Abstract
One of the major difficulties in paleontology is the acquisition of fossil data from the 10% of Earth’s terrestrial surface that is covered by thick glaciers and ice sheets. Here we reveal that DNA and amino acids from buried organisms can be recovered from the basal sections of deep ice cores and allow reconstructions of past flora and fauna. We show that high altitude southern Greenland, currently lying below more than two kilometers of ice, was once inhabited by a diverse array of conifer trees and insects that may date back more than 450 thousand years. The results provide the first direct evidence in support of a forested southern Greenland and suggest that many deep ice cores may contain genetic records of paleoenvironments in their basal sections.
The environmental histories of high latitude regions such as Greenland and Antarctica are poorly understood because much of the fossil evidence is hidden below kilometer thick ice sheets (1-3). Here, we test the idea that the basal sections of deep ice cores can act as archives for ancient biomolecules and show that these molecules can be used to reconstruct significant parts of the past plant and animal life in currently ice covered areas.
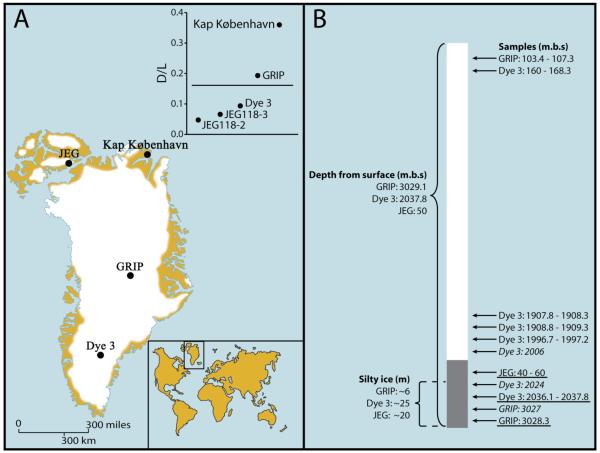
The samples studied come from the basal impurity rich (silty) ice sections of the 2km long Dye 3 core from south-central Greenland (4), the 3km long GRIP core from the summit of the Greenland ice sheet (5), and the Late Holocene John Evans Glacier on Ellesmere Island, Nunavut, northern Canada (Fig. 1A,B). The latter sample was included as a control to test for potential exotic DNA because the glacier has recently overridden a land surface with a known vegetation cover (6). As an additional test for long-distance atmospheric dispersal of DNA, we included five control samples of debris-free Holocene and Pleistocene ice taken just above the basal silty samples from the Dye 3 and GRIP ice cores (Fig. 1B). Finally, our analyses included sediment samples from the Kap København Formation from the northernmost part of Greenland, dated to 2.4 million years before present (Ma BP) (1, 2).
Fig. 1.
Sample location and core schematics. (A) Map showing the locations of the Dye 3 (65°11′N, 45°50′W) and GRIP (72°34′N, 37°37′W) drilling sites and the Kap København Formation (82° 22′ N, W21°14′ W) in Greenland as well as the John Evans Glacier (JEG; 79°49′ N, 74°30′ W) on Ellesmere Island (Canada). The insert shows the ratio of D to L aspartic acid, a measure of the extent of protein degradation; more highly degraded samples (above the line) failed to yield amplifiable DNA. (B) Schematic drawing of ice core/icecap cross-section, with depth (in meters below surface, m.b.s.) indicating the depth of the cores and the positions of the Dye 3, GRIP, and JEG samples analyzed for DNA, DNA/amino acid racemization/luminescence (underlined), and 10Be / 36Cl (italic, the control GRIP samples are not shown). The lengths (in meters) of the silty sections are also shown.
The silty ice yielded only few pollen grains and no macrofossils (7). However, the Dye 3 and John Evans Glacier silty ice samples showed low levels of amino acid racemization (Fig. 1A, insert), indicating good organic matter preservation (8). Therefore, following previous success with permafrost and cave sediments (9-11), we attempted to amplify ancient DNA from the ice. This was done following strict criteria to secure authenticity (12-14), including covering the surface of the frozen cores with plasmid DNA to control for potential contamination that may have entered the interior of the samples through cracks or during the sampling procedure (7). PCR products of the plasmid DNA were obtained only from extracts of the outer ice scrapings but not from the interior, confirming that sample contamination had not penetrated the cores.
We could reproducibly PCR amplify short amplicons (59-120 base-pairs, bp) of the chloroplast DNA (cpDNA) rbcL gene and trnL intron from about 50g of the interior ice melts from the Dye 3 and the John Evans Glacier silty samples. From Dye 3, we also obtained 97bp amplicons of invertebrate cytochrome oxidase subunit I (COI) mitochondrial DNA (mtDNA). Attempts to reproducibly amplify DNA from the GRIP silty ice and from the Kap København Formation sediments were not successful. These results are consistent with the amino acid racemization data demonstrating superior preservation of biomolecules in the Dye 3 and John Evans silty samples, which is likely due to these samples being colder (Dye 3) or younger (John Evans) than the GRIP sample (Fig. 1A, insert). We also failed to amplify DNA from the five control samples of Holocene and Pleistocene ice taken just above the silty samples from the Dye 3 and GRIP ice cores (volumes 100g to 4kg, Fig. 1B, (7)). None of the samples studied yielded putative sequences of vertebrate mtDNA.
A previous study has shown that simple comparisons of short DNA sequences to GenBank sequences using BLAST Search make misidentification likely (15). Therefore, we assigned the sequences obtained to the taxonomic levels of order, family, or genus using a new rigorous statistical approach (7). In brief, this Bayesian method calculates the probability that each sequence belongs to a particular clade by considering its position in a phylogenetic tree based on similar GenBank sequences. In the calculation of these probabilities, uncertainties regarding phylogeny, models of evolution and missing data are taken into account. Sequences with >90% posterior probability of membership to a taxonomic group were assigned to that group. Additionally, a given plant taxon was only considered genuine if at least two sequences assigned to that taxon were found to be 100% identical and reproducibly obtained in separate analyses (for Dye 3, by independent laboratories and for the John Evans Glacier control sample within laboratory). This strict criterion of authenticity obviously dismisses many putative taxa that are present at low abundance or have heterogeneous distributions, as is typical of environmental samples (16), but efficiently minimizes the influence of possible low-level contamination and misidentifications due to DNA damage (17).
Approximately 31% of the sequences from the John Evans Glacier silty sample were assigned to plant taxa passing the authentication and identification criteria. These belong to the Rosales (an order of flowering plants, including nine families such as the rose family Rosaceae), the Salicaceae (willow) family, and the genus Saxifraga (Table 1). This result is consistent with the John Evans Glacier forming no more than a few thousand years ago in a high Arctic environment (18), characterized by low plant diversity and sparse vegetation cover similar to that currently surrounding the glacier which consists mainly of Arctic willow (family Salicaceae), purple saxifrage (genus Saxifraga), Dryas (order Rosales), and Arctic poppy (19). Thus, by confirming the expected result, the John Evans Glacier study can be regarded as a positive control showing that DNA data from silty ice reliably record the local ecology.
Table 1.
Plant and insect taxa obtained from the John Evans Glacier (JEG) and Dye 3 silty ice samples. For each taxon (assigned to order, family, or genus level), the genetic markers (rbcL, trnL, or COI), number of clone sequences supporting the identification (no. clones), and the probability support (in percentage) are shown. Sequences have been deposited in GenBank under accession numbers XXXXXXXX-XXXXXXXX
| Sample | Order | Marker/ no. clones/ support | Family | Marker/ no. clones/ support | Genus | Marker/ no. clones/ support |
|---|---|---|---|---|---|---|
| JEG | Rosales | rbcL/ 3/ 90-99 | ||||
| Malpighiales |
rbcL/ 2/ 99-100 trnL/ 5/ 99-100 |
Salicaceae | trnL/ 4/ 100 | |||
| Saxifragales | rbcL/ 3/ 92-94 | Saxifragaceae | rbcL/ 2/ 92 | Saxifraga | rbcL/ 2/ 91 | |
| Dye 3 | Coniferales |
rbcL/ 44/ 97-100 trnL/ 27/ 100 |
Pinaceae |
rbcL/ 20/ 100 trnL/ 25/100 |
Picea
Pinus |
rbcL/ 20/ 99-100 trnL/ 17/ 90-99 |
| Taxaceae |
rbcL/ 23/ 91-98 trnL/ 2/ 100 |
|||||
| Poales |
rbcL/ 67/ 99-100 trnL/ 17/ 97-100 |
Poaceae |
rbcL/ 67/ 99-100 trnL/ 13/ 100 |
|||
| Asterales |
rbcL/ 17/ 90-100 trnL/ 27/ 100 |
Asteraceae |
rbcL/ 2/ 91 trnL/ 27/ 100 |
|||
| Fabales |
rbcL/ 10/ 99-100 trnL/ 3/ 99 |
Fabaceae |
rbcL/ 10/ 99-100 trnL/ 3/ 99 |
|||
| Fagales |
rbcL/10/ 95-99 trnL/ 12/ 100 |
Betulaceae |
rbcL/ 7/ 93-97 trnL/ 11/ 98-100 |
Alnus |
rbcL/ 7/ 91-95 trnL/ 9/ 98-100 |
|
| Lepidoptera | COI/ 12/ 97-99 |
In contrast to the John Evans Glacier silty sample, the 45% of the Dye 3 DNA sequences that could be assigned to taxa reveal a community very different from that of Greenland today. The taxa identified include trees such as alder (genus Alnus), spruce (genus Picea), pine (genus Pinus), and members of the yew family (Taxaceae) (Table 1). Their presence indicates a northern boreal forest ecosystem rather than today’s Arctic environment. The other groups identified, including Asteraceae, Fabaceae, and Poaceae, are dominated by herbaceous plants and are represented by many species found in northern regions at present (Table 1). The presence of these herb-dominated families suggests an open forest, allowing heliophytes to thrive. Additionally, we recorded taxa that are common in the Arctic and/or Boreal regions but lacked 100% sequence identity between independent laboratories. These are yarrow (Achillea), birch (Betula), chickweed (Cerastium), fescue (Festuca), rush (Luzula), plantain (Plantago), bluegrass (Poa), saxifrage (Saxifraga), snowberry (Symphoricarpos), and aspen (Populus). Although not independently authenticated at the sequence level the presence of these taxa adds further support to the conclusion of a northern boreal forest ecosystem at Dye 3.
To date, the youngest well-dated fossil evidence of native forest in Greenland is from macrofossils in the deposits of the Kap København Formation from the northernmost part of Greenland and dates back to around 2.4Ma (1, 2). Other less well-dated traces of forests in Greenland include wood at two other late Cenozoic sites in northern Greenland (20), pollen spectra of unknown age in marl concretions found in a late glacial moraine, and wood and spruce seeds in eastern Greenland (21). Dye 3, almost exactly 2000km to the southwest of the Kap København Formation (Fig. 1A), therefore provides the first direct evidence of a forested southern-central Greenland.
The invertebrate sequences obtained from the Dye 3 silty ice are related to beetles (Coleoptera), flies (Diptera), spiders (Arachnida), brushfoots (Nymphalidae), and butterflies and moths (Lepidoptera) (probability supports between 50% and 90%). However, only sequences of the latter two are supported by more than 90% significance (Table 1). Thus, although detailed identifications of the COI sequences are in general not strongly supported, the results show that DNA from a variety of invertebrates can be obtained from sediments even in the absence of macrofossils as was previously shown for plants, mammals, and birds (9-11).
Several observations suggest that the DNA sequences we obtained from the Dye 3 ice are of local origin and not due to long-distance dispersal. The reproducible retrieval of diverse DNA from the silty basal ice but not from similar or larger volumes of the overlying “clean” ice largely precludes long-distance atmospheric dispersal of microfossils as a source of the DNA. Although pollen grains are found in the Greenland ice sheet, including the Dye 3 silty ice (7), the concentrations are in general too low (0.3-15 grains/liter (22), Bourgeois pers. comm.) for them to be present in the sample volumes studied. Furthermore, long-term survival of DNA in pollen has proved fairly poor (23) and the vast majority of angiosperm pollen does not contain cpDNA (24). These factors effectively exclude pollen as the general source of the silty ice plant DNA. Moreover, the Dye 3 silty ice appears to have originated as solid precipitation without going through stages of superimposed ice and most likely formed by mixing in the absence of free water (25), effectively excluding subsurface transportation. As explained in (26) the ice is believed to be predominantly of local origin having been shielded from participating in the large-scale glacier-flow by a bedrock trough, in agreement with the solid ice-mixing hypothesis (25). Thus, being of local origin, the DNA sequences from the Dye 3 silty ice must derive from the plants and animals that inhabited this region the last time it was ice free, as possible older DNA records from previous ice-free periods will vanish with the establishment of a new ecosystem, or at least be out-competed during PCR by DNA from the most recent record. Interestingly, the plant taxa suggest that this period had average July temperatures that exceeded 10°C and winter temperatures not colder than −17°C, which are the limits for northern boreal forest and Taxus, respectively (1). Allowing for full recovery of the isostatic depression that is produced by two-kilometers of ice, Dye 3 would have been about a thousand meters above sea level. In combination, these factors suggest that a high altitude boreal forest at Dye 3 may date back to a period considerably warmer than present.
There are no established methods for dating basal ice, and it remains uncertain whether the overlying clean ice of Dye 3 is temporally contiguous with the lower silty section. Therefore, in order to obtain a tentative age estimate for the Dye 3 silty ice and its forest community, we applied a series of dating techniques; 10Be/36Cl isotope ratios, single grain luminescence measurements, amino acid racemization coupled with modeling of the basal ice temperature histories of GRIP and Dye 3, and maximum likelihood estimates for the branch length of the invertebrate COI sequences (7). All four dating methods suggest that the Dye 3 silty ice and its forest community predate the Last Interglacial (LIG, ~130-116Ka) (Fig 2), which contrasts with the results of recent models suggesting that Dye 3 was ice-free during this period (27, 28). Indeed, all four dating methods give overlapping dates for the silty ice between 450Ka and 800Ka (Fig. 2), exceeding the current record of long-term DNA survival from Siberian permafrost of 300-400Ka (9). However, due to the many assumptions and uncertainties connected with the interpretation of the age estimates (7), we cannot rule out the possibility of a LIG age for the Dye 3 basal ice.
Fig. 2.
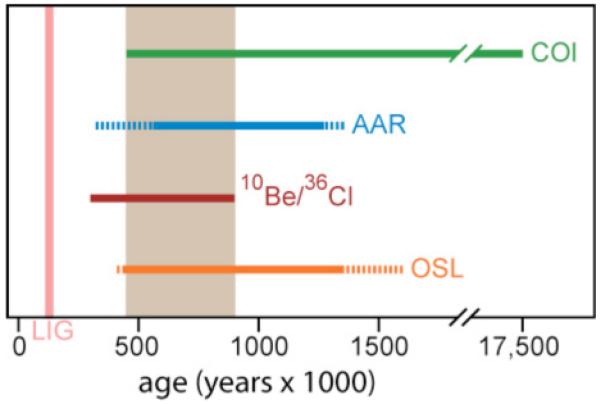
Summary of dating results for the silty ice from Dye 3. From top to bottom, the bars indicate: maximum likelihood estimates for the branch length of the invertebrate COI sequences (COI); amino acid racemization results using alternative activation energies, models of racemization behavior, and basal temperature histories (AAR); age estimate from 10Be/36Cl measurements in silty ice; minimum ages based on single grain luminescence results (OSL). The time span covered by all dating methods (450-800Ka) is marked in gray. LIG = Last Interglacial. Stippled lines represent the results of less likely models. It should be noted that the maximum age estimate for the invertebrate COI sequences is based on an unlikely slow substitution rate. For details see main text and (6).
In conclusion, our results reveal that ancient biomolecules from basal ice offer a novel means for environmental reconstruction from ice covered areas and can yield new insights into the climate and the ecology of communities from the distant past. As many deep ice cores exist from both hemispheres and further drillings are planned, this new approach may be used on a larger scale. Excitingly, basal ice at even lower temperatures than Dye 3 may contain an archive of genetic data of even greater antiquity.
Supplementary Material
Acknowledgments
We thank S. Funder, P. Hartvig, J. Bourgeois, O. Seberg, J. J. Böcher, K. Høegh, J. W. Leverenz, and S. Y. W. Ho for helpful discussions and R. Bailey, N. Belshaw, N. Charnley, C. Doherty and D. Peat for technical assistance and advice. EW, TB, MBH were supported by the Carlsberg Foundation, DK, and the National Science Foundation. EW and KP were both supported by Wellcome Trust Bioarchaeology Fellowships. NERC supported KP and MC. EC was in receipt of a Marie Curie Intra European Fellowship, grant number 501340. EW and MC acknowledge support from Europe (MEST-CT-2004-007909). MB and HNP were supported by NSERC grant #299103-2004 and McMaster University. MS and JB were supported by NSERC and the Polar Continental Shelf Project. MH was supported by the Max Planck Society.
References and Notes
- 1.Bennike O. Meddelelser om Grønland, Geoscience. 1990;23:85. [Google Scholar]
- 2.Funder S, et al. Bulletin of the Geological Society of Denmark. 2001;48:117. [Google Scholar]
- 3.Francis JE, Hill RS. Palaios. 1996;11:389. [Google Scholar]
- 4.Dansgaard W, et al. Science. 1982;218:1273. doi: 10.1126/science.218.4579.1273. [DOI] [PubMed] [Google Scholar]
- 5.Dansgaard W, et al. Nature. 1993;364:218. [Google Scholar]
- 6.Copland L, Sharp M. J. Glaciology. 2001;47:232. [Google Scholar]
- 7.Supporting Online Material.
- 8.Collins MJ, Riley M. In: Perspectives in Amino Acid and Protein Geochemistry. Goodfriend GA, et al., editors. Oxford University Press; N.Y.: 2000. p. 120. [Google Scholar]
- 9.Willerslev E, et al. Science. 2003;300:791. doi: 10.1126/science.1084114. [DOI] [PubMed] [Google Scholar]
- 10.Hofreiter M, et al. Curr. Biol. 2003;13:R693. doi: 10.1016/j.cub.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 11.Haile J, et al. Mol. Biol. Evol. 2007;24:982. doi: 10.1093/molbev/msm016. [DOI] [PubMed] [Google Scholar]
- 12.Willerslev E, et al. Trends. Ecol. Evol. 2004;19:141. doi: 10.1016/j.tree.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Hebsgaard MB, et al. Trends Microbiol. 2005;13:212. doi: 10.1016/j.tim.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Willerslev E, Cooper A. Proc. Royl. Soc. Lon. B. 2005;272:3. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofreiter M, et al. Mol. Ecol. 2000;12:1975. doi: 10.1046/j.1365-294x.2000.01106.x. [DOI] [PubMed] [Google Scholar]
- 16.Von Wintzingerode F, et al. Microbiol. Rev. 1997;21:213. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 17.Binladen J, et al. Genetics. 2006;172:733. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blake W., Jr. Radiocarbon. 1989;31:570. [Google Scholar]
- 19.Bergsma BA, et al. Arctic. 1984;37:49. [Google Scholar]
- 20.Bennike O. Geology of Greenland Survey Bulletin. 2000;186:29. [Google Scholar]
- 21.Bennike O, et al. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;186:1. [Google Scholar]
- 22.Bourgeois J, et al. J. Geophys. Res. 2001;106:5255. [Google Scholar]
- 23.Parducci L, et al. Mol. Ecol. 2005;14:2873. doi: 10.1111/j.1365-294X.2005.02644.x. [DOI] [PubMed] [Google Scholar]
- 24.Shi-yi H. Acta Botanica Sinica. 1997;391:363. [Google Scholar]
- 25.Souchez RA, et al. Geophys. Res. Lett. 1998;25:1943. [Google Scholar]
- 26.Gudmundsson GH. J. Glaciology. 1997;43:80. [Google Scholar]
- 27.Overpeck JT, et al. Science. 2006;311:1747. doi: 10.1126/science.1115159. [DOI] [PubMed] [Google Scholar]
- 28.Otto-Bliesner BL, et al. Science. 2006;311:1751. doi: 10.1126/science.1120808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.