Abstract
Background
The human zinc finger protein 191 (ZNF191) is a member of the SCAN domain family of Krüppel-like zinc finger transcription factors. ZNF191 shows 94% identity to its mouse homologue zinc finger protein 191(Zfp191), which is the most highly conserved among the human-mouse SCAN family member orthologues pairs. Zfp191 is widely expressed during early embryogenesis and in adult organs. Moreover, Zfp191-/- embryos have been shown to be severely retarded in development and die approximately at embryonic day E7.5. ZNF191 can specifically interact with the widespread TCAT motif which constitutes the HUMTH01 microsatellite in the tyrosine hydroxylase (TH) gene. Allelic variations of HUMTH01 have been stated to have a quantitative silencing effect on TH gene expression and to correlate with quantitative and qualitative changes in the binding by ZNF191. In addition, ZNF191 displays a suppressive effect on the transcription; however, little downstream targets have identified.
Results
We searched for ZNF191 target genes by using a transient overexpression and knockdown strategy in the human embryo kidney (HEK293) cells. Microarray analyses identified 6094 genes modulated by overexpression of ZNF191 and 3332 genes regulated by knockdown of ZNF191, using a threshold of 1.2-fold. Several interested candidate genes, validated by real time RT-PCR, were correlated well with the array data. Interestingly, 1456 genes were identified in both transient overexpression and transient knockdown strategies. The GenMAPP and MappFinder software packages were further used for pathway analysis of these significantly altered genes. Several gene pathways were found to be involved in processes of the regulation of kinase activity, transcription, angiogenesis, brain development and response to DNA damage.
Conclusion
Our analysis reveals for the first time that ZNF191 is a pleiotropic factor that has a role in hematopoiesis, brain development and cancers.
Background
The regulation of gene expression in response to intrinsic and extrinsic cues is a fundamental cellular process in the growth and development of organisms. Critical to the control of gene expression are transcription factors that bind to specific DNA sequences and subsequently modulate gene transcription [1]. A significant number of transcription factors use a conserved zinc finger domain to bind their target DNAs. Zinc finger factors participate in a variety of cellular activities, such as development and differentiation, and play a role in human disease. In fact, the human genome encompasses approximately 600 to700 genes that contain a particular C2H2-type of zinc finger, which employs two cysteine and two histidine amino acid residues to coordinate the single zinc atom in the finger-like structure [2-4]. In the C2H2-type of zinc finger proteins, there is a highly conserved consensus sequence TGEKP(F/Y)X (X representing any amino acid) between adjacent zinc finger motifs. The zinc finger proteins containing this specific structure are termed Krüppel-like zinc finger proteins because the structure was first found in Drosophila Krüppel protein [5,6]. Many Krüppel-like factors exhibit diverse regulatory functions in cell growth, proliferation, differentiation, and embryogenesis [7-10]. The SCAN (SRE-ZBP, Ctfin 51, AW-1, and Number 18) [11,12], which is also known as the leucine rich region (LeR) [12], KRAB (Krüppel-associated box) [10,13], and POZ (poxvirus and zinc finger) [14] domains are highly conserved modules found in the N-terminus of C2H2 zinc finger transcription factors that have been demonstrated to mediate specific protein-protein interactions [15-18]. The SCAN domain is a conserved motif that appears to control the association of SCAN-containing proteins into non-covalent complexes, and may be the primary mechanism underlying partner choice in the dimerization of these transcription factors [16,19-22]. The genes encoding SCAN domains are clustered, often in tandem arrays, in both the human and mouse genomes and are capable of generating isoforms that may affect the function of family members. Although the function of most of the family members is not known, some of the SCAN domain family members play roles in cell survival and differentiation or development [16,23-25]. Like the KRAB motif, the SCAN domain-containing C2H2 zinc finger proteins are unique to the vertebrate lineage [2,3].
Human zinc finger protein 191 (ZNF191, also known as ZNF24 and KOX(17) [10]) is a member of the SCAN domain family of Krüppel-like zinc finger transcription factors [15]. This gene is initially named as RSG-A (for retinoic acid suppressed gene-A) because its mRNA can be amplified by homologous RT-PCR only in retinoic acid-untreated but not in retinoic acid-treated acute promyelocytic leukemia NB4 cells[26]. ZNF191 shows 94% identity to its mouse homologue zinc finger protein 191(Zfp191, also called ZF-12), which is the most highly conserved among the human-mouse SCAN family member orthologues pairs[16]. Tissue mRNA analysis showed that ZNF191 gene was ubiquitously expressed [26,27]. The mouse Zfp191 has been previously isolated from chondrocytic and mesenchymal precursor cell lines using a subtractive hybridization screening. Zfp191 mRNA is expressed during embryonic development and in different organs in adult, including rib cartilage, suggesting that Zfp191 may have a role in cartilage differentiation and in basic cellular processes[28].
A study demonstrated that ZNF191 can specifically bind to the TCAT repeats (HUMTH01) in the first intron of the human tyrosine hydroxylase (TH) gene, which encodes the rate-limiting enzyme in the synthesis of catecholamines [27,29]. Allelic variations of HUMTH01 have a quantitative silencing effect on TH gene expression in vitro, and correlate with quantitative and qualitative changes in the binding by ZNF191 [27]. ZNF191 mRNA levels was present at high levels in catecholaminergic tissues including the substantia nigra, the hypothalamus and the olfactory bulb. In the rat embryo, the mRNA was present at high levels in total embryonic head, which may reflect a role in brain development [27]; however, the molecular mechanisms remain unknown.
The ZNF191 gene is located on chromosome 18q12.1[10,26], a region frequently deleted in colorectal carcinomas, suggesting a possible role in the negative regulation of tumor growth[30,31]. ZNF191 contains four continuous typical C2H2 zinc fingers in its C-terminus, and one SCAN domain in its N-terminus [10,26]. Biochemical binding study shows the SCAN domain of ZNF191 as a selective oligomerization domain[20]. The SCAN domain of ZNF191 displays a suppressive effect on the transcription in CHO and NIH3T3 cells[26]; however, little else is known regarding its role in gene expression.
In the present study, we transiently overexpressed and silenced ZNF191 gene in the human embryo kidney (HEK293) cells and then applied microarray assay to identify possible ZNF191 target genes. Expression of interested candidate genes was also examined by real time RT-PCR, and these data correlated well with the array data. In general, our data reveal that ZNF191 modulates the expression of a large number of genes that are implicated in regulation of kinase activity, regulation of transcription, angiogenesis, brain development and response to DNA damage. Our analysis reveals for the first time that ZNF191 is a pleiotropic factor that has a role in hematopoiesis, brain development, tumor growth.
Results
Data analysis: general features
To identify ZNF191 target genes, a combined approach of transient overexpression and transient knockdown (KD) are used to identify genes modified by the ZNF191 transcription factor with 44 K Whole-Genome 60-mer oligonucleotide microarrays, using a cellular model (HEK293). Plasmids overexpressing ZNF191 cDNA (pcDNA3-ZNF191) or control empty pcDNA3 vectors were transiently transfected into HEK293 cells for 24 hours. As expected, introduction of pcDNA3-ZNF191 in HEK293 cells resulted in higher expression of ZNF191 mRNA than in cells transfected with control empty plasmid or untreated cells (Figure 1A). Real-time PCR results showed that the ZNF191 expression level was increased > 30 times in cells transfected with pcDNA3-ZNF191 compared with control cells (Figure 1B). For the reduction of ZNF191 expression, three hairpin siRNA expression vectors against ZNF191 were generated [32] and the knock-down of ZNF191 was assessed 48 hours after transfection of the HEK293 cells. Figure 2 showed that the three siRNA expression vectors were found to efficiently eliminate the target mRNA. The reduction of ZNF191 mRNA levels in transfected cells appears to be approximately 60% at the best compared to the control level. Due to no greater inhibition of ZNF191 mRNA at longer times than 48 hours, transfection of the HEK293 cells with the most efficient vector for 48 h was used in subsequent experiments.
Figure 1.
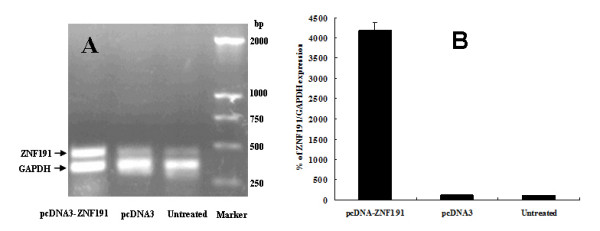
RT-PCR analysis of the ZNF191 gene expression in HEK293 cells transfected with pcDNA3-ZNF191 or pcDNA3 and untreated cells. (A) Semiquantitative RT-PCR analysis of the ZNF191 gene expression in the transfected cells and untreated cells. Total cellular RNA was isolated from pcDNA3-ZNF191 or pcDNA3 transfectants and untreated cells, and, after reverse transcription, PCR was performed with ZNF191- and GAPDH- specific primers. The up-regulation of the 481-bp ZNF191-specific band was detected in cells transfected with pcDNA3-ZNF191 plasmid. (B) Quantitative RT-PCR analysis of the relative ZNF191/GAPDH expression in the transfected cells and untreated cells.
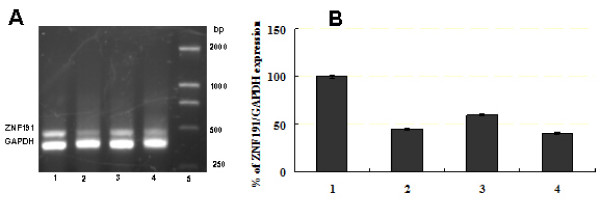
Figure 2.
RT-PCR analysis of the ZNF191 gene expression in HEK293 cells transfected with ZNF191 siRNA expression vectors or empty plasmid. (A) Semiquantitative RT-PCR analysis of the ZNF191 gene expression in the transfected cells. RT-PCR and gel electrophoresis of amplified products. The sizes of the amplified products are 481 bp (ZNF191) and 381 bp (GAPDH). Lane 1, the control cells (transfected with empty vector). Lane 2, lane 3, or lane 4, HEK293 cells transfected with pSZNF191-1, -2, or -3 (ZNF191 siRNA expression vectors), respectively. Lane 5, DNA size marker. (B) Quantitative RT-PCR analysis of the relative ZNF191/GAPDH expression in the transfected cells. Lane 1, the control cells (transfected with empty vector). Lane 2, lane 3, or lane 4, HEK293 cells transfected with pSZNF191-1, -2, or -3, respectively.
RNA samples were prepared from the transient overexpression and transient knockdown cells as well as control cells. Probe labelling, hybridization, and scanning were done by Shanghai Biochip Co., Ltd (China). The complete list of genes with differential expression can be found at the website http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14273. Microarray showed that 3273 genes were up-regulated more than 1.2-fold, while 3631 were down-regulated by the overexpression of the ZNF191 gene (see Additional file 1), while 2111 were up-regulated by the KD and 1221 were down-regulated, using the same threshold (see Additional file 2). Interestingly, 1456 genes were identified in both situations, and 1344 (92%) behaved consistently between the two experiments (i.e. induced by the overexpression and repressed by the KD, or the other way around) (see Additional file 3). These identified genes were subsequently used in pathway analysis with MappFinder.
Using a threshold of 2 for data analysis, 592 genes were found to be regulated by the overexpression with 143 genes up-regulated and 449 down-regulated (see Additional file 1, highlighted data) while 172 genes were regulated by the KD with 83 genes up-regulated and 89 down-regulated (see Additional file 2, highlighted data). The overlap was found between transient overexpression and transient knockdown with 69 shared genes(see Additional file 3, highlighted data). Thus, ZNF191 appears to modulate multiple transcriptional events, acting both as an activator and a repressor.
Pathway analysis of the ZNF191 transient overexpression and knockdown signature
To further evaluate our microarray data, GenMAPP and MAPPFinder software http://www.genmapp.org were used to organize gene expression data into MAPPs (microarray pathway profiles) that represent specific biological pathways and functionally grouped genes, based on the GO system. Three main classes of branches in the GO tree are biological processes, molecular functions and cellular components [33].
We focused on microarray gene expression data of 1456 genes identified in both transient overexpression and transient knockdown treatment. Using gene-association files from the GO Consortium, MAPPFinder assigns the thousands of genes in the dataset to numerous GO terms. Our analysis was limited to the GO terms that comprise more than eight genes in a class. Several significant (i.e., z score > 2) functional MAPPs were revealed with MAPPFinder and were presented in Table 1. Gene pathways that are involved in the regulation of kinase activity and angiogenesis were significantly up-regulated. In contrast, processes associated with response to DNA damage stimulus and brain development were down-regulated. Many up-regulated processes are localized to cellular component (endoplasmic reticulum) while down-regulated processes belong mainly to the cellular components (ribosome, extracellular matrix and cytoplasmic membrane-bound vesicle). Figure 3, 4, 5 and 6 and Table 2 showed the ZNF191 overexpression and ZNF191 knockdown differentially expressed genes involved in these signaling pathways.
Table 1.
Identification of significantly changed molecular pathways by mappfinder analysis of the significantly altered genes in both transient overexpression and transient knockdown of the ZNF191 gene.
| GOID | Pathway Name/GO Term | NO. genes | % Changed | Z-score |
| Up-regulated in overexpression and down-regulated in KD | ||||
| 43549 | regulation of kinase activity | 12 | 60 | 2.73 |
| 45859 | regulation of protein kinase activity | 12 | 60 | 2.73 |
| 165 | MAPKKK cascade | 8 | 66.7 | 2.60 |
| 51338 | regulation of transferase activity | 12 | 57.1 | 2.51 |
| 65008 | regulation of biological quality | 15 | 53.6 | 2.51 |
| 22402 | cell cycle process | 21 | 47.7 | 2.32 |
| 1525 | angiogenesis | 8 | 61.5 | 2.31 |
| 6952 | defense response | 13 | 52 | 2.19 |
| 6508 | proteolysis | 12 | 52.2 | 2.12 |
| 48514 | blood vessel morphogenesis | 8 | 57.1 | 2.04 |
| 7264 | small GTPase mediated signal transduction | 10 | 52.6 | 1.96 |
| 5515 | protein binding | 108 | 36.9 | 2.38 |
| 5525 | GTP binding | 9 | 60 | 2.36 |
| 30246 | carbohydrate binding | 10 | 55.6 | 2.18 |
| 19001 | guanyl nucleotide binding | 9 | 56.3 | 2.11 |
| 5783 | endoplasmic reticulum | 15 | 48.4 | 2.01 |
| Down-regulated in overexpression and up-regulated in KD | ||||
| 6066 | alcohol metabolic process | 14 | 100 | 2.56 |
| 45449 | regulation of transcription | 95 | 77.9 | 2.45 |
| 9719 | response to endogenous stimulus | 17 | 94.4 | 2.40 |
| 6974 | response to DNA damage stimulus | 17 | 94.4 | 2.40 |
| 19219 | regulation of nucleobase\, nucleoside\, nucleotide and nucleic acid metabolic process | 95 | 77.2 | 2.30 |
| 7420 | brain development | 11 | 100 | 2.27 |
| 6355 | regulation of transcription\, DNA-dependent | 89 | 77.4 | 2.25 |
| 6350 | transcription | 96 | 76.8 | 2.21 |
| 3735 | structural constituent of ribosome | 15 | 100 | 2.65 |
| 46872 | metal ion binding | 153 | 75.7 | 2.63 |
| 43167 | ion binding | 154 | 75.1 | 2.43 |
| 43169 | cation binding | 140 | 75.3 | 2.32 |
| 3676 | nucleic acid binding | 129 | 75.4 | 2.25 |
| 46914 | transition metal ion binding | 103 | 76.3 | 2.17 |
| 35091 | phosphoinositide binding | 10 | 100 | 2.16 |
| 8270 | zinc ion binding | 94 | 76.4 | 2.09 |
| 4930 | G-protein coupled receptor activity | 9 | 100 | 2.05 |
| 16023 | cytoplasmic membrane-bound vesicle | 11 | 100 | 2.27 |
| 5830 | cytosolic ribosome (sensu Eukaryota) | 11 | 100 | 2.27 |
| 5578 | proteinaceous extracellular matrix | 23 | 88.5 | 2.23 |
| 31012 | extracellular matrix | 23 | 88.5 | 2.23 |
| 5840 | ribosome | 15 | 93.8 | 2.20 |
| 5842 | cytosolic large ribosomal subunit (sensu Eukaryota) | 10 | 100 | 2.16 |
| 15934 | large ribosomal subunit | 10 | 100 | 2.16 |
| 44420 | extracellular matrix part | 9 | 100 | 2.05 |
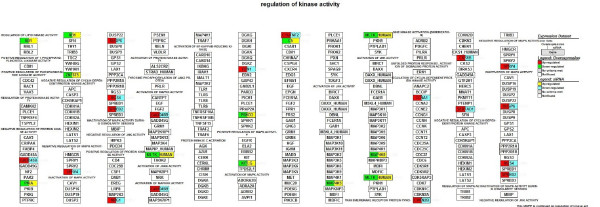
Figure 3.
Gene list involved in the regulation of kinase activity pathway generated by GenMAPP. The color on the left side of gene box illustrates the gene changes by ZNF191 overexpression; the color on the right indicates the gene changes by siRNA-mediated knockdown of ZNF191. Red and green indicates up-regulated and down-regulated in ZNF191 overexpression experiment, respectively. Yellow and blue indicates up-regulated and down-regulated in siRNA experiment, respectively. Grey indicates that selection criteria were not met but the gene was represented in the array. White boxes indicate that the gene was not present in the chip.
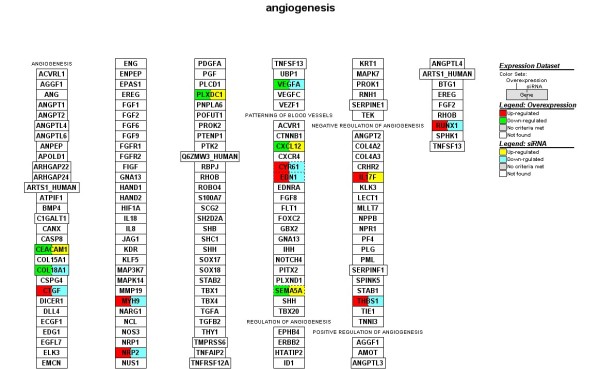
Figure 4.
Gene list involved in the angiogenesis pathway generated by GenMAPP. The color on the left side of gene box illustrates the gene changes by ZNF191 overexpression; the color on the right indicates the gene changes by siRNA-mediated knockdown of ZNF191. Red and green indicates up-regulated and down-regulated in ZNF191 overexpression experiment, respectively. Yellow and blue indicates up-regulated and down-regulated in siRNA experiment, respectively. Grey indicates that selection criteria were not met but the gene was represented in the array. White boxes indicate that the gene was not present in the chip.
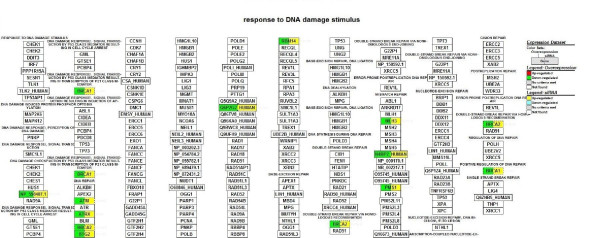
Figure 5.
Gene list involved in the response to DNA damage stimulus pathway generated by GenMAPP. The color on the left side of gene box illustrates the gene changes by ZNF191 overexpression; the color on the right indicates the gene changes by siRNA-mediated knockdown of ZNF191. Red and green indicates up-regulated and down-regulated in ZNF191 overexpression experiment, respectively. Yellow and blue indicates up-regulated and down-regulated in siRNA experiment, respectively. Grey indicates that selection criteria were not met but the gene was represented in the array. White boxes indicate that the gene was not present in the chip.
Figure 6.
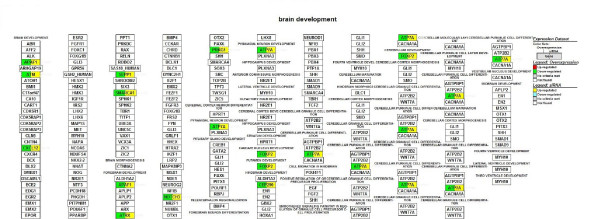
Gene list involved in the brain development pathway generated by GenMAPP. The color on the left side of gene box illustrates the gene changes by ZNF191 overexpression; the color on the right indicates the gene changes by siRNA-mediated knockdown of ZNF191. Red and green indicates up-regulated and down-regulated in ZNF191 overexpression experiment, respectively. Yellow and blue indicates up-regulated and down-regulated in siRNA experiment, respectively. Grey indicates that selection criteria were not met but the gene was represented in the array. White boxes indicate that the gene was not present in the chip.
Table 2.
List of genes involved in the signal pathways
| Fold change | |||||
| Gene symbol | ProbeName | GenBank | Description | Overexpression | Knockdown |
| Regulation of kinase activity | |||||
| RB1 | A_23_P204850 | NM_000321 | Retinoblastoma 1 (including osteosarcoma) | 0.74 | 1.53 |
| GADD45B | A_23_P142506 | NM_015675 | Growth arrest and DNA-damage-inducible, beta | 1.45 | 0.68 |
| PKIA | A_23_P31765 | NM_006823 | Protein kinase (cAMP-dependent, catalytic) inhibitor alpha | 0.68 | 1.41 |
| TIZ/ZNF675 | A_24_P931755 | D70835 | TRAF6-inhibitory zinc finger protein | 0.71 | 1.38 |
| SPRY4 | A_24_P269062 | NM_030964 | Sprouty homolog 4 (Drosophila) | 2.12 | 0.57 |
| DUSP6 | A_23_P139704 | NM_001946 | Dual specificity phosphatase 6 | 1.56 | 0.74 |
| RGS4 | A_23_P200737 | BC000737 | Regulator of G-protein signalling 4 | 1.83 | 0.65 |
| SPRED2 | A_32_P225854 | NM_181784 | Sprouty-related, EVH1 domain containing 2 | 1.52 | 0.72 |
| NRG1 | A_23_P315815 | NM_004495 | Neuregulin 1 | 1.51 | 0.74 |
| ZAK/MLTK_HUMAN | A_23_P318300 | NM_133646 | Sterile alpha motif and leucine zipper containing kinase AZK | 0.69 | 1.56 |
| EDN1 | A_23_P214821 | NM_001955 | Endothelin 1 | 2.33 | 0.49 |
| PRKD3 | A_24_P165656 | NM_005813 | Protein kinase D3 | 0.74 | 1.56 |
| KITLG | A_24_P133253 | NM_000899 | KIT ligand | 0.68 | 1.51 |
| C1QTNF6 | A_24_P211565 | NM_031910 | C1q and tumor necrosis factor related protein 6 | 0.47 | 1.54 |
| C5 | A_23_P71855 | NM_001735 | Complement component 5 | 0.42 | 2.62 |
| MAP4K5 | A_23_P205646 | BC036013 | Mitogen-activated protein kinase kinase kinase kinase 5 | 0.74 | 1.39 |
| CCNA1 | A_23_P48414 | NM_003914 | Cyclin A1 | 2.51 | 0.54 |
| CDKN2B | A_24_P360674 | NM_078487 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 1.81 | 0.71 |
| CKS1B | A_32_P175864 | BG205572 | CDC28 protein kinase regulatory subunit 1B | 1.45 | 0.64 |
| SERTAD1 | A_23_P218463 | NM_013376 | SERTA domain containing 1 | 1.51 | 0.75 |
| Angiogenesis | |||||
| CEACAM1 | A_24_P382319 | NM_001712 | Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 0.60 | 1.78 |
| COL18A1 | A_23_P211212 | NM_030582 | Collagen, type XVIII, alpha 1 | 0.74 | 0.66 |
| CTGF | A_23_P19663 | NM_001901 | Connective tissue growth factor | 3.88 | 0.24 |
| MYH9 | A_23_P57497 | BC011915 | Myosin, heavy polypeptide 9, non-muscle | 1.34 | 0.75 |
| NRP2 | A_24_P50801 | AF280546 | Neuropilin 2 | 1.77 | 0.59 |
| PLXDC1 | A_23_P3911 | BC036059 | Plexin domain containing 1 | 0.63 | 1.38 |
| VEGF | A_23_P70398 | NM_003376 | Vascular endothelial growth factor | 0.75 | 1.31 |
| CXCL12 | A_23_P202448 | U19495 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 0.73 | 1.37 |
| CYR61 | A_23_P46426 | Z97068 | Cysteine-rich, angiogenic inducer, 61 | 1.74 | 0.44 |
| EDN1 | A_23_P214821 | NM_001955 | Endothelin 1 | 2.33 | 0.49 |
| SEMA5A | A_23_P213415 | NM_003966 | Sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and sh | 0.28 | 2.26 |
| IL17F | A_23_P167882 | NM_052872 | Interleukin 17F | 1.78 | 2.01 |
| RUNX1 | A_24_P34155 | X90980 | Runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 1.48 | 0.74 |
| THBS1 | A_23_P206212 | X04665 | Thrombospondin 1 | 1.97 | 0.59 |
| Brain development | |||||
| APAF1 | A_23_P36611 | NM_181861 | Apoptotic protease activating factor | 0.58 | 1.36 |
| ATM | A_23_P35916 | NM_000051 | Ataxia telangiectasia mutated (includes complementation groups A, C and D) | 0.63 | 1.35 |
| CXCL12 | A_23_P202448 | U19495 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 0.73 | 1.37 |
| SEPP1 | A_23_P121926 | NM_005410 | Selenoprotein P, plasma, 1 | 0.66 | 1.36 |
| SMARCA1 | A_23_P44244 | NM_003069 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | 0.43 | 1.40 |
| ATRX | A_24_P348660 | NM_000489 | Alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 0.60 | 1.49 |
| NOTCH3 | A_32_P42895 | A_32_P42895 | Notch homolog 3 (Drosophila) | 0.68 | 1.46 |
| PRKG1 | A_23_P136041 | NM_006258 | Protein kinase, cGMP-dependent, type I | 0.43 | 1.34 |
| ATP7A | A_23_P217737 | NM_000052 | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | 0.48 | 1.52 |
| FOXP2 | A_24_P16559 | AF467257 | forkhead box P2 | 0.67 | 1.40 |
| CEP290 | A_23_P36865 | AB002371 | Centrosome protein cep290 | 0.70 | 1.43 |
| Response to DNA damage stimulus | |||||
| ZAK/NP_598407.1 | A_23_P318300 | NM_133646 | Sterile alpha motif and leucine zipper containing kinase AZK | 0.69 | 1.56 |
| BRCA1 | A_23_P207400 | NM_007294 | Breast cancer 1, early onset | 0.71 | 1.3 |
| ATM | A_23_P35916 | NM_000051 | Ataxia telangiectasia mutated (includes complementation groups A, C and D) | 0.63 | 1.35 |
| ATRX | A_24_P348660 | NM_000489 | Alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 0.60 | 1.49 |
| BRCA2 | A_23_P99452 | NM_000059 | Breast cancer 2, early onset | 0.62 | 1.3 |
| BTG2 | A_23_P62901 | NM_006763 | BTG family, member 2 | 0.72 | 1.35 |
| Q6P2G2_HUMAN | A_24_P361457 | NM_173627 | Hypothetical protein FLJ35220 (FLJ35220) | 0.63 | 1.49 |
| RBM14 | A_23_P161644 | NM_032886 | RNA-binding motif protein 14 | 0.62 | 1.47 |
| MLH3 | A_24_P61520 | NM_014381 | MutL homolog 3 (E. coli) | 0.59 | 1.45 |
| N4BP2_HUMAN | A_24_P222114 | AK001542 | Nedd4 binding protein 2 | 0.52 | 1.47 |
| PMS1 | A_23_P40059 | NM_000534 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) | 0.65 | 1.34 |
Quantitative Real-Time RT-PCR
Twenty-one interested genes were selected and subjected to real-time PCR analysis to confirm our microarrays results. As shown in Figure 7, the direction of regulation of ATP7A, RECK, PDGFRB, BMPR2, RB1, BRCA1, BRCA2, ATM, ATRX, IFI16, CCNB2, MYO6, GADD45B, SEMA5A, NRP2, CTGF, C5, VEGF, THBS1, KITLG and FOXP2 (expect for CCNB2) by the overexpression and knockdown of ZNF191 was consistent with the microarray results.
Figure 7.
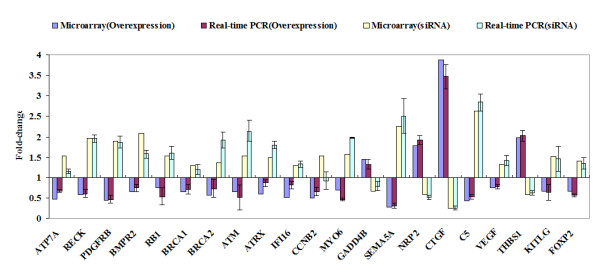
Quantitative real-time PCR confirmation of the microarray results. qPCR was performed on 21 genes that showed differential regulation in response to ZNF191 overexpression and knockdown by siRNA. Gene expression levels are shown as the mean normalized to the expression of the housekeeping gene GAPDH. Each sample was measured in triplicate. Columns, mean of three independent experiments; bars, SD. Comparison of fold change produced by microarray with relative expression ratio obtained from real-time PCR, with good concordance.
Discussion
In this study, we identify genes modified by the ZNF191 transcription factor with a combined strategy of transient overexpression and transient knockdown (KD) in a cellular model (HEK293), using oligonucleotide microarray technology. Several gene pathways were revealed by MAPPfinder to be involved in processes of the regulation of kinase activity, transcription, angiogenesis, brain development and response to DNA damage.
Pathway of regulation of kinase activity was significantly affected (Z-score 2.73). This pathway had a large number of expression changes, mostly due to the regulation of 12 genes (GADD45B, SPRY4, DUSP6, RGS4, SPRED2, NRG1, EDN1, CCNA1, CDKN2B, CKS1B, SERTAD1 and DUSP6), which were up-regulated in the ZNF191-overexpressed cells and down-regulated in the ZNF191 knockdown cells. In additional, 8 genes (KITLG, PKIA, RB1, ZAK, PRKD3, C1QTNF6, C5 and MAP4K5) were down-regulated in the ZNF191-overexpressed cells and up-regulated in the ZNF191 knockdown cells. A map of the genes involved in regulation of kinase activity was shown in Figure 3. GADD45B, originally termed MyD118, is first cloned as a myeloid differentiation primary response gene. It can be induced in the absence of protein synthesis following treatment of M1 myeloblastic leukemia cells with differentiation inducers[34], suggesting that GADD45B play a role in hematopoiesis. KITLG is a pleiotropic factor that acts in utero in germ cell and neural cell development, and hematopoiesis[35]. Accordingly, ZNF191 has been isolated from bone marrow and promyelocytic leukemia cell lines [26]. These data infer that ZNF191 may play a role in hematopoiesis.
Angiogenesis was another pathway markedly affected by ZNF191 (Z-score 2.31). As shown in Figure 4, CTGF, CYR61, EDN1, MYH9, NRP2, RUNX1, THBS1 were up-regulated in the ZNF191-overexpressed cells, and down-regulated in the knockdown cells. In addition, CEACAM1, PLXDC1, CXCL12, SEMA5A and VEGF were down-regulated in the ZNF191-overexpressed cells, and up-regulated in the knockdown cells. Angiogenesis, the growth of new blood vessels, is required for a variety of normal proliferative processes. Furthermore, angiogenesis is well established as also playing an important role in neoplastic growth and metastasis. VEGF is a potent stimulator of angiogenesis. ZNF191 has been reported to be up-regulated in angiogenic tumor nodules where VEGF expression is significantly decreased compared with preangiogenic nodules[36]. In this study, our result in HEK293 cells is consistent with the findings that in human breast carcinoma cells overexpression of ZNF191 results in a significant down-regulation of VEGF, whereas silencing of ZNF191 with small interfering RNA leads to increased VEGF expression as well as the same inverse correlation between ZNF191 and VEGF observed in malignant tissues from human colon and breast biopsies [36]. In addition, thrombospondin-1 (THBS1/TSP-1) has been shown to inhibit angiogenesis through direct effects on endothelial cell migration and survival, and through effects on vascular endothelial cell growth factor bioavailability. Aside from the inhibitory activity of angiogenesis, THBS1 also suppresses tumor growth by activating transforming growth factor beta and affects tumor cell function through interaction with cell surface receptors and regulation of extracellular proteases[37]. The data in this study revealed that overexpression of ZNF191 resulted in a significant up-regulation of THBS1, whereas silencing of ZNF191 led to decreased THBS1 expression in HEK293 cells. Taken together, these data suggest that ZNF191 may participate in negative regulation of angiogenesis. Furthermore, BMPR2 is overexpressed in the majority of human lung carcinomas independent of cell type[38,39]. PDGFR expression in colorectal cancer significantly correlates with lymphatic dissemination[40]. MYO6 (myosin VI) is critical in maintaining the malignant properties of the majority of human prostate cancers diagnosed today[41]. The finding that overexpression of ZNF191 significantly down-regulated BMPR2, PDGFR and MYO6, whereas silencing of ZNF191 increased BMPR2, PDGFR and MYO6 expression suggests a tumor suppressor role for ZNF191 in human cancers.
The response to DNA damage node was significantly affected (Z-score 2.4), mostly due to the regulation of BRCA1, BRCA2, ATM, BTG2 and ATRX (Figure 5). The most obvious explanation for this induction was that it was related to initiation of DNA replication. Indeed, DNA repair function is complementary to DNA replication, as the latter process is not error-free and produces mismatches that need to be repaired. However, we reported here that overexpression or elimination of ZNF191 affected the upstream regulators of the DNA damage pathway, such as ATM, ATRX, BRCA2 and BTG2. These data suggest that ZNF191 may be involved in the control of DNA damage response in addition to the regulation of DNA replication and concurrent DNA repair. Microarray and real-time PCR showed that RB1, ATM, ATRX and BRCA1 genes were down-regulated in the ZNF191-overexpressed cells (Figure 7). Accordingly, RB1 knockdown by siRNA caused the down-regulation of ATM, ATRX and BRCA1[42]. RB1 mRNA level was down-regulated in MCF-7 breast carcinoma cells by the overexpression of ZNF191 (data not published).
Mice Zfp191 transcript is detected early during embryogenesis in ectodermal, endodermal, mesodermal and extra-embryonic tissues. It is particularly observed in the developing central nervous system (CNS)[43]. In rat, ZNF191 mRNA is present at high levels in total embryonic head, and abundant in catecholaminergic tissues including substantia nigra, hypothalamus and olfactory bulb as well as peripheral catecholaminergic tissue adrenal medulla[27]. In human, ZNF191 transcript is also detected in brain[27]. These studies demonstrate an important role of ZNF191 in brain development. Interestingly, in support with previous studies, our results showed a node affected by ZNF191 was the brain development node (Z-score 2.27). 11 genes (ATP7A, FOXP2, APAF1, ATM, CXCL12, SEPP1, SMARCA1, ATRX, NOTCH3, PRKG1, and CEP290) down-regulated in the ZNF191-overexpressed cells and up-regulated in the ZNF191 knockdown cells (Figure 6). The human copper-transporting ATP7A is essential for dietary copper uptake, normal development and function of the CNS, and regulation of copper homeostasis in the body[44]. In addition, studies on Alzheimer's disease (AD) suggest an important role for copper in the brain, with some AD therapies focusing on mobilising copper in AD brains[45]. The transport of copper into the brain is complex and involves numerous players, including amyloid precursor protein, A beta peptide and cholesterol[45,46]. Interestingly, in a yeast two-hybrid experiment using the SCAN domain of ZNF191 as bait, we identified that the zinc finger transcription factor ZNF191 can interact with NAE1(NEDD8 activating enzyme E1 subunit 1; amyloid beta precursor protein binding protein 1, 59 kDa) (unpublished data). FOXP2 mutations in humans are associated with a disorder that affects both the comprehension of language and its production, speech[47]. ZNF191 transcript was abundant in the mesencephalon, a structure that contains the major rat embryonic catecholaminergic tissues[27]. Accordingly, ZNF191 can specifically interact with the widespread TCAT motif which constitutes the HUMTH01 microsatellite in the tyrosine hydroxylase (TH) gene (encoding the rate-limiting enzyme in the synthesis of catecholamines)[27]. Allelic variations of HUMTH01 are known to have a quantitative silencing effect on TH gene expression and to correlate with quantitative and qualitative changes in the binding by ZNF191[27]. In this study, overexpression of ZNF191 resulted in a significant down-regulation of TH, whereas knockdown of ZNF191 resulted in a non-significant change of TH level. The expression level of the TH gene is indistinguishable between the wild-type, Zfp191+/-, and Zfp191-/- mice[32]. These data suggest that other genes may compensate at least in part for the loss of ZNF191 activity, ameliorating the effect of the ZNF191 knockdown. Taken together, these data suggest that ZNF191 may be involved in the brain development and the neuropsychiatric diseases.
We have to point out that in the knockdown experiments we can only deduce the level of ZNF191 protein by the level of mRNA of ZNF191 because no antibody against the ZNF191 factor is commercially available and the turn-over of the protein is still unknown. So there is no mean to ascertain whether and when the ZNF191 protein is reduced significantly. In addition, in this study HEK293 cells were used as a cellular model to identify genes modified by the ZNF191 transcription factor. Some other cells could give at least partially over transcriptome modification. Furthermore, the transcriptional response found upon alteration of ZNF191 levels does not necessarily imply that this transcription factor regulates the differentially expressed genes. Many of the observed changes could be downstream and not direct effects of ZNF191 regulation. A proof of direct regulation would require a promoter analysis or some kind of DNA-protein binding experiment or in silico analysis.
Conclusion
We applied overexpression and siRNA-mediated gene silencing coupled with microarray screening and systematic pathway analysis to obtain insights into the pathways controlled by the target genes. We found that ZNF191 regulated the expression of a large group of genes involved in regulation of kinase activity, angiogenesis, brain development and response to DNA damage. Our analysis reveals that ZNF191 is a pleiotropic factor in hematopoiesis, brain development, and human cancers. Our approach can be used in functional genomics to elucidate the role of other transcription factors.
Methods
Cell culture and transfection
HEK-293 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) in an atmosphere of 95% air, 5% CO2 at 37°C. Cells were plated into 60 mm dishes without antibiotics 24 h prior to transfection. The recombinant plasmids, or control vector, were transfected into HEK-293 cells using Lipofectamine 2000 reagents according to the manufacturer's instructions. HEK-293 cells transfected with each cDNA or siRNA were lysed after 24 h or 48 h to isolate total RNA.
Plasmids
The cDNA clone of ZNF191 was provided by Prof. Long Y. (Genetics Institute, Fudan University, Shanghai, China). ZNF191 cDNA was cloned into pcDNA3 (Invitrogen) by using HindIII and XhoI restriction sites. The cloning was verified by DNA sequencing. The hairpin siRNA expression vectors against ZNF191 were previously generated [32].
RNA isolation
Total RNA was extracted using the Trizol reagent (Invitrogen) and purified on RNeasy columns (Qiagen). The amount and quality of RNA preparations were evaluated on the Agilent 2100 Bioanalyzer with RNA6000 Nano Reagents and Supplies (Agilent).
Microarray hybridization
Five hundred nanograms of RNA were reverse-transcribed to cDNA during which process a T7 sequences was introduced into cDNA. T7 RNA polymerase-driven RNA synthesis was used for the preparation and labeling of cRNA with Cy3 (the reference sample) and Cy5 (the treated sample), respectively. A Qiagen RNeasy Mini Kit (Qiagen Inc., Valencia, CA) was used to purify fluorescent cRNA probes. An equal amount (1 μg) of Cy3 and Cy5 labeled probes were mixed and used for hybridization on one Agilent 4 × 44 K Human Whole-Genome 60-mer oligonucleotide microarrays(Agilent Techonologies Inc., Santa Clara, CA) following the protocol provided by the manufacturer.
Data acquisition and processing
Microarrays were scanned with a dynamic autofocus microarray scanner (Agilent dual laser DNA microarray scanner, Agilent technologies, Palo Alto, CA, USA), using Agilent-provided parameters (Red and Green PMT were each set at 100%, and scan resolution was set to 10 μm). The Feature Extraction Software v7.5 (Agilent Technologies) was used to extract and analyse the signals. Agilent-provided settings were used except for subtraction of the local background and adjustment of the global background. Poor quality features that were either saturated in the two channels (50% of pixels > saturation threshold) or non-uniform were flagged. Only those features with a signal-to-noise ratio (SNR) of up to 2.6 in at least one channel and significantly different from the local background (two sided Student's t-test < 0.05) were used for further analysis. The mean signal ratio of the two fluorescent intensities (Cy-5 treated cRNA/Cy-3 reference cRNA) is expressed as a logarithm (base 2), providing a relative quantitative gene expression measurement between two samples. For subsequent analysis, we used mean-centered log2 of the normalized (linear & lowess method) sample: reference ratio. Microarray data meeting a criterion (> 1.2-fold change, Flag = 0, S/N > 2.6 and pValueLogRatio < 0.05) were considered to be significant and used for further analysis.
Pathway analysis
The GeneMapp and MappFinder software packages http://www.genmapp.org were used for pathway analysis of the significantly altered genes identified in both transient overexpression and transient knockdown of the ZNF191gene. The MAPPFinder works with GenMAPP and uses the annotations from the Gene Ontology (GO) Consortium to identify global biological trends in gene expression data. MAPPFinder relates microarray data to each term in the GO hierarchy, calculates the percentage of genes changed within each GO term, and generates a gene expression profile at the level of biological processes, cellular components, and molecular functions that allows for identification of specific biological pathways. MAPPFinder then calculates the total number of genes changed within a "parent GO term" and all of its "children" (local MAPPs) using Fisher's exact test. The results are expressed as a "z score" for a particular pathway, and values greater than 2 were considered to be significant. GenMAPP was also used to view and analyze microarray data on biological pathways[33].
Quantitative real-time PCR
Twenty-one interested genes were selected for validation by real time PCR. cDNAs were prepared from 1 μg of total RNA using AMV reverse transcriptase (Promega) and random primers according to the manufacturer's protocol. For each gene, PCR reactions were run three times on one sample. PCR was performed on Chromo4 Multicolor Real-Time Detection System (Bio-Rad, Inc.) in 20 μl reactions by using the SYBR™ Green PCR Master Mix (TaKaRa). The reaction was repeated for 40 cycles; each cycle consisted of denaturing at 95°C for 15 s, annealing and synthesis at 60°C for 1 min as per manufacturer's instructions. The relative amounts of transcript of the tested genes were normalized by GAPDH endogenous control primers set. The average ratios of relative amounts of transcript in the ZNF191-overexpressed cells or ZNF191 knockdown cells versus empty vector cells from three replicate runs were calculated. Quantitative values are obtained from the cycle number (Ct value) using Opticon Monitor 3.1 (Bio-Rad) according to the manufacturer's instructions. The threshold cycle (CT) values were averaged from the values obtained from each reaction, and each gene was normalized to GAPDH level. Each gene of interest and GAPDH were tested in triplicates to determine the Ct-difference. These Ct values were averaged and the difference between the GAPDH Ct (Avg) and the gene of interest Ct (Avg) was calculated (Ct-diff). The fold change was calculated by the 2-ΔCT, where ΔΔCT = (CT-treated-CT-contrl). CT-treated means the signal of an individual gene expressed in ZNF191-overexpressed or knockdown samples, while CT-control means the signal of the same gene expressed in control samples. The relative expression of the gene of interest was analyzed using the 2-ΔΔCT method [48]. Because SYBR Green binding is not sequence specific, careful design and validation of each primer pair, as well as cautious manipulation of RNA were undertaken to ensure that only target gene sequence-specific, non-genomic products were amplified by real-time PCR. To achieve this, primers were designed to either span or flank introns. PCR primers are listed in Additional file 4. A dissociation curve analysis was performed at the end of the amplification process in order to verify the specificity of the PCR products. The same PCR products were also evaluated by agarose gel electrophoresis. Data analysis and graphics were the results (mean ± SEM) of three different experiments.
Semi-quantitative PCR
Semi-quantitative PCRs were performed with Zfp191 specific primers (forward: 5'-ATCAGCGGTGGCCACATCAA-3' and reverse: 5'-GATGGGCCCAACCCAATATAT-3'), which yielded 481 bp products. To serve as an internal control, a 381 bp fragment of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was co-amplified with primers (forward, 5'-AACTTTGGTATCGTGGAAGGA-3'; reverse, 5'-GGAGGAGTGGGTGTCGCTGT-3'). The PCRs were carried out for 25–40 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min with a final extension of 5 min at 72°C. All RT-PCR experiments were repeated at least twice and the products were electrophoresed on 1.5% agarose gels containing ethidium bromide.
Authors' contributions
JL was involved in all experiments, experimental design and bioinformatics analyses. XC and YL co-drafted the manuscript, performed Real RT-PCR. XG and HF assisted in bioinformatics analyses and uploaded data to Gene Expression Omnibus. LQ and ZH co-drafted the manuscript and participated in cell culture. JZ co-drafted the manuscript, made revisions critically for important intellectual content.
Supplementary Material
Regulated genes by ZNF191 overexpression. Table S1 consists of the regulated probes by ZNF191 overexpression, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio(smaller than 0.05)for these genes.
Regulated genes by ZNF191 knockdown. Table S2 consists of the regulated probes by ZNF191 knockdown, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio(smaller than 0.05)for these genes.
Commonly altered genes by ZNF191 overexpression and knockdown. Table S3 consists of the regulated probes by ZNF191 overexpression and knockdown, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio (smaller than 0.05) for these genes.
List of primers used for RT-PCR. Abbreviations: see table 2.
Acknowledgments
Acknowledgements
We thank Professor Long Yu for providing the cDNA clone of ZNF191.This work was supported in part by grants from the National Natural Science Foundation of China (No. 30871353 and 30400269).
Contributor Information
Jianzhong Li, Email: lijianzhong1234@yahoo.com.
Xia Chen, Email: chenxia1978@yahoo.com.
Xuelian Gong, Email: witous2002@hotmail.com.
Ying Liu, Email: liuchanger1984@163.com.
Hao Feng, Email: fenghsmmu2001@yahoo.com.cn.
Lei Qiu, Email: quilei72@hotmail.com.
Zhenlin Hu, Email: zhenlinhu@hotmail.com.
Junping Zhang, Email: jpzhang08@hotmail.com.
References
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- Schuh R, Aicher W, Gaul U, Cote S, Preiss A, Maier D, Seifert E, Nauber U, Schroder C, Kemler R, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. Faseb J. 1995;9:597–604. [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/S1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Merck MF, Huebner K, Berger R, Thiesen HJ. Chromosomal localization of two human zinc finger protein genes, ZNF24 (KOX17) and ZNF29 (KOX26), to 18q12 and 17p13-p12, respectively. Genomics. 1991;9:154–161. doi: 10.1016/0888-7543(91)90233-5. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Khachigian LM, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J Biol Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- Pengue G, Calabro V, Bartoli PC, Pagliuca A, Lania L. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 1994;22:2908–2914. doi: 10.1093/nar/22.15.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid EJ, Poncelet DA, Lecocq PJ, Revelant O, Martial JA. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci USA. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/S0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JR, Maki JL, Blacklow SC, Collins T. The SCAN domain of ZNF174 is a dimer. J Biol Chem. 2002;277:5448–5452. doi: 10.1074/jbc.M109815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, et al. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- Sander TL, Haas AL, Peterson MJ, Morris JF. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J Biol Chem. 2000;275:12857–12867. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Blacklow SC, Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol Cell Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry JJ, Rutkoski NJ, Burke TL, Carter BD. A functional interaction between the p75 neurotrophin receptor interacting factors, TRAF6 and NRIF. J Biol Chem. 2004;279:16646–16656. doi: 10.1074/jbc.M309209200. [DOI] [PubMed] [Google Scholar]
- Adachi H, Tsujimoto M. Characterization of the human gene encoding the scavenger receptor expressed by endothelial cell and its regulation by a novel transcription factor, endothelial zinc finger protein-2. J Biol Chem. 2002;277:24014–24021. doi: 10.1074/jbc.M201854200. [DOI] [PubMed] [Google Scholar]
- Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, Barde YA. The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. Embo J. 1999;18:6050–6061. doi: 10.1093/emboj/18.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZG, Zhang QH, Ye M, Kan LX, Gu BW, He KL, Shi SL, Zhou J, Fu G, Mao M, et al. Molecular cloning of six novel Kruppel-like zinc finger genes from hematopoietic cells and identification of a novel transregulatory domain KRNB. J Biol Chem. 1999;274:35741–35748. doi: 10.1074/jbc.274.50.35741. [DOI] [PubMed] [Google Scholar]
- Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 2001;10:1785–1792. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- Prost JF, Negre D, Cornet-Javaux F, Cortay JC, Cozzone AJ, Herbage D, Mallein-Gerin F. Isolation, cloning, and expression of a new murine zinc finger encoding gene. Biochim Biophys Acta. 1999;1447:278–283. doi: 10.1016/s0167-4781(99)00157-8. [DOI] [PubMed] [Google Scholar]
- Meloni R, Albanese V, Ravassard P, Treilhou F, Mallet J. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum Mol Genet. 1998;7:423–428. doi: 10.1093/hmg/7.3.423. [DOI] [PubMed] [Google Scholar]
- Kern SE, Fearon ER, Tersmette KW, Enterline JP, Leppert M, Nakamura Y, White R, Vogelstein B, Hamilton SR. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] Jama. 1989;261:3099–3103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Li J, Chen X, Yang H, Wang S, Guo B, Yu L, Wang Z, Fu J. The zinc finger transcription factor 191 is required for early embryonic development and cell proliferation. Exp Cell Res. 2006;312:3990–3998. doi: 10.1016/j.yexcr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahi A, Lord KA, Hoffman-Liebermann B, Liebermann DA. Sequence and expression of a cDNA encoding MyD118: a novel myeloid differentiation primary response gene induced by multiple cytokines. Oncogene. 1991;6:165–167. [PubMed] [Google Scholar]
- Wehrle-Haller B. The role of Kit-ligand in melanocyte development and epidermal homeostasis. Pigment Cell Res. 2003;16:287–296. doi: 10.1034/j.1600-0749.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- Harper J, Yan L, Loureiro RM, Wu I, Fang J, D'Amore PA, Moses MA. Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 2007;67:8736–8741. doi: 10.1158/0008-5472.CAN-07-1617. [DOI] [PubMed] [Google Scholar]
- Kazerounian S, Yee KO, Lawler J. Thrombospondins in cancer. Cell Mol Life Sci. 2008;65:700–712. doi: 10.1007/s00018-007-7486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenfeld EM, Bojnowski J, Perone J, Langenfeld J. Expression of bone morphogenetic proteins in human lung carcinomas. Ann Thorac Surg. 2005;80:1028–1032. doi: 10.1016/j.athoracsur.2005.03.094. [DOI] [PubMed] [Google Scholar]
- Langenfeld EM, Langenfeld J. Bone morphogenetic protein-2 stimulates angiogenesis in developing tumors. Mol Cancer Res. 2004;2:141–149. [PubMed] [Google Scholar]
- Wehler TC, Frerichs K, Graf C, Drescher D, Schimanski K, Biesterfeld S, Berger MR, Kanzler S, Junginger T, Galle PR, et al. PDGFRalpha/beta expression correlates with the metastatic behavior of human colorectal cancer: a possible rationale for a molecular targeting strategy. Oncol Rep. 2008;19:697–704. [PubMed] [Google Scholar]
- Dunn TA, Chen S, Faith DA, Hicks JL, Platz EA, Chen Y, Ewing CM, Sauvageot J, Isaacs WB, De Marzo AM, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169:1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semizarov D, Kroeger P, Fesik S. siRNA-mediated gene silencing: a global genome view. Nucleic Acids Res. 2004;32:3836–3845. doi: 10.1093/nar/gkh714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfallah O, Faucon-Biguet N, Nardelli J, Meloni R, Mallet J. Expression of the transcription factor Zfp191 during embryonic development in the mouse. Gene Expr Patterns. 2008;8:148–154. doi: 10.1016/j.gep.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Gupta A, Burkhead JL, Zuzel V. Cellular multitasking: the dual role of human Cu-ATPases in cofactor delivery and intracellular copper balance. Arch Biochem Biophys. 2008;476:22–32. doi: 10.1016/j.abb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macreadie IG. Copper transport and Alzheimer's disease. Eur Biophys J. 2008;37:295–300. doi: 10.1007/s00249-007-0235-2. [DOI] [PubMed] [Google Scholar]
- Kong GK, Miles LA, Crespi GA, Morton CJ, Ng HL, Barnham KJ, McKinstry WJ, Cappai R, Parker MW. Copper binding to the Alzheimer's disease amyloid precursor protein. Eur Biophys J. 2008;37:269–279. doi: 10.1007/s00249-007-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff C, Haesler S. An evolutionary perspective on FoxP2: strictly for the birds? Curr Opin Neurobiol. 2005;15:694–703. doi: 10.1016/j.conb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regulated genes by ZNF191 overexpression. Table S1 consists of the regulated probes by ZNF191 overexpression, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio(smaller than 0.05)for these genes.
Regulated genes by ZNF191 knockdown. Table S2 consists of the regulated probes by ZNF191 knockdown, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio(smaller than 0.05)for these genes.
Commonly altered genes by ZNF191 overexpression and knockdown. Table S3 consists of the regulated probes by ZNF191 overexpression and knockdown, containing the assigned designations (Accession number and sequence name), the fold of changed expression and PValueLogRatio (smaller than 0.05) for these genes.
List of primers used for RT-PCR. Abbreviations: see table 2.