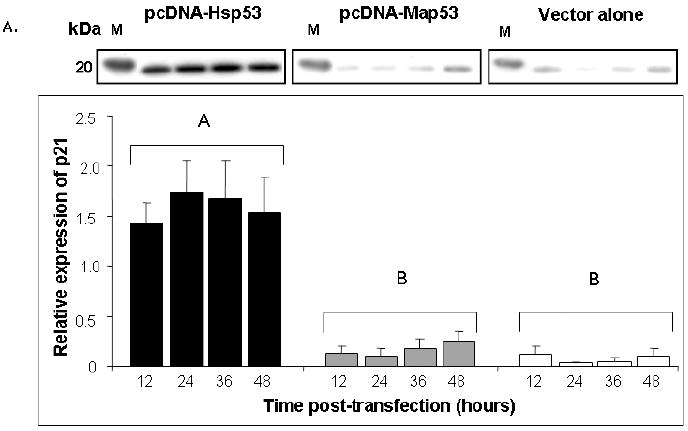
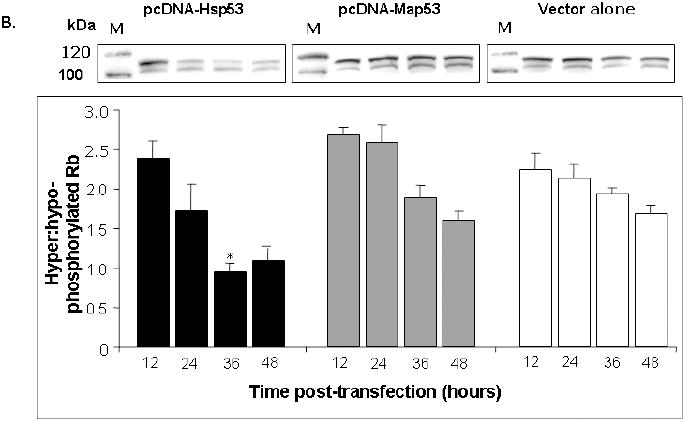
Hsp53 elicits expression of downstream markers of growth arrest in H1299 cells, but Map53 does not. (A) Representative western blot and summary densitometry data show that Hsp53 (black bars) elicits a robust p21 response that remains elevated over the 48-hour observation period. Neither Map53 (gray bars) nor vector alone (white bars) altered p21 expression. There are no significant differences in p21 expression over time within any treatment group (P > 0.05; N=3). Values were pooled for between-treatment comparisons; different letters denote significant differences; M, marker lane. (B) A second indicator of growth arrest, a decrease in the ratio of hyper- to hypo-phosphorylated pRb, was observed in Hsp53-H1299 cells (black bars), but not in those expressing Map53 (gray bars) or vector-only controls (white bars). M, marker lane; * = significantly lower ratio; values are expressed as the mean ± standard error; N=4.
Cells were also analyzed for activated caspase-3, an indicator of apoptotic activity. Although western blot analysis demonstrated a strong signal for the inactive, 32kDa caspase-3 proenzyme, expression of the cleaved, activated 12 and 17 kDa subunits was not detected (data not shown).
Biochemical studies were validated by flow cytometry (Fig. 7). As expected, a significantly greater proportion of the Hsp53-H1299 cells were found in G1 growth arrest at T≥ 24 hours post-transfection compared with Map53-H1299 or cells treated with the vector alone (Fig. 7). The fraction of cells in the sub-G1 phase (hypodiploid cells, a reflection of cell debris often associated with apoptosis) was also significantly higher in Hsp53-H1299 cells as compared to the other treatments at T≥ 24 hours post-transfection. Numbers of cells in sub-G1, however, remained low relative to those in G1 (Fig. 7). There was no significant difference in the cell cycle profile between cells expressing Map53 and the controls, suggesting that Map53 did not induce growth arrest or apoptosis in this cell line under non-stress conditions.
Fig. 7.
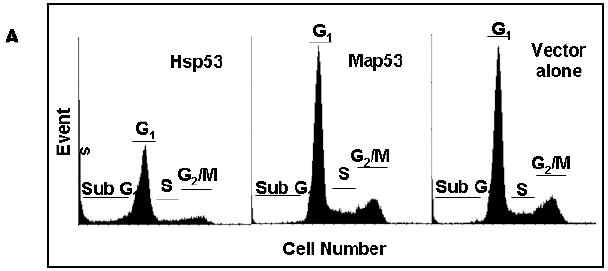
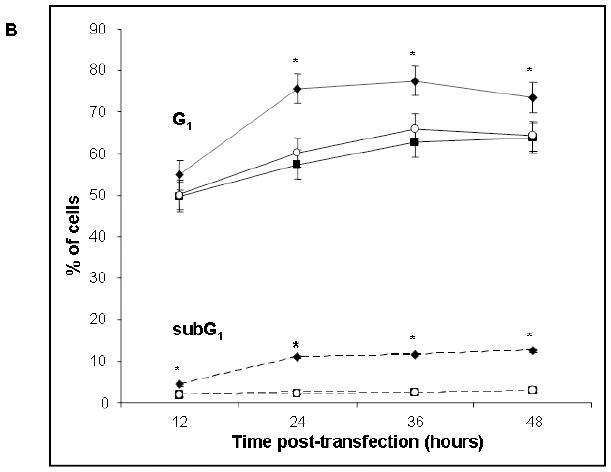
Flow cytometry reveals Hsp53-mediated growth arrest and apoptosis. (A) Representative histograms of Hsp53-H1299, Map53-H1299 and vector-alone transfected H1299 cells, at 24-hours post-transfection. Sub-G1, apoptotic cells; G1, Gap1 or growth-arrested cells; S, S-phase; G2/M, Gap2 or M phase. (B) Analysis of flow cytometry data shows that significantly more (*) Hsp53-H1299 cells (-◆-) remain in G1 (solid lines) than do Map53-H1299 (-■-) or vector-alone cells (-○-). In addition to an increased growth arrested cell population, significantly more (*) Hsp53-H1299 cells reside in the sub-G1 phase (dashed lines) as compared with Map53-H1299 or vector alone cells. Values are expressed as the mean ± the standard error; N=3.
3.4 Hsp53 and Map53 proteins localize to the nucleus
Subcellular localization was determined for both human and clam p53 to test the hypothesis that aberrant nuclear localization contributed to the observed functional differences. Greater than 90% of the p53 proteins in Hsp53- and Map53-H1299 cells localized to the nuclear fraction (Fig 8), eliminating cytoplasmic sequestration of Map53 as a possible cause of the functional differences.
Fig. 8.
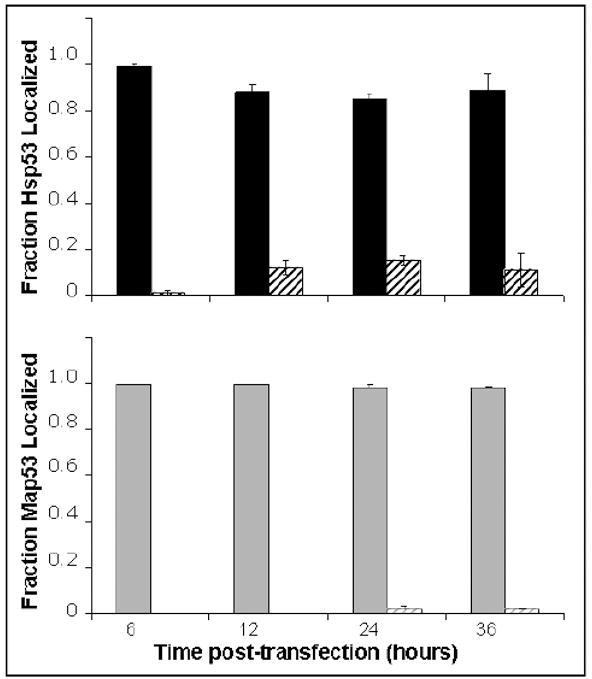
Densitometry scans of western blots show the vast majority of Hsp53 (solid black bars) and Map53 proteins (solid gray bars) localized to the nuclear fraction. Striped bars represent cytosolic proteins. Values are expressed as the mean ± the standard error; N=3.
4. Discussion
4.1 Potential differences in post-translational modifications may alter p53 activity
There are a number of potential explanations for the functional differences observed between Hsp53 and Map53 in H1299 cells. These include logistical artifacts, such as cell line-specific characteristics or aberrant regulation of Map53 expression in a non-homologous system. The use of different cell lines and differences in cell treatment, e.g., non-stressed vs. various cellular stresses, have yielded variable, and sometimes conflicting, information on p53 expression and activity (Michalak et al., 2005; Bensaad and Vousden, 2007). As such, selection of a different cell line, or perhaps the H1299 cells under stress conditions (UV or chemically induced DNA damage), might reveal additional functional similarities between Hsp53 and Map53.
Activity of Map53 in a non-homologous system may also be influenced by differences in post-translational modifications (PTM). P53 protein activity is highly regulated by a number of specific PTMs, including phosphorylation, acetylation, methylation, sumoylation, neddylation, ubiquitination and glycosylation (reviewed by Lavin and Gueven, 2006). Such modifications are responsible for regulating protein/protein interactions, p53 stability, localization, and transcription. It is hypothesized that expression in a non-homologous system may alter PTM of clam p53. Supporting this hypothesis is the observation of large differences between the expected molecular mass, based on the predicted amino acid sequence, compared to observed migrations on SDS-PAGE gels (data not shown). Map53 expressed in H1299 cells is approximately 7% larger than predicted, whereas native Map53 protein exhibits a mass approximately 14% larger than predicted. The observed size of Hsp53-expressed protein in H1299 cells is approximately 30% larger than the predicted amino acid sequence. Based upon the large discrepancy between predicted and observed mass, one may conclude: (1) it is likely that different modifications occur on Map53 and Hsp53 proteins when expressed in their native environment; (2) Map53 and Hsp53 acquire different PTMs in the H1299 cells. It is unclear if these differences in PTM are in response to expression in a non-homologous system or a reflection of species-specific differences in p53 regulation.
4.2 In vitro results may reflect species-specific differences
Alternatively, the observed functional differences between Map53 and Hsp53 may reflect true species-specific differences. Such differences have been well documented for the D. melanogaster p53 homologue (Dmp53) that shares sequence similarity in several functional domains with mammalian p53 (reviewed by Sutcliffe et al., 2003). Dmp53 plays an important role in the activation of apoptotic pathways in response to DNA damaging events. However, neither Dmp53 nor cep-1 (the C. elegans p53 homologue) play a role in damage-induced growth arrest in vivo (de Nooij and Hariharan, 1995; Derry et al., 2001). If the clam p53 functions in a manner similar to other invertebrates, then one might predict that the Map53 would also lack the capacity to activate the mammalian p21 pathway, consistent with the observations of this study. This is in agreement with Ollmann et al., (2000) who suggested that more recent evolutionary events led to the targeting of p21 by p53, linking p53 with cell cycle arrest in vertebrates.
Differences in primary structure may explain in part the inability of Map53 to initiate transactivation of downstream pathways in this cell system. A critical step in mammalian p53 function is the formation of a homodimer and a homotetramer. A comparison of Hsp53 and Map53 primary structures suggest that hydrophobic interactions important for dimer formation (McCoy et al., 1997; Ho et al., 2006) are conserved in the clam. Protein structural modeling revealed the loss in Map53 of four symmetrical charge pairs that are present in Hsp53 (Fig. 9). The Hsp53 D352/ R337 pair is replaced by alanine/asparagine in Map53. In addition to losing charge interactions, the substitution of two significantly smaller residues in Map53 might be expected to reduce packing in the area. The impact of the loss of these charge pairs on the oligomerization potential of Map53 depends on the solvent exposure of the charged residues before and after tetramerization. If the individual charged residues in Hsp53 are first solvated in the monomer and then transferred into a more hydrophobic environment during tetramer formation, these charge pairs could be expected to have a large stabilizing effect. Their loss may cause these ionic interactions to be decreased or lost in Map53 tetramerization, ultimately affecting p53/DNA interaction.
Fig. 9.
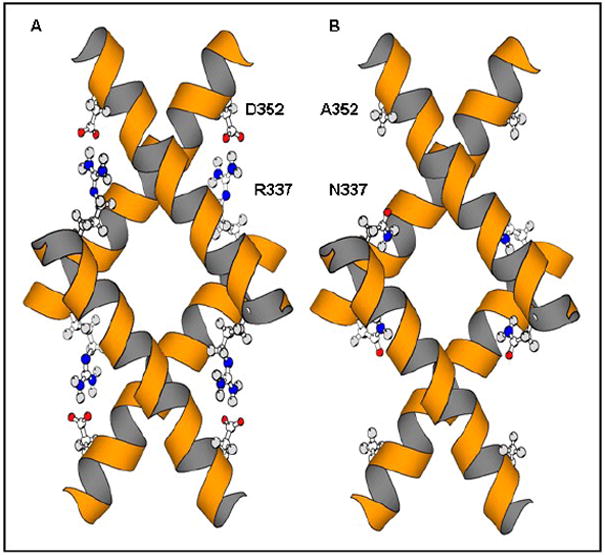
Comparison of predicted tetramerization domains of Hsp53 and Map53 proteins. (A) Hsp53 tetramerization domain showing D352/R337 charge pairs; (B) Map53 showing substitutions at the equivalent positions (A352, N337) with the loss of four symmetrical charge pairs. Modified from Clore et al. (1995) using MolScript (Kraulis, 1991).
4.3 Summary
Although sequence conservation between human and clam p53 within key functional domains suggested the potential for functional similarities, comparison of Map53 and Hsp53 activities in a human p53-null cell line, however, did not uncover similarities in the activation of either the p53-mediated growth arrest or apoptotic pathways. Subcellular localization studies indicated that clam p53, like the human p53, localizes predominantly to the nucleus where it is predicted to act as a functional transcription factor. In addition, it appears that Map53 has the capacity to interact with the human Mdm2 ubiquitin ligase, implying some functional similarity in p53 regulation under non-stressed conditions. We hypothesize the inability of the clam p53 to completely rescue p53 function in the human p53-null cells is likely due to differences in primary structure and/or post-translational modifications. This study does not rule out the possibility that clam p53 acts differently in the environment of the bivalve cell where it may function in a manner analogous to mammalian p53. Future studies will examine the effect of cell-stress conditions and cell backgrounds on Map53 function and may reveal more information on the evolution of this important gene family.
Acknowledgments
We thank Wendy Morrill and Rachel Shropshire for technical assistance and Dr. W. Halteman for help with statistical analysis. We also thank Drs. B. Vogelstein and K. Kinzler for donation of the human wild-type p53 plasmid and Dr. S. Pelsue and D. Swett from the University of Southern Maine Cellular and Molecular Imaging Core Facility for their help with the flow cytometry. This work was supported by grants from the NIH (R010RR08774/ ES012066) and the Maine Agriculture and Forestry Experiment Station (ME08509) to RJVB.
Abbreviations
- D21and D352
aspartic acid 21and 352, respectively
- Dmp53
Drosophila melanogaster p53 gene
- E20
glutamic acid 20
- ECL
enhanced chemiluminescence
- F19
phenylalanine 19
- G1
gap 1 phase of the cell cycle
- H1299
human small cell lung carcinoma cell line
- Hsp53
Homo sapiens p53
- Hsp-H1299
H1299 cells expressing human p53 protein
- L22
leucine 22
- M
M phase of the cell cycle
- Map53
Mya arenaria p53
- Map-H1299
H1299 cells expressing Mya arenaria p53 protein
- MDM2
mouse double-minute 2 (used interchangeably with human homologue, HDM2)
- MES
2-(N-morpholino) ethanesulfonic acid
- p21
WAF1/CIP1
- PTM
post-translational modification
- PVDF
polyvinylidene fluoride
- R337
arginine 337
- pRb
retinoblastoma protein
- RNAi
RNA interference
- S
S-phase (synthesis) of the cell cycle
- S20
serine 20
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Sub-G1
apoptotic cell fraction identified by flow cytometry
- T
time
- W23
tryptophan 23
- Y21
tyrosine 21
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 Network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- Alarcon-Vargas D, Ronai Z. p53-Mdm2 – the affair that never ends. Carcinogenesis. 2002;23:541–547. doi: 10.1093/carcin/23.4.541. [DOI] [PubMed] [Google Scholar]
- Barber BJ. Neoplastic diseases of commercially important marine bivalves. Aquat Living Resour. 2004;17:449–466. [Google Scholar]
- Barry M, Yevich PP. The Ecological, Chemical and Histopathological Evaluation of an Oil Spill Site, Part III: Histopathological Studies. Mar Pollut Bull. 1975;6:171–173. [Google Scholar]
- Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Böttger SA, Jerszyk EC, Low BE, Walker CW. Genotoxic stress-induced expression of p53 and apoptosis in leukemic clam hemocytes with cytoplasmically sequestered p53. Cancer Res. 2008;68:777–782. doi: 10.1158/0008-5472.CAN-06-0968. [DOI] [PubMed] [Google Scholar]
- Butler RA, Kelley ML, Gardner GR, Olberding KE, Van Beneden RJ. Aryl hydrocarbon receptor (AHR)-independent effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on soft-shell clam (Mya arenaria) reproductive tissue. Comp Biochem Physiol C. 2004;138:375–381. doi: 10.1016/j.cca.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Ernst J, Clubb R, Omichinski JG, Poindexter-Kennedy WM, Sakaguchi K, Appella E, Gronenborn AM. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SMJ, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Ho WC, Fitzgerald MX, Marmorstein R. Structure of the p53 core domain dimer bound to DNA. J Biol Chem. 2006;281:20494–20502. doi: 10.1074/jbc.M603634200. [DOI] [PubMed] [Google Scholar]
- Kannan K, Kaminski N, Rechavi G, Jakob-Hirsch J, Amariglio N, Givol D. DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene. 2001;20:3449–3455. doi: 10.1038/sj.onc.1204446. [DOI] [PubMed] [Google Scholar]
- Kelley ML, Winge P, Heaney JD, Stephens RE, Farell JH, Van Beneden RJ, Reinisch CL, Lesser MP, Walker CW. Expression of homologues for p53 andp73 in the softshell clam (Mya arenaria), a naturally occurring model for human cancer. Oncogene. 2001;20:748–758. doi: 10.1038/sj.onc.1204144. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallography. 1991;24:947–950. [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006;13:909–912. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- McCoy M, Stravridi ES, Waterman JLF, Wieczorek AM, Opella SJ, Halazonetis JD. Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain. EMBO J. 1997;16:6230–6236. doi: 10.1093/emboj/16.20.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak E, Villunger A, Erlacher M, Strasser A. Death squads enlisted by tumour suppressor p53. Biochem Biophys Res Commun. 2005;331:786–798. doi: 10.1016/j.bbrc.2005.03.183. [DOI] [PubMed] [Google Scholar]
- Moll UM, Petrenko O. The Mdm2-p53 Interaction. Mol Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker F, Demsky M, Fisher WW, Buchman A, Duyk G, Friedman L, Prives C, Kopczynski C. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Perry ME. Mdm2 in the Response to Radiation. Mol Cancer Res. 2004;2:9–19. [PubMed] [Google Scholar]
- Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2; fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- Sayle R, Milner-White EJ. RasMol: Biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Shengkan J, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JE, Korenjak M, Brehm A. Tumour suppressors – a fly’s perspective. Eur J Cancer. 2003;39:1355–1362. doi: 10.1016/s0959-8049(03)00263-6. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmostein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Critical role for Ser 20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999a;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T, Vogt Sionov R, Moallem E, Lee CL, Howley PM, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999b;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH. Activation of the p53 tumor suppressor protein. Biochim Biophys Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Walker CW, Böttger SA. A naturally occurring cancer with molecular connectivity to human diseases. Cell Cycle. 2008;7(15):2286–2289. doi: 10.4161/cc.6366. [DOI] [PubMed] [Google Scholar]
- Walker C, Böttger S, Low B. Mortalin-based cytoplasmic sequestration of p53 in a non-mammalian cancer model. Amer J Pathol. 2006;168:1526–1530. doi: 10.2353/ajpath.2006.050603. [DOI] [PMC free article] [PubMed] [Google Scholar]


