Abstract
Genetic diseases are recognized to be one of the major categories of human disease. Traditionally genetic diseases are subdivided into chromosomal (numerical or structural aberrations), monogenic or Mendelian diseases, multifactorial/polygenic complex diseases and mitochondrial genetic disorders. A large proportion of these conditions occur sporadically. With the advent of newer molecular techniques, a number of new disorders and dysmorphic syndromes are delineated in detail. Some of these conditions do not conform to the conventional inheritance patterns and mechanisms are often complex and unique. Examples include submicroscopic microdeletions or microduplications, trinucleotide repeat disorders, epigenetic disorders due to genomic imprinting, defective transcription or translation due to abnormal RNA patterning and pathogenic association with single nucleotide polymorphisms and copy number variations. Among these several apparently monogenic disorders result from non-allelic homologous recombination associated with the presence of low copy number repeats on either side of the critical locus or gene cluster. The term ‘disorders of genome architecture’ is alternatively used to highlight these disorders, for example Charcot-Marie-Tooth type IA, Smith-Magenis syndrome, Neurofibromatosis type 1 and many more with an assigned OMIM number. Many of these so called genomic disorders occur sporadically resulting from largely non-recurrent de novo genomic rearrangements. Locus-specific mutation rates for genomic rearrangements appear to be two to four times greater than nucleotide-specific rates for base substitutions. Recent studies on several disease-associated recombination hotspots in male-germ cells indicate an excess of genomic rearrangements resulting in microduplications that are clinically underdiagnosed compared to microdeletion syndromes. Widespread application of high-resolution genome analyses may offer to detect more sporadic phenotypes resulting from genomic rearrangements involving de novo copy number variation.
Keywords: Genome, Genome architecture, Genomic disorder, Genomic rearrangements, Mendelian disease, Sporadic disease, De novo mutations, Malformation syndrome
Introduction
Developments in genetics and molecular biology have provided a vast amount of data and information to support the view that most human diseases have a significant genetic component. Characterization of the genetic determinants of disease provides remarkable opportunities for clinical medicine through an improved understanding of pathogenesis, diagnosis and therapeutic options. An understanding of the genetic basis of human disease has opened the way forward for a new taxonomy of human disease that will be free from limitations and bias in developing diagnostic criteria related to events which are often secondary and peripheral to its cause (Bell 2003). For instance, genetic information has allowed us to identify distinct forms of diabetes mellitus, defining an auto-immune form associated with highly diverse and complex human leukocyte antigens [HLA] and other factors that affect both expression and modification of gene products in mediating the adult form of the disease (Cardon and Bell 2001). Similarly, a number of genetically determined molecules and pathways have been characterized that are crucial in the pathogenesis of bronchial asthma. It is now widely believed that a clearer understanding of the mechanisms and pathways of a disease will assist us in delineating distinct disease subtypes, and may resolve many questions relating to variable disease symptoms, progression and response to therapy. This might help in revising the current diagnostic criteria. Eventually, genetics may contribute to a new taxonomy of human disease for clinical practice.
Although genetics is acknowledged to be an important aspect in understanding the pathogenesis of disease, genetic classification of human disease has not yet received full recognition. There is ample evidence in support of the argument that genetic factors are probably associated with all human diseases except probably for trauma. It is argued that the outcomes to trauma might be influenced by inherited factors such as genetically determined inflammatory markers, host-response to infection and tissue damage. Various categories of genetic disorders are considered to be rare with a tendency to be included under the broad title of organ-system diseases. Often these are listed as simply etiological factors rather than a distinct disease category. This concept and approach is now rapidly being outdated and replaced with new classes of diseases. This progress is seriously hampered by the lack of formal education at all levels and integration of appropriate technologies into the modern medical diagnostic and therapeutic infrastructure.
Traditionally, genetic diseases are classified as chromosomal (numerical or structural), Mendelian or single-gene disorders, multifactorial/polygenic complex diseases or congenital anomalies and diseases associated with specific mitochondrial gene mutations. Apart from chromosomal disorders, essentially all genetic disorders result from some form of alteration or mutation occurring in a specific gene (single gene diseases) or involving multiple loci spread across the human genome (polygenic disorders). The major impact of chromosomal disorders occurs before birth and carries a serious health burden throughout childhood and during the early years of life. On the other hand single gene diseases can pose a real medical and health burden from the perinatal period to adult age with a peak around mid-childhood. In contrast, the polygenic/multifactorial diseases tend to present late, except for developmental anomalies requiring active multi-disciplinary care during early life.
Recent advances in molecular genetics have enabled us to identify specific groups of disorders that result from characteristic mechanisms involving specific areas of the human genome. Often these do not conform to the standard basic principles of genetics. A broad term ‘genomic disorders’ has been coined to describe these conditions (Table 1). A number of hereditary disorders present with complex genetic pathology that do not follow the conventional principles of inheritance. There is now overwhelming evidence within these disorders that indicates unusual mechanisms suggesting ‘non-traditional inheritance’. The mechanisms involve certain genomic regions that directly or indirectly influence regulation and expression of one or more genes manifesting in complex phenotypes. Currently some of these disorders are either listed as chromosomal or single-gene disorders.
Table 1.
Classification of genomic disorders
| Disorders of genome architecture (genomic rearrangements) |
| Disorders of genome architecture (genomic rearrangements) |
| Tri-nucleotide repeats disorders (genomic instability) |
| Chromosome breakage disorders (genomic instability) |
| Non-dysjunction disorders (genomic instability) |
| Complex genomic diseases (genomic variation-SNPs/CNVs) |
Phenotypes of disorders of genome architecture
Recent completion of the human genome project and sequencing of the total genomes of yeast and other bacterial species have enabled investigators to view genetic information in the context of the entire genome. As a result it is now possible to recognize mechanisms of some genetic diseases at the genomic level. The evolution of the mammalian genome has resulted in the duplication of genes, gene segments and repeat gene clusters (Lupski 1998). This aspect of genome architecture provides recombination hot spots between nonsyntenic regions of chromosomes that are distributed across the whole genome. These genomic regions become susceptible to further DNA rearrangements that may be associated with an abnormal phenotype. Such disorders are collectively grouped under the broad category of ‘genome architecture disorders’.
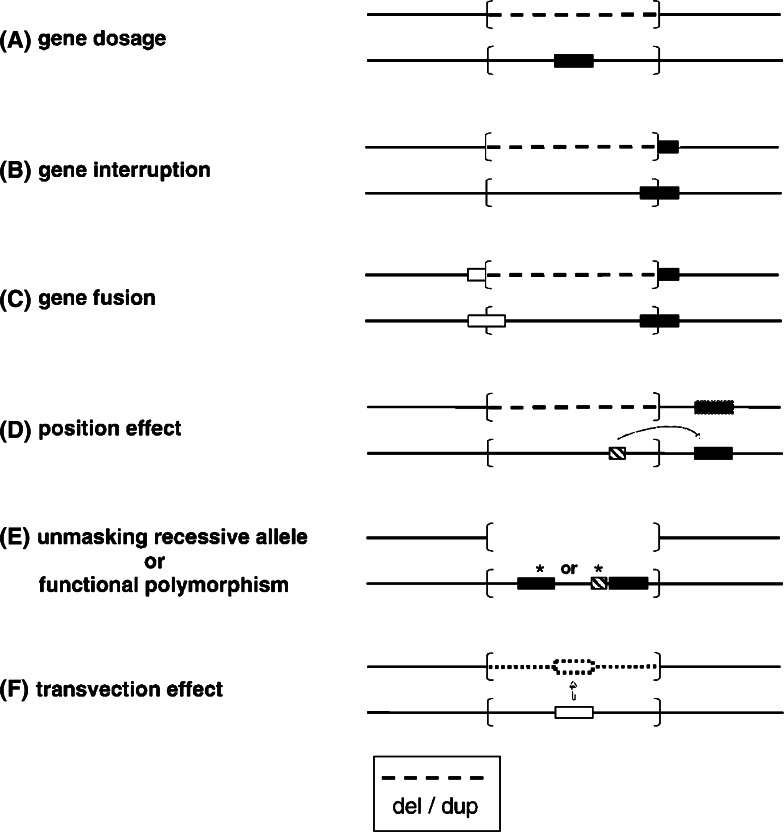
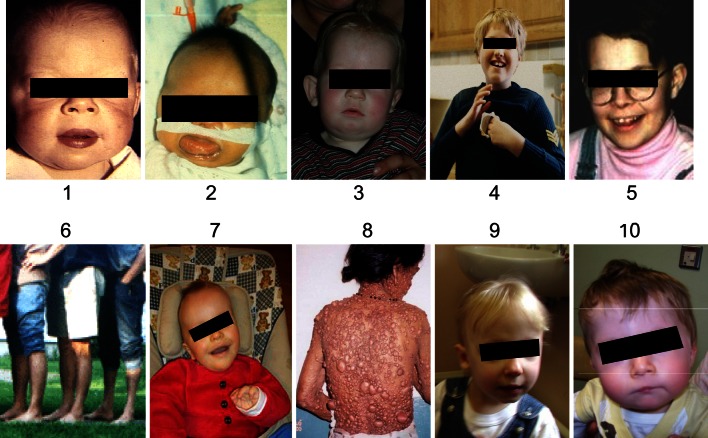
The term ‘genome architecture disorder’ refers to a disease that is caused by an alteration of the genome that results in complete loss, gain or disruption of the structural integrity of a dosage sensitive gene(s) (Shaw and Lupski 2004; Lupski and Stankiewicz 2005). Disruption in the function of dosage sensitive gene may result from number of mechanisms including gene interruption, gene fusion, position effect, unmasking of a recessive allele, presence of a functional polymorphism and gene transvection effect (Fig. 1). Notable examples include a number of micro-deletion/duplication syndromes (Table 2). In these conditions, there is a critical rearranged genomic segment flanked by large (usually >10 kb), highly homologous low copy repeat [LCR] structures that can act as recombination substrates. Meiotic recombination between non-allelic LCR copies, also known as non-allelic homologous recombination, can result in deletion or duplication of the intervening segment. The phenotype in some of these disorders is distinct recognizable with distinguishing clinical and facial dysmorphic features (Fig. 2).
Fig. 1.
Molecular mechanisms for genomic disorders (Lupski and Stankiewicz 2005)—dashed lines indicate either deleted or duplicated region; the rearranged genomic interval is shown in brackets; gene is depicted by filled horizontal rectangle; regulatory gene is shown as horizontal hach-marked rectangle; asterisks denote point mutations
Table 2.
Selected disorders of genome architecture
| Disorder | Inheritance | Locus | Gene | Rearrangement | Repeat size (kb) | Reference | |
|---|---|---|---|---|---|---|---|
| Type | Size (kb) | ||||||
| Williams-Beuren syndrome | AD | 7q11.23 | ELN | del/inv | 1600 | >320 } | Somerville et al. (2005) |
| Dup7(q11.23) syndrome | ? | 7q11.23 | ? | dup | } | ||
| Prader-Willi syndrome | AD | 5q11.2q13 | ? | del | 3500 | >500 } | |
| Angelman syndrome | AD | 15q11.2q13 | UBE3A | del | 3500 | >500 } | Long et al. (1998) |
| Dup(15)(q11.2q13) | AD? | 15q11.2q13 | ? | dup | 3500 | >500 } | |
| Triplication 15q11.2q13 | AD? | 15q11.2q13 | ? | trip | >500 } | ||
| Smith-Magenis syndrome | AD? | 17p11.2 | RA13 | del | 4000 | ~250 } | Potocki et al. (2000) |
| Potocki-Luspki syndrome | AD | 17p11.2 | ? | dup | } | ||
| CMT1A | AD | 17p11.2 | PMP22 | dup | 4000 | ~250 } | Chance et al. (1994) |
| HNPP | AD | 17p11.2 | PMP22 | del | } | ||
| Neurofibromatosis type 1 | AD | 17q | NF1 | del | De Luca et al. (2007) | ||
| DiGoerge/VCFS | AD | 22q11.2 | TBX1 | del | 3000 | ~400 } | Edelman et al. (1999) |
| Dup 22(q11.2q11.2) syndrome | ?AD | 22q11.2 | ? | dup | } | ||
| Sotos syndrome | AD | NSD1 | del | Rauch and Dörr (2007) | |||
| Male infertility | YL | Yq11.2 | DBY | del | 800 | ~10 } | Blanco et al. (2000) |
| AZFa-HERV microdeletion | USP9Y | del | |||||
| AZFc microdeletion | YL | Yq11.2 | RBMY DAZ? | del | 3500 | ~220 } | Boch and Jobling (2003) |
del, deletion; dup, duplication; inv, inversion
Fig. 2.
Selected disorders of the genome architecture-Williams-Beuren syndrome (1); Beckwith-Wiedemann syndrome (2); Prader-Willi syndrome (3); Angelman syndrome (4); Smith-Magenis syndrome (5); CMTIA (6); Sotos syndrome (7);Neurofibromatosis-1 (8); 22q deletion syndrome (9,10)
Similarly, other chromosomal rearrangements (Table 3), including reciprocal, Robertsonian, and jumping translocations, inversions, isochromosomes, and small marker chromosomes, may also involve susceptibility to rearrangement related to genome structure or architecture. In several cases, LCRs, A–T rich palindromes and pericentromeric repeats are located at such rearrangement breakpoints. This susceptibility to genomic rearrangements is not only implicated in disease etiology, but also in primate genome evolution (Shaw and Lupski 2004).
Table 3.
Genomic diseases resulting from recurrent genomic rearrangements
| Rearrangement | Type | Recombination substrates | |||
|---|---|---|---|---|---|
| Repeat size | Identity (%) | Orientation | Type | ||
| Inv dup(15)(q11q13) | Inverted dup | >500 | C | ||
| Inv dup(22)(q11.2) | Inverted dup | ~225–400 | 97–98 | C | |
| Idic(X)(p11.2) | Isodicentric | I? | |||
| Inv dup(8)(pterp23.1::p23.2pter); Del(8)(p23.1p23.2) | inv/dup/del | ~400 | 95–97 | I | Olfactory receptor gene cluster |
| dup(15)(q24q26) | dup | ~13–60 | ? | ||
del, deletion; dup, duplication; inv, inversion; D, direct; C, complex; I, inverted
An increasing number of Mendelian diseases (Table 3) are recognized to result from recurrent inter and intra chromosomal rearrangements involving unstable genomic regions facilitated by low-copy repeats [LCRs]. These genomic regions are predisposed to non-allelic homologous recombination [NAHR] between paralogous genomic segments. LCRs usually span approximately 10–400 kb of genomic DNA, share 97% or greater sequence identity, and provide the substrates for NAHR, thus predisposing to rearrangements. LCRs have been shown to facilitate meiotic DNA rearrangements associated with several multiple malformation syndromes and some disease traits (Table 2). Seminal examples (Fig. 3) include microdeletion syndromes [Williams-Beuren syndrome (7q11del), DiGoerge syndrome (22q11del)], autosomal dominant Charcot-Marie-Tooth disease type 1A [PMP22 gene duplication], Hereditary Neuropathy of Pressure Palsy [HNPP: PMP22 gene deletion] mapped to 17p11.2 and Smith-Magenis, a contiguous gene syndrome [CGS] with del (17)(p11.2p11.2). Dominantly inherited male infertility related to AZF gene deletion follows a similar mechanism. In addition this LCR-based complex genome architecture appears to play a major role in the primate karyotype evolution, pathogenesis of complex traits and human carcinogenesis (Frank et al. 2007).
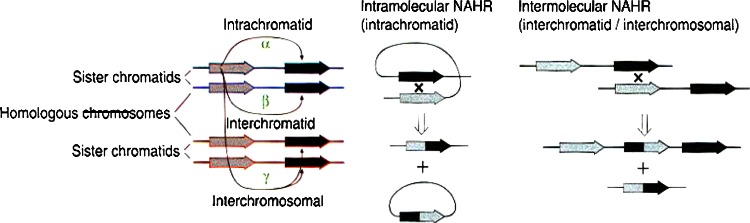
Fig. 3.
Mechanisms for non-allelic homologous recombination [NAHR] (Reproduced with permission from Nature Genetics, Turner et al. 2008)
Most commonly NAHR occurs between highly similar duplicated sequences. NAHR between duplicated sequences in direct orientation results in deletion or duplication ofintervening sequences. In contrast, inversions result from NAHR between sequences aligned in inverted orientation (Turner et al. 2008). The predominant pathogenic mechanism for the genomic disorders associated with deletions and duplications is altered copy number of dosage sensitive genes (Lupski and Stankiewicz 2005). In addition, NAHR is probably the most prevalent mechanisms contributing to non-pathogenic genomic variation (Redon et al. 2006). The breakpoints of genomic rearrangements caused by NAHR have been shown to cluster in defined hot spots within duplicated sequences, in a manner similar to allelic recombination hot spots (Turner et al. 2008). NAHR can occur in a number of ways. A simple model suggests three mechanisms—between paralogs on the same chromatid (intrachromatid), between sister chromatids (interchromatid), and between homologous chromosomes (interchromosomal). Deletion or duplication products result from the latter two mechanisms (Fig. 3). According to this model of NAHR, the relative rate of deletion and duplication depends on frequency of intrachromatid NAHR during meiosis. It is estimated that the rate of duplication never exceeds that of deletion (Turner et al. 2008).
A notable example includes genetically heterogeneous Charcot-Marie-Tooth disease [CMTD]. The disorder is also known as hereditary motor and sensory neuropathy [HMSN] by virtue of being a peripheral neuropathy due to either involvement of the axonal or myelinated segments of the peripheral nerve. Genetically autosomal dominant, autosomal recessive and X-linked dominant types are recognized. The disorder is not uncommon affecting approximately 1 in 2,500 of the adult population. This could be an underestimate since medically the condition is benign often not requiring any medical and/or surgical intervention. However, some affected individuals experience increasingly progressive neuro-muscular weakness of distal muscles of lower legs, feet, distal forearms, and hands with onset in early teens and causing severe loco-motor restrictions.
An affected person usually presents late with relative hypertrophy of the upper calf muscles described as ‘inverted Champagne bottle’ associated with pes cavus due to wasting of the small muscles of the feet. Similarly, wasting of the small muscles of hand leads to ‘claw-hands’. Neurophysiological studies remain an essential method of differentiating the two major types of CMTD. A reduced motor nerve conduction velocity of less than 35 m/sec helps in differentiating type 1 CMTD from type 2 CMTD, in which the motor nerve conduction velocity is usually normal but the sensory nerve conduction is often slow. Whilst this distinction is undoubtedly helpful in the clinical management, application for genetic counseling is limited because both types are genetically heterogeneous. For instance, molecular characterization and gene mapping have confirmed the existence of at least four types of type 1 CMTD, autosomal dominant types 1a, 1b, and 1c and the X-linked type [XCMT]. Similarly there are distinct genetic types within the type 2 CMTD group.
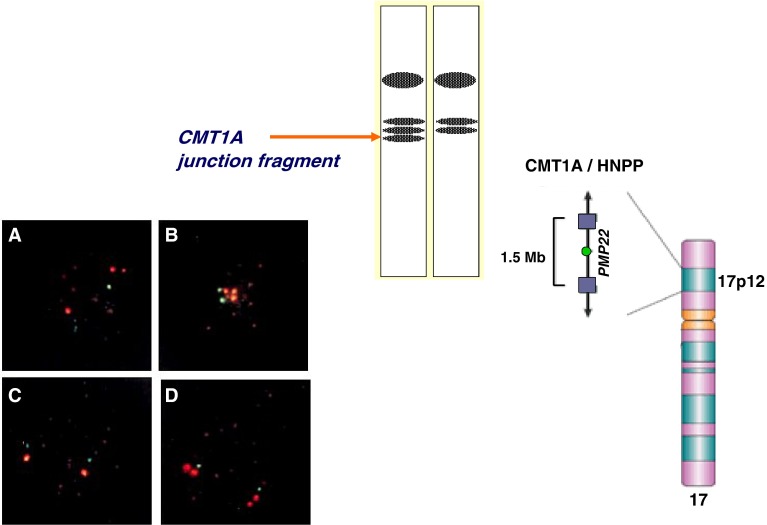
Approximately two-thirds of cases of CMT1 have a detectable 1.5 Mb duplication within a proximal chromosomal segment of the short arm of chromosome 17 (17p12) (Lupski et al. 1991). This duplicated chromosomal segment contains a gene for peripheral myelin protein called PMP22. This duplication results in disruption of the gene leading to abnormal myelination of the peripheral nerves, an essential molecular pathological step resulting in the CMT1 phenotype designated as CMT1A. The CMT1A duplication was visualized by multiple molecular methods (Patel and Lupski 1994), including fluorescence in situ hydridisation [FISH], pulsed-field gel eletrophoresis [PFGE], and dosage differences of heterozygous alleles by restriction-fragment-length polymorphisms [RFLPs] (Fig. 4). This finding led to further molecular studies on the origin of the 1.5 Mb duplicated 17p12 segment (Lupski 2003).
Fig. 4.
The 1.5 Mb duplicated chromosomal region of 17p12 including the PMP22 gene—note 500 kb junction fragment allele flanking the CMT1A gene detected by PFGE and Southern analysis. Note additional 17p segment (red colour) by metaphase (top two pictures) and interphase (lower two pictures) FISH (Reproduced with permission from Oxford University Press, New York)
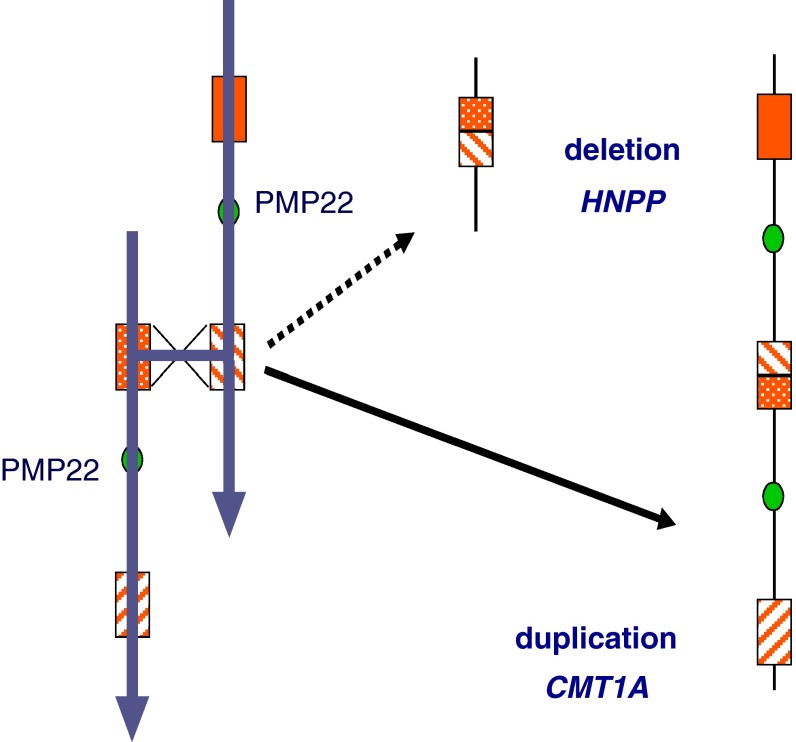
Studies by several investigators have revealed a significant variation in the size of marker alleles flanking the duplicated 17p12 region. It soon became apparent that a 500 kb allele co-segregated with 17p duplication in all affected individuals. This suggested a stable mutation and followed a precise recombination mechanism. However, in de novo duplication, the presence of repeated flanking marker alleles indicated the mechanism of unequal-crossing over leading to duplication. Indeed, this was confirmed when a highly homologous >20 kb size repeat sequence was confirmed flanking the 17p duplication. It was appropriately termed “CMT1A-REP.” As predicted by the unequal crossing-over model, CMT1A-REP was found to be present in three copies on the CMT1A duplication-bearing chromosome (Pentao et al. 1992). Interestingly, the presence of only one copy was soon demonstrated in another peripheral nervous system disorder known as hereditary neuropathy with liability to pressure [HNPP] (Chance et al. 1994; Reiter et al. 1996). The affected individuals with this disorder present with mild to moderate episodic weakness of the lower limbs and occasionally of upper limbs when subjected to prolonged pressure, such as sitting and sleeping. The disorder is dominantly inherited in an autosomal dominant manner. This is generally a clinically mild and benign hereditary neuropathy. The presence of only one copy results from a reciprocal deletion following unequal crossing-over involving the CMT1A-REP repeat (Fig. 5).
Fig. 5.
The unequal meiotic recombination (crossing-over) resulting in duplication [CMT1A] and deletion [HNPP]
Similar observations were also made in relation to Smith-Magenis syndrome (SMS), a contiguous gene syndrome associated with a microdeletion of 17p11.2 segment (Greenberg et al. 1991). Affected children present with facial dysmorphic features, severe speech delay and behavioural problems with signs of self-harm. A specific junction fragment was detected by PFGE (SMS-REP) that was involved in recurrent rearrangement resulting in either SMS or reciprocal 17p11.2 duplication. Pathogenic mutations in RAI1 gene, mapped to the 17p11.2 chromosomal region, are now shown to be etiologically linked with SMS (Slager et al. 2003). It is also possible to have both duplication and deletion at the same time, resulting from DNA rearrangements on both homologues of chromosome 17. This was demonstrated in a patient with mild delay and a family history of autosomal dominant carpel-tunnel syndrome, designated as Potocki–Lupski syndrome (Potocki et al. 1999, 2007). The occurrence of both the 17p11.2 duplication and HNPP deletion in this patient reflects the relatively high frequency at which these abnormalities arise and the underlying molecular characteristics of the genome in this region.
It is perfectly reasonable to accept the argument that similar molecular mechanisms apply in causing several other apparently Mendelian disorders often occurring sporadically (Table 4) (Lee et al. 2007). The human genome has evolved an architecture that may make us as a species more susceptible to de novo rearrangements causing genomic disorders (Lupski 2003, 2007). Several questions remain to be answered. To what extent de novo mutations result in sporadic traits? What is the difference in mutations rates for locus-specific point mutation or genomic rearrangement resulting in clinically indistinguishable phenotype? Is there any difference in male-germ cells meiotic genomic rearrangements resulting in either microdeletions or microduplications? Recent studies point to an excess of genomic rearrangement hot spots in male germ cells. Deletions are generated at a relatively higher rate that reciprocal duplications in the male germline (Turner et al. 2008).
Table 4.
Genomic rearrangement in selected Mendelian disorders
| Disorders | Inheritance | Chromosome location | Gene | Rearrangement | Recombination substrates | |||
|---|---|---|---|---|---|---|---|---|
| Type | Size(kb) | Repeat Size (kb) | Identity(%) | Orientation | ||||
| Bartter syndrome type III | AD | 1p36 | CLCNKA/8 | del | 11 | 91 | D | |
| Gaucher disease | AR | 1q21 | GBA | del | 16 | 14 | D | |
| Familial juvenile nephronophthisis | AR | 2q13 | NPHP1 | del | 290 | 45 | >97 | D |
| Facioscapulohumeral muscular dystrophy | AD | 4q35 | FRG1? | Del | 25–222 | 3.3 | D | |
| Spinal muscular dystrophy | AR | 5q13.2 | SMN | inv/dup | 500 | I | ||
| Congenital adrenal hyperplasia-21 hydroxylase deficiency | AR | 6p21.3 | CYP21 | del | 30 | 96–98 | D | |
| Glucorticoid remediable aldosteronism | AD | 8q21 | CYP11B1/2 | dup | 45 | 10 | 95 | D |
| β-Thalassemia | AR | 11p15.5 | β-globin | del | 4,(7)? | D | ||
| α-Thalassemia | AR | 16p13.3 | α-globin | del | 3,7,4.2? | 4 | D | |
| Polycystic kidney disease type 1 | AD | 16p13.3 | PKD1 | 50 | 95 | |||
| Charcot-Marie-Tooth (CMT1A) | AD | 17p12 | PMP22 | dup | 1400 | 24 98 | 7 | D |
| Hereditary neuropathy with liability to pressure palsy(HNPP) | AD | 17p12 | PMP22 | del | 1400 | 24 | 98.7 | D |
| Neurofibromatosis type 1(NF1) | AD | 17q11.2 | NF1 | del | 1500 | 85 | D | |
| Pituitary dwarfism | AR | 17q23.3 | GH1 | del | 6.7 | 2.24 | 99 | D |
| CYP2D6-phramcogenetic trait | AR | 22q13.1 | CYP2D6 | del/dup | 9.3 | 2.8 | ||
| Ichthyosis | XL | Xq28 | STS | del | 1900 | 20 | D | |
| Red–green colour blindness | XL | Xq28 | RCP/GCP | del | 0 | 39 | 98 | D |
| Incontinentia pigmenti | XL | Xq28 | NEMO | del | 10 | 0.870 | D | |
| Hemophilia A | XL | Xq28 | F8 | inv | 300-500 | 9.5 | 99.9 | I |
| Emery-Dreifuss muscular Dystrophy (EMD) | XL | Xq28 | Emerin/FLN1 | del/dup/inv | 48 | 11.3 | 99.2 | |
| Hunter syndrome | XL | Xq28 | IDS | inv/del | 20 | 3 | >88 | |
del, deletion; dup, duplication; inv, inversion; D, direct; C, complex; I, inverted
Rapid developments in this direction, for example commercially available opportunities for individual whole human genome sequencing or whole genome scan using a 1,000,000-probe SNP chip might offer to answer some of these questions. Currently probably the best results are likely to be achieved through multi-institutional efforts for developing better arrays for high-resolution genome analyses and detection of copy-number variation in conjunction with robust data bases (Lupski 2007), for example ‘Database of Genomic Variants’ (http://projects.tcag.ca/variation/) and DECIPHER-database for chromosomal imbalance and phenotype in humans using Ensembl (www.ensembl.org) resources (http://www.sanger.ac.uk/PostGenomics/decipher/). In conclusion, for many apparent sporadic diseases, and perhaps for multi-factorial and complex traits, one must consider the possibility of de novo genomic rearrangement as the potential molecular mechanism (Lupski 2007). Several research groups are engaged in looking for more genomic recombination hot spots that could reveal more disorders informing both clinical genetics and other clinicians (Osborne 2008). There is now ample evidence for the existence of distinct clinical conditions that result from a number of genomic mechanisms related to either disruption in the genome architecture/function or both. Term ‘genomic disorders’ is probably appropriate to designate these conditions as a new class of human disease included in the taxonomy for human disease.
Acknowledgments
The author is grateful for permission to all concerned for allowing pictures and clinical photographs appearing in Fig. 3. Oxford University Press for permission to reproduce figures and tables from chapter number 6 ‘Genetic and genomic approaches to taxonomy for human disease’ In ‘Genomics and Clinical Medicine’ Eds Dhavendra Kumar and Sir David Weatherall, published Oxford University Press, New York, 2008.
References
- Bell JI. The double helix in clinical practice. Nature. 2003;421(6921):414–416. doi: 10.1038/nature01402. [DOI] [PubMed] [Google Scholar]
- Blanco P, et al. Divergent outcomes of intrachromosomal recombination on the human Y chromosome male infertility and recurrent polymorphisms. J Med Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch E, Jobling MA. Duplication of the AZFa region of the human Y chromosome are mediated by homolgous recombination between HERVs and are compatible with male fertility. Hum Mol Genet. 2003;12:341–347. doi: 10.1093/hmg/ddg031. [DOI] [PubMed] [Google Scholar]
- Cardon IR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lesch MW, et al. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet. 1994;3:223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- De Luca A, Bottillo I, Dasdia MC, et al. Deletions of NF1 gene and exons detected by multiplex ligation dependent probe amplification. J Med Genet. 2007;44:800–808. doi: 10.1136/jmg.2007.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman L, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Frank B, Bermejo JL, Hemminki K, et al. Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis. 2007;28(7):1442–1445. doi: 10.1093/carcin/bgm033. [DOI] [PubMed] [Google Scholar]
- Greenberg F, Guzzeta V, Montes de Oca-Luna R, Magenis RE, Smith ACM, et al. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous gene syndrome associated with del(17)(p11.2) Am J Hum Genet. 1991;49:1207–1218. [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR (2007) A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 28;131(7):1235–1247 [DOI] [PubMed]
- Long FL, Duckett DP, Billam LJ, et al. Triplication of 15q11–q13 with inv dup(15) in a female with developmental delay. J Med Genet. 1998;35:425–428. doi: 10.1136/jmg.35.5.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–420. doi: 10.1016/S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: recombination-based disease resulting from genome architecture. Am J Hum Genet. 2003;72:246–252. doi: 10.1086/346217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Montes de Oca-Luna R, Slaugenhaupt S, Pentao L, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- Osborne LR. Genomic rearrangements in the spotlight. Nat Genet. 2008;40(1):6–7. doi: 10.1038/ng0108-6. [DOI] [PubMed] [Google Scholar]
- Patel P, Lupski JR. Charcot-Marie-Tooth disease: a new paradigm for the mechanism of inherited disease. Trends Genet. 1994;10:128–133. doi: 10.1016/0168-9525(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, et al. DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet. 1999;64:471–478. doi: 10.1086/302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, et al. Molecular mechanisms for duplication 17p11.2-the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Potocki L, et al. Characterization of the Pottocki-Lupski syndrome [dup(17)(p11.2p11.3) and delineation of dosage-sensitive critical interval that can convey autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Dörr HG. Chromosome 5q subtelomeric deletion syndrome. Am J Med Genet C Semin Med Genet. 2007;145(4):372–376. doi: 10.1002/ajmg.c.30151. [DOI] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, et al. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;13(1):R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAII associated with Smith-Magenis syndrome. Nat Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- Somerville MJ, et al. Severe expressive-language delay related to duplication of the William-Beuren locus. N Eng J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40(1):90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]