Abstract
Structures of intact receptors with single-pass transmembrane (TM) domains are essential to understand how extracellular and cytoplasmic domains regulate association and signaling through TM domains. A chemical and computational method to determine structures of the membrane regions of such receptors on the cell surface is developed here and validated with glycophorin. An integrin heterodimer structure reveals association over most of the lengths of the α and β TM domains, and that the principles governing association of hetero and homo TM dimers differ. A turn at the Gly of the juxtamembrane (JM) GFFKR motif caps the α TM helix, and brings the two Phe of GFFKR into the α/β interface. A JM Lys residue in β also has an important role in the interface. The structure is incompatible with previous NMR JM/cytoplasmic complex structures, and together with NMR structures of isolated α and β TM domains, shows how TM association/dissociation regulates integrin signaling. A joint ectodomain and membrane structure shows that substantial flexibility between the extracellular and TM domains is compatible with TM signaling.
Introduction
Membrane proteins with a single transmembrane (TM) domain per monomer are important in transmitting signals between the extracellular and cytoplasmic environments. For example, signal transmission in receptor kinases generally involves ligand-triggered association of monomeric single-TM proteins into homodimers or heterodimers (Zhang et al., 2006).
Integrins, adhesion receptors that transmit signals bidirectionally across membranes, are another heavily studied class of signaling receptors (Luo et al., 2007; Wegener and Campbell, 2008). Integrins have two subunits which constitutively heterodimerize through their large, ligand-binding extracellular domain. Reversible association through the single-pass α and β subunit TM domains mediates TM signaling. The cytoplasmic domains are short and can bind cytoskeletal proteins.
Structures are known in both active and inactive conformations for receptor kinase extracellular and cytoplasmic domain fragments (Zhang et al., 2006), and for integrin extracellular ligand-binding fragments (Luo et al., 2007) and cytoplasmic domains (Vinogradova et al., 2002). However, none of these structures include TM domains, and how signals are transmitted between these domains and across the membrane is largely unknown. In contrast, structural work on signaling proteins that span the membrane six or more times, such as channels and G-protein coupled receptors, is far more advanced.
Thus far, our understanding of how single-TM proteins associate in the membrane is confined to a limited number of experimental studies on isolated TM domains. Structures of the constitutively associating TM domains of the erythrocyte glycoprotein glycophorin A (GPA) have been solved in detergent and in pelleted vesicles (MacKenzie et al., 1997; Smith et al., 2001). A constitutively disulfide-linked TM dimer was also structurally defined in detergent by NMR (Call et al., 2006). Bicelles, made of a mixture of lipids and short-chain, detergent-like lipids, are closer mimics of bilayers than detergents. Two further dimeric TM fragment structures (Bocharov et al., 2008a; Bocharov et al., 2008b), and monomeric αIIb and β3 TM domain structures (Lau et al., 2008a; Lau et al., 2008b), have recently been determined in bicelles. However, since regulated TM domain association is driven by domains outside the membrane, structures of such associated TM domain fragments beg the question of whether they are physiologically relevant, and if so, whether they correspond to resting or active conformations.
The biological importance of integrins, and the tractability of studying regulated TM and cytoplasmic domain association in the absence of ligand, in contrast to most other signaling receptors, has made them an attractive model system for studying association of their TM and cytoplasmic domains (Wegener and Campbell, 2008). In integrins, the α and β subunit TM domains associate in the resting state, driven by the close proximity of the C-termini of α and β subunit ectodomains in the bent conformation. TM and cytoplasmic domain separation, induced by binding of the β subunit cytoplasmic domain through talin to the actin cytoskeleton, drives integrin extension, and shifts the ligand-binding α/β headpiece to an open, high-affinity conformation (Luo et al., 2007; Wegener and Campbell, 2008; Zhu et al., 2008). Disulfide cross-linking of portions of the TM domains in the outer membrane leaflet demonstrates helicity and the approximate orientations between the αIIb and β3 helices in the resting state, and an absence of association in the active state (Luo et al., 2004). Various TM domain orientations have been proposed based on these data or mutagenesis (Gottschalk, 2005; Li et al., 2005; Partridge et al., 2005). A comprehensive NMR complex structure showed how the integrin cytoplasmic domains associated, and provided a model for integrin activation by showing that the cytoskeletal protein talin dissociated the complex (Vinogradova et al., 2002). However, different α/β complex structures, or a lack of association have also been described (Ulmer et al., 2001; Vinogradova et al., 2004; Weljie et al., 2002). A unique feature of integrins is a highly conserved GFFKR motif following the α subunit TM domain. Mutation of any of the residues in this motif activates integrins by destabilizing association of αand β subunit TM domains (Hughes et al., 1996; O’Toole et al., 1994).
Disulfide cross-linking has been used to obtain structural information about membrane proteins, especially to probe TM domain organization and conformational change in bacterial chemoreceptors (Bass et al., 2007). However, development of this technique has not proceeded to the level of describing three dimensional structures. Rosetta uses knowledge-based as well as physicochemical potentials and an efficient method of sampling conformational space to find low energy structures. Furthermore, the potentials in Rosetta can be combined with experimental restraints (Das and Baker, 2008).
Here, we use disulfide-based distance restraints with Rosetta Membrane (Barth et al., 2007; Barth et al., 2009) to characterize the structure of the TM and juxtamembrane (JM) domains in intact integrins on the cell surface. The juxtamembrane GFFKR motif in and a juxtamembrane Lys in β form an important part of the intersubunit interface. The conclusions are supported by mutational studies. Joining an αIIbβ3 ectodomain crystal structure to the membrane structure yields insights about integrin orientation on the cell surface, and the finding that ectodomain flexibility is compatible with transmembrane signaling.
Results
Method Development
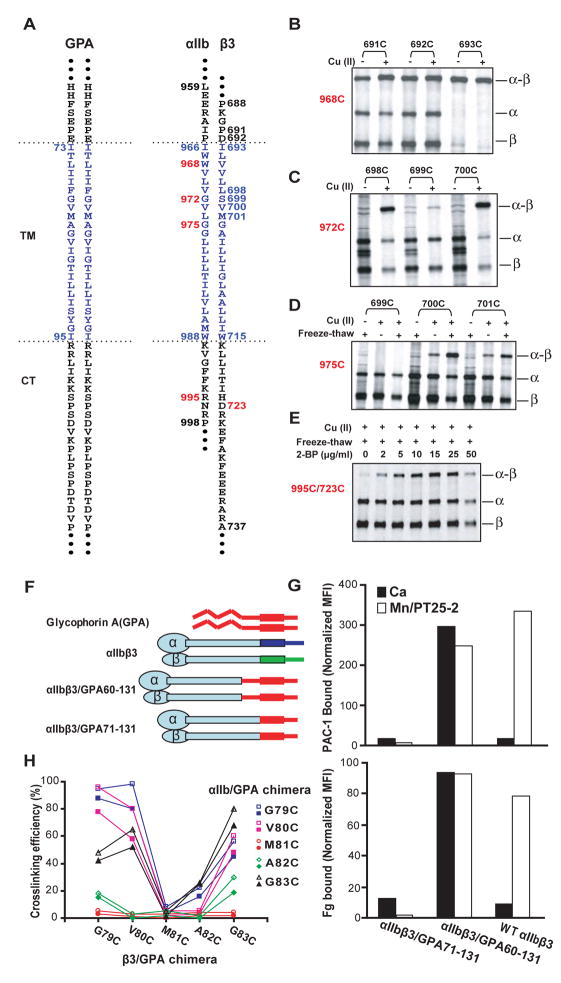
We extended previous work on disulfide crosslinking of the first 9 integrin TM residues (Luo et al., 2004) across the membrane and into the cytoplasm. Residues in the JM and TM segments of αIIb, β3, and GPA (Fig. 1A) were individually mutated to cysteine. αIIb and β3 mutants were co-expressed in 293T transfectants. Cells were treated with or without Cu (II)-o-phenanthroline (Cu-phenanthroline) to catalyze disulfide bond formation, quenched with N-ethylmaleimide, and lysed with detergent. [35S] integrins were immunoprecipitated and subjected to nonreducing SDS-PAGE (Fig. 1B–D), and disulfide-linked heterodimer was quantitated as % of total integrin. Disulfide bond formation within the first few TM residues was not increased further by Cu-phenanthroline (Fig. 1B). Disulfide formation in this region and external to the membrane was also quantitated in redox buffers and after reduction with dithiothreitol followed by Cu-phenanthroline (Methods and Fig. S1). By the seventh TM residue, disulfide formation was almost completely Cu-phenanthroline dependent (Fig. 1C) (Luo et al., 2004), whereas by the tenth TM residue, Cu-phenanthroline alone had little effect (Fig. 1D). However, disulfide cross-linking in the middle and cytoplasmic side of the membrane was made possible by freeze-thawing cells, which enables access of Cu-phenanthroline and oxygen to the otherwise reducing cytoplasmic environment (Fig. 1D and E). Freeze-thawing had no effect on crosslinking of more exofacial residues (Fig. S2). Cysteines near the membrane/cytoplasm interface may be palmitylated and thus unavailable for crosslinking (Kovalenko, 2004). Blocking palmitylation with 2-bromopalmitate (2-BP) markedly enhanced cross-linking efficiency (Fig. 1E).
Figure 1. Disulfide crosslinking in native cell membrane.
A. Sequences of GPA and αIIbβ3 integrin TM and cytoplasmic domains. Numbers in red show positions tested in panels B–E. B–E. Disulfide crosslinking of αIIbβ3 with indicated cysteine mutations in 293T transfectants, with or without Cu-phenanthroline, freeze-thaw, and 2-BP treatment as indicated. Immunoprecipitated 35S-labeled material was subjected to nonreducing SDS-PAGE and autoradiography. Positions of αIIb (α), β3 (β) and αIIbβ3 heterodimer (α-β) are shown. F. Integrin αIIbβ3/GPA chimeras. G. Ligand binding by chimeras. Binding of ligand mimetic PAC-1 (IgM) (upper panel) or FITC-labeled fibrinogen (Fg) (lower panel) to 293T transfectants was measured in the presence of 1mM Ca2+ or 1mM Mn2+ plus 10 μg/ml activating mAb PT25-2. Binding is expressed as mean fluorescence intensity (MFI) of Fg or PAC-1 relative to MFI of Cy3-labled anti- β3 mAb AP3. H. αIIbβ3/GPA60–131 (closed symbols) and αIIbβ3/GPA71–131 (open symbols) chimeras show similar disulfide crosslinking.
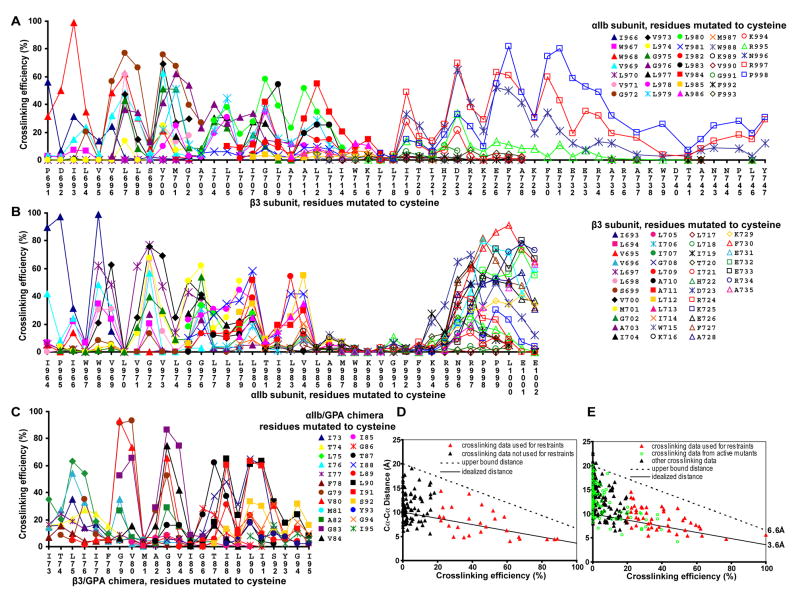
Standard cross-linking conditions thus employed 15 μg/ml 2-BP, a 1.5h pulse with [35S] methionine and cysteine, a 17h chase, freeze-thaw, and treatment with Cu-phenanthroline for 10 min at 0°C. Kinetics showed that cross-linking was maximal at 10 min., even with cysteine pairs showing only partial cross-linking (Fig. S3). In cysteine crosslinking, it is thought that a thiol peroxide (S-OH) equivalent to sulfenic acid is the first oxidation product, and that nucleophilic attack on the oxidized cysteine by a second, unoxidized cysteine SH group results in disulfide formation, with release of water. We hypothesize that crosslinking by the second, unoxidized cysteine is in competition with oxidation of the second cysteine, which would prevent crosslinking to the first cysteine and account for the lack of further crosslinking after 10 min. The dependence of disulfide formation on kinetics of oxidation of the first cysteine, and its limitation by the kinetics of oxidation of the second cysteine, may make the extent of cross-linking relatively insensitive to kinetic differences in SH oxidation at different depths within the membrane, because the first and second cysteines must be nearby and at similar depths. This hypothesis agrees with the lack of correlation between depth in the membrane and extent of crosslinking (Fig. 2).
Figure 2. Disulfide crosslinking efficiency and correlation with Cα-Cα atom distance.
Disulfide crosslinking in transfectants between subunits with indicated cysteine mutations. As described in Methods, data is with 2-BP, freeze-thaw, and Cu-phenanthroline, except GPA residues 73–80 are with DTT and Cu-phenanthroline. A and B show the same αIIbβ3 disulfide crosslinking data, plotted against β3 (A) or αIIb sequence position (B). C. GPA TM domain disulfide crosslinking in αIIbβ3/GPA71–131 chimera. D and E show the same crosslinking data as in A–C, plotted against Cα-Cα distance. D. Crosslinking efficiency in αIIbβ3/GPA71–131 chimera is plotted against Cα-Cα distance in the GPA solid state NMR structure (Smith et al., 2001). E. Crosslinking efficiency in αIIbβ3 is plotted against Cα-Cα distance in the final integrin structure. Solid lines show the assumed relationship between crosslinking efficiency and Cα-Cα distance, and dashed lines show the distance, above which, restraint violation was penalized in Membrane Rosetta.
Dimerization through the GPA TM domains is constitutive, and is not dependent on the extracellular, O-glycosylated domain (MacKenzie et al., 1997). The αIIb and β3 ectodomains were fused to GPA residues 71–131, comprising the TM and cytoplasmic domains, or GPA residues 60–131, which include an additional 11 extracellular residues (Fig. 1F). The use of αIIb GPA/β3 GPA heterodimers enabled measurement of crosslinking not only between identical GPA TM residues (e.g. 80 with 80), but also between nonidentical residues (e.g. 80 with 81). Compared to wild-type αIIbβ3, the αIIbβ3/GPA 60–131 heterodimer was constitutively active, whereas αIIbβ3/GPA 71–131 was resistant to activation (Fig. 1G). These results are consistent with the importance of close association between the C-termini of integrin α and β subunit ectodomains in stabilizing the resting state (Luo et al., 2007), and stronger association between the GPA than the integrin TM domains. The αIIbβ3/GPA60–131 and αIIbβ3/GPA71–131 chimeras gave essentially identical disulfide crosslinking (Figure 1H), showing that GPA TM domain association was unaffected by the length of GPA extracellular domain included in the chimera. Crosslinking between non-identical GPA transmembrane residues was also independent of the integrin subunit fusion partner (Fig. S4).
Crosslinking and Restraints
The integrin and GPA TM domains show α-helical periodicity with crosslinking peaks every 3–4 residues, suggesting helix against helix packing (Fig. 2A–C). Similar periodic peaks in the β3 cytoplasmic domain at residues I719, D723, F727, and F730 (Fig. 2A) suggest that this region is α-helical as well, and interacts with the Lys and Arg residues of the GFFKR motif and several following residues in the α subunit.
The GPA crosslinking data fit the GPA NMR structure well (Fig. 2D) with the assumption that maximal crosslinking of 100% would be seen with maximal contact between Cys Cα atoms at the van der Waals distance of 3.6 Å, and that crosslinking would linearly fall to 0% when this distance was increased to 10 Å (solid line, Fig. 2D). Because distance is not the only factor that could affect crosslinking efficiency, it was important not to over-restrain the distance between crosslinked residues. Therefore, an upper distance bound was chosen, below which no penalty would be applied (dashed lines, Fig. 2D and E). Restraints were used only for residue pairs with ≥20% crosslinking (Fig. 2DE) and only for residues that did not activate ligand binding by αIIbβ3 when individually mutated to cysteine (see below). Furthermore, to compensate for inherent flexibility or structural perturbations introduced by cysteine substitution, the lowest crosslinking value within two residues was subtracted prior to restraint calculation (Fig. S5).
Structure refinement
Rosetta has an efficient method for sampling structural diversity by assembling 3 and 9-residue fragments from the protein data bank into protein structures. Fragments are selected that are similar in sequence to the target sequence, and are enriched for (but do not all have) the predicted secondary structure of the target structure. Fragments are selected from high-resolution crystal structures irrespective of whether they are membrane or water-soluble proteins. Structures are built using a Monte Carlo fragment insertion protocol, minimizing an energy function that favors hydrophobic burial, strand pairing, and other low-resolution properties of protein structures. A subsequent high-resolution refinement stage optimizes van der Waals packing, hydrogen bonding, and solvation interactions (Methods).
Membrane Rosetta uses a model of a membrane with three slabs, a central hydrophobic slab corresponding to the lipid alkyl groups, and two intermediate slabs corresponding to the interface regions between the alkyl groups and the polar headgroups. Both the low resolution (Barth et al., 2009) and high resolution (Barth et al., 2007) potentials differ from water-soluble proteins in these slabs, with exposure of non-polar groups favored rather than disfavored. The potentials for the head group regions and the external regions are the same as for water-soluble proteins. The embedding (depth and orientation) of proteins in the membrane slab is optimized to give the lowest energy. Disulfide distance restraints are incorporated as an additional potential term used to evaluate model energy; a distance greater than the upper bound shown in Fig. 2DE raises energy. This energy term is evaluated in every Monte Carlo step, and thus affects the course of model building.
Building the integrin TM and juxtamembrane domains was done in several stages, as detailed in Methods and the Supplement. In the final stage, all models with the 10% lowest energy were clustered by structural similarity (Fig. S6, S7). The vast majority of low energy structures have similar structural features (Fig. S7). Structure assessment below uses the structures at the center of the largest (top) GPA and integrin clusters (referred to as final structures), the structural ensembles within the top clusters (Table 1), and the centers of the top five clusters (Fig. S7).
Table 1.
| Table 1A. Experimental restraints and structure statistics | ||
|---|---|---|
| Experimental restraints | ||
| GPA | Integrin | |
| Number of residues | 46 | 90 (EC,12; TM, 46; CT, 32)a |
| Number of restraints | 54 (symmetrized) | 48 (TM, 33; CT, 15)a |
|
| ||
| Structure statistics | ||
| GPA (20 models) |
Integrin (52 models) |
|
| TM (46 residues) | TM + CT (78 residues)b | |
| Cα-RMSD (Å) to cluster center model | 0.68±0.23 | 1.94±0.42 |
| Distance restaints RMS violation (Å) | 0.01±0.02 (0.00)c | 0.06±0.04 (0.00)c |
| Angle between TM helix axes (°) | −45.4±1.8 (−43.1)c | −37.5±3.1 (−36.9)c |
| Distance between TM helix axes (Å) | 7.3±0.49 (7.1)c | 8.2±0.77 (7.8)c |
| RMSDs from idealized geometry | ||
| Bonds (Å) | 0.0067±0.008 | 0.012±0.005 |
| Angles (°) | 0.28±0.0015 | 0.57±0.10 |
| Ramachandran statisticsd | ||
| Ramachandran favored | 100.0% | 99.8±0.4% |
| Ramachandran outliers | 0.0% | 0.02±0.15% |
| PDB code | ||
| Table 1B. Comparison of solution NMR structure (1AFO) and disulfide/ROSETTA GPA structures to solid state NMR structure | ||||||
|---|---|---|---|---|---|---|
| I73-I95 (46 residues) |
I76-I88 (26 residues) |
|||||
| Atoms | Cα | Backbone | All heavy | Cα | Backbone | All heavy |
| RMSD (Å) | ||||||
| 1AFO (20 models) | 0.97±.0.10 | 1.14±0.11 | 1.69±0.08 | 0.67±0.10 | 0.64±0.09 | 1.09±0.18 |
| Disulfide/ROSETTA (20 models) | 0.96±0.15 | 1.09±0.13 | 1.59±0.19 | 0.61±0.12 | 0.59±0.12 | 1.27±0.20 |
| Largest cluster center model | 0.71 | 0.84 | 1.28 | 0.44 | 0.43 | 0.91 |
EC, extracellular region; TM, transmembrane region; CT, juxtamembrane and cytoplasmic region.
Only TM (I966-W988 of αIIb subunit and I693-W715 of β3 subunit) and CT (K989-P998 of αIIb subunit and K716-A737 of β3 subunit) residues are included.
Values are for the cluster center structure.
Ramachandran statistics were measured by MolProbity [Davis, 2007 #18419].
Structure Assessment
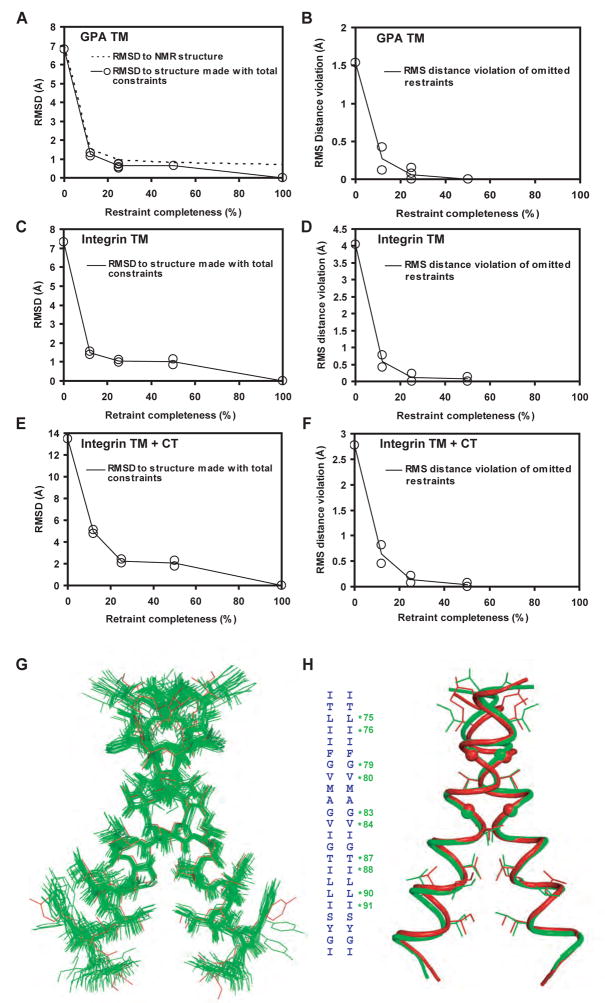
Cross-validation with omitted restraints is used for the validation of crystal and NMR structures (Brunger et al., 1993). GPA and integrin models generated without restraints were far from the GPA NMR and integrin final models (Fig. 3 and S8). When 12.5% restraints were used, models were much closer. When 25 and 50% restraints were used, the models approached a plateau in Cα RMSD for integrin of 1 Å for TM and 1.8 Å for TM + cytoplasmic domains and 0.8 Å for GPA (Fig. 3A,C,E). This provides one estimate of structure accuracy. Crystal structure accuracy is evaluated by omitting data from refinement, and checking the agreement between the omitted data and the final structure (Rfree). Similarly, we checked structures for violations of restraints that were not used in structure generation (Fig. 3B,D,E). When 50% of restraints were used in structure generation, there was little violation of the other 50% of omitted restraints (Fig. 3B,D,E). These results suggest that at least for the regions of the structures that are well covered by restraints, i.e. the TM regions, the structures match the actual structure well.
Figure 3. Estimation of structure accuracy and validation of the GPA cell surface structure.
A–F. Effect of restraint omission. Models were generated using different subsets of 12.5, 25, or 50% of the total restraints as described in the Supplement. A, C and E. Models were compared over Cα atoms to the final structure made with all restraints, and in the case of GPA, also to the GPA NMR structure. By definition, models made with 100% of restraints are identical to the final structure. B, D, and F. Models generated with a subset of restraints were scored for violation of the omitted restraints, i.e., those not used in model generation. The per residue RMS distance violation, i.e., distance above the upper bound (see Fig. 2D and E), is shown. How well the models satisfy omitted restraints is a measure of model accuracy similar to the Rfree value in crystallography. G. The ensemble of 20 GPA Disulfide/Rosetta structures, showing all heavy atoms (green) superimposed on the solid state NMR structure (red). H. Cartoon of the central GPA Disulfide/Rosetta structure (green) superimposed on the solid state NMR structure (red) with sidechains (or spheres for Gly) shown for residues in crosslinking peaks (asterisked in the sequence insert).
The solid state GPA NMR structure, which fits the solution state restraints better than the solution NMR structure (Smith et al., 2001), fits the solution state NMR and Disulfide/Rosetta structures, respectively, with Cα RMSD of 0.97±0.10 and 0.96±0.15 Å (see Table 1B for more details). These results show that the Disulfide/Rosetta structure is as similar to the solid state NMR structure as the latter is to the solution state NMR structure. The similarities between the Disulfide/Rosetta and NMR structures extend to details including sidechain rotamers (Fig. 3G and H). For example, sidechains in cross-linking peaks have identical rotamers, except for Leu-90 and Ile-91 in one of the two Disulfide/Rosetta TM helices (Fig. 3H), which in contrast to the solid state NMR structure, were not constrained to be symmetric.
The final αIIbβ3 membrane structure showed no restraint violation (Fig. 2E) and the ensemble showed little restraint violation (Table 1A). Furthermore, crosslinking data from activating cysteine mutations that were omitted from structure calculation also did not violate restraints and trended well with the idealized crosslinking-distance relationship (Fig. 2E and Table S1). The apparent lack of effect on crosslinking by these activating mutations may result from the use of 0°C and 37°C in crosslinking and activation assays, respectively.
αIIbβ3 Membrane structure and comparison to mutational results
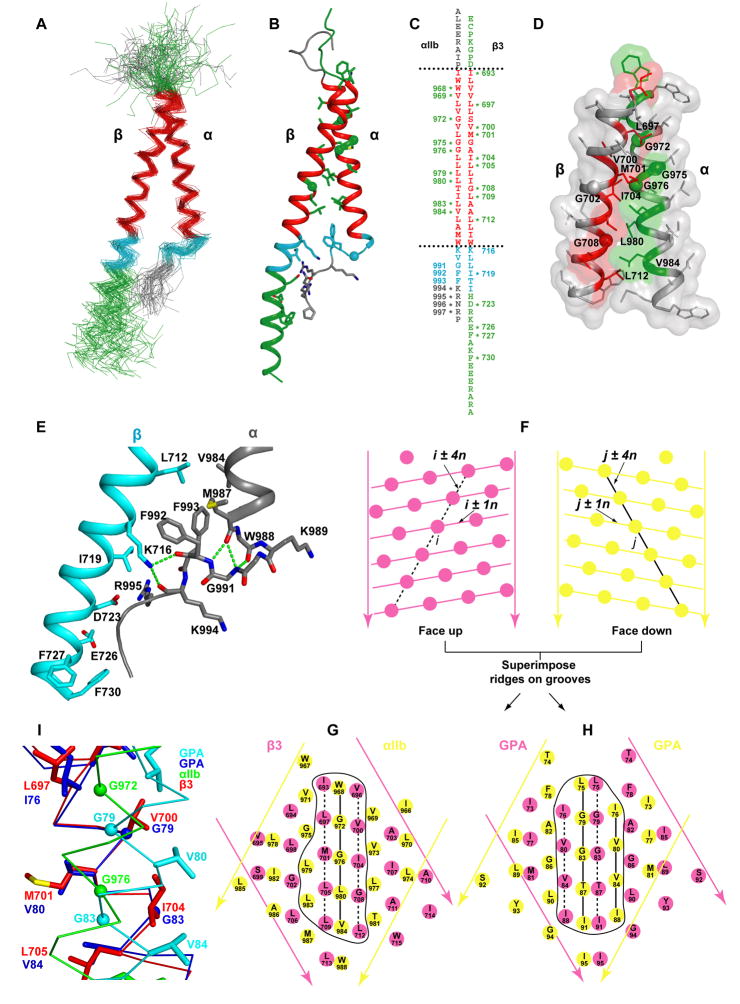
All low energy αIIbβ3 structures have the same interhelical interface, and almost all have the Gly of GFFKR in a left-handed helix conformation that caps the αIIb α-helix and brings the two Phe of GFFKR into the interface with the β3 α-helix (Fig. 4A–E and S7A, B). These key features are seen in all 5 top clusters with clustering at 2.0 Å (Fig. S7A) and 4 of 5 of the top clusters at 3.0 Å (Fig. S7B). The sole exception, Cluster 3 with clustering at 3.0 Å (Fig. S7B), has the highest disulfide restraint violation (Fig. S6B), and can be ruled out because it is incompatible with mutational data described below that show that the αIIb GFFKR motif and β3 Lys-716 have an important role in the αIIb/β3 interface. The β3 TM α-helix continues as α-helix into the cytoplasm (Fig. 4B), and in some clusters the cytoplasmic portion of the β3 TM α-helix is divided into two segments separated by a turn near Glu-726 (Fig. S7)
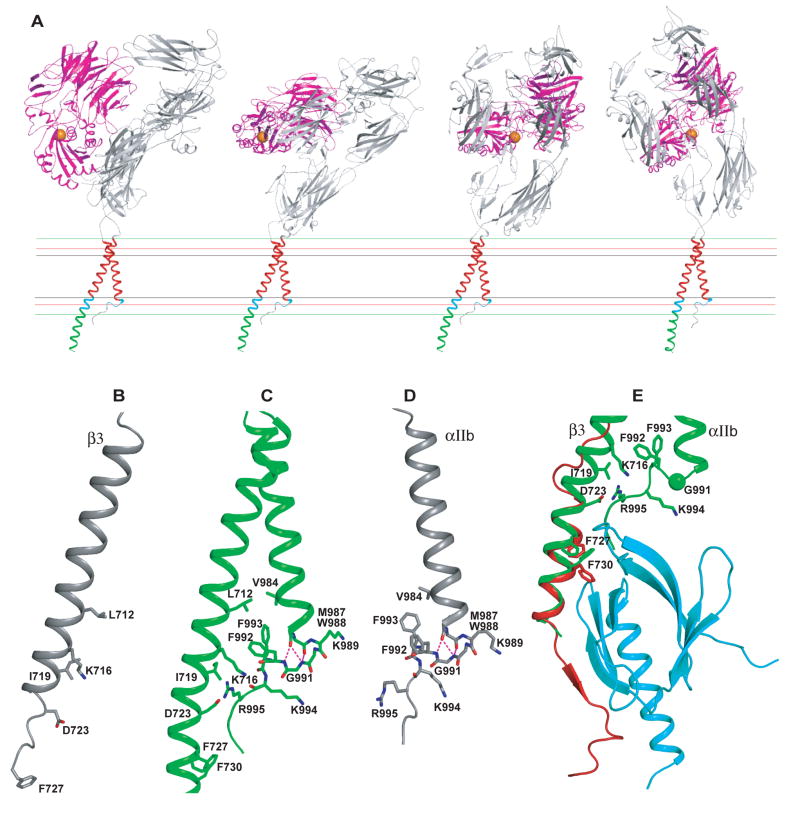
Figure 4. Structure of the membrane region of an integrin.
A. The disulfide/Rosetta structural ensemble superimposed on the 46-residue TM segments. B. The cluster-center structure. C. The sequences, with residues in crosslinking peaks asterisked. D. Transparent molecular surfaces for αIIb and β3 TM segments with all sidechains shown. In A–D, sidechains and Gly Cα spheres in the crosslinking peaks in the 23 residue hydrophobic segment are shown in red or green. Residues in the cytoplasmic juxtamembrane interface region (18 to 12 Å from the membrane center) are cyan. E. A blow-up of the lower TM, JM, and cytoplasmic segments. Hydrogen bonds are dashed green lines. Nitrogens, oxygens, and sulfurs are blue, red and yellow, respectively. F–H. Crick helical-net diagrams (Chothia et al., 1981). Cylindrical helical surfaces are cut at one position along the circumference, unrolled, and aligned at the helical interfaces. I. Superposition of the integrin and GPA TM helices, on residues that form the two ridges forming the groove in β3 and one GPA monomer in which the GXXXG motifs nestle.
The integrin TM domains associate with a right-handed crossing angle of −37° compared to −41 to −45° in GPA (Table 1) (MacKenzie et al., 1997; Smith et al., 2001). The most prominent features of α-helical surfaces are ridges formed by the sidechains of every fourth residue in sequence (i+4n), and grooves in between (Fig. 4F). In a common packing mode between adjacent α-helices, observed for α-helices that cross at angles of −50° ± 20°, a ridge formed by residues i+4n on one helix fits into a groove on another helix between the ridges formed by residues j+4n and j+1+4n (Chothia et al., 1981). This has been termed 4-4 packing. The αIIb and β3 TM α-helices show precisely this packing mode (Fig. 4F and G). The ridge is on αIIb, and is formed by residues W968, G972, G976, and L980. The groove is on β3 between residues V696 and L697, V700 and M701, I704 and L705; G708 and L709 (Fig. 4G). Residues in crosslinking peaks are found at the interface (Fig. 4C, D, and G). Since the ridges and grooves spiral on the side of the cylindrical α-helix, they are convex. The G972-XXX-G976 motif in αIIb decreases the height of the ridge at the center of the packing interface, enabling the two helical cylinders to pack closer together.
In the NMR and cell surface GPA structures, the TM helices pack most closely together at a GXXXG motif (MacKenzie et al., 1997) (Fig. 3H and 4I). Two differences in interhelical orientation with integrins are apparent when the grooves in β3 and one of the GPA monomers are superimposed (Fig. 4I). 1) Although packing in GPA resembles ridge in groove (MacKenzie et al., 1997), the two low ridges bearing G79 and G80 move closer to one another than in integrin (horizontally in Fig. 4H), providing more separation from the high ridges bearing V80 and V84. 2) The ridge in the integrin slides in its groove relative to GPA (vertically in Fig. 4G), so that the spacing of residues in the interface differs (Fig. 4G–H).
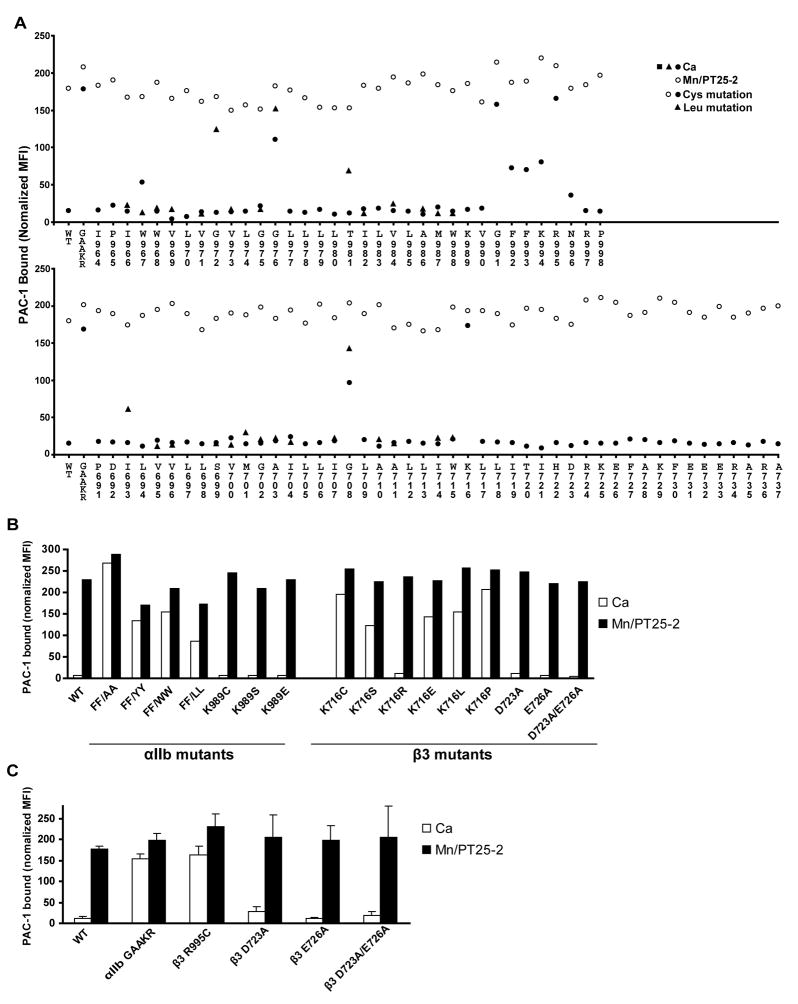
Mutational studies are in excellent agreement with the inter-helical integrin interface identified here. Cysteine and leucine scanning show that the most activating, and hence structurally disruptive mutations within the TM domains, are of the three interfacial Gly residues, αIIb Gly-972 and Gly-976, and β3 Gly-708 (Fig. 5A) (Luo et al., 2005). By contrast, mutations of αIIb Gly-975 and β3 Gly-702, which are not interfacial (Fig. 4D and G), are not activating (Fig. 5A). These results correlate with the observations that the interfacial, but not the non-interfacial Gly residues, are invariantly small (Gly, Ala, or Ser) residues in integrin TM domains (Fig. S10).
Figure 5. Ligand binding by αIIbβ3 integrin mutants.
A. Binding of ligand-mimetic PAC-1 IgM by cysteine-scanning mutants in αIIb (upper panel) and β3 (lower panel). Results in absence (Ca) and presence of activation (Mn/PT25-2) are shown, along with wild-type (WT) and GFFKR/GAAKR mutant controls. Leucine-scanning results from a previous study (Luo et al., 2005) are also plotted. B and C. Effects of other mutations in 293T (B) and CHO-K1 (C) transient transfectants.
The GFFKR motif has a structural role in αIIbβ3 association consonant with its long-known functional importance. The αIIb TM α-helix extends beyond the 23-residue TM hydrophobic segment through Lys-989 and Val-990. However, at Gly-991 of the GFFKR motif, the backbone adopts a left-handed rather than right-handed α-helical conformation (Fig. 4E). Gly in left-handed conformation at the C-termini of α-helices is the most common C-cap motif, and is responsible for the marked over-representation of Gly at the +1 position following α-helices (Richardson and Richardson, 1989). Almost all (91%) low energy models, and all but one outlier cluster (Fig. S7), have a turn at the Gly (Fig. S11). Many of these models, including the final structure, have Phi/Psi angles for this Gly that are disallowed for non-Gly residues (Fig. S11), potentially explaining why mutation of this residue is so destabilizing. The turn at the Gly brings the two Phe residues of the GFFKR motif into the interface between the αIIb and β3 TM α-helices (Fig. 4E). The αIIb and β3 TM α-helices are further apart near this interface than anywhere else in the membrane, and Phe-993 in particular occupies space between the two helices and is central in the interface. Phe-993 contacts on the β3 subunit Leu-712 and is nearby the aliphatic portion of Lys-716. In the αIIb subunit, Phe-993 abuts Met-987 and Phe-992. Both Phe sidechains orient toward the hydrophobic core of the membrane. A meander in the αIIb cytoplasmic domain containing the Lys and Arg of the GFFKR motif and three more C-terminal residues abuts the β3 cytoplasmic α-helix.
Mutational data support the juxtamembrane αIIb structure. Cysteine substitutions in the GFFKR motif are activating (Fig. 5A), emphasizing the important role of this motif in the juxtamembrane interface between the αIIb and β3 subunits. Mutation of FF of GFFKR to AA, YY, WW, or LL demonstrated that even the aromatic substitutions YY and WW are activating (Fig. 5B). These results are consistent with the important packing role, particularly of Phe-993, in the αIIbβ3 interface, and not with the general role that aromatic residues, particularly Trp, often play in the hydrophobic/polar membrane interface.
In the β3 juxtamembrane domain, residues Lys-716, Ile-719, and Asp-723 are in the interface with the αIIb GFFKR motif (Fig. 2A and 4E). Remarkably, the Lys-716 ε-amino group hydrogen bonds to the αIIb backbone (Fig. 4E). Hydrogen bonds to either or both of the αIIb Phe-992 and Lys-994 carbonyls, with Phe-992 predominating, are found in 31% of all low energy models, 65% of the cluster 1 ensemble, and in 5 of 9 of the cluster center models shown in Fig. S7 that have the turn at Gly-991.
Cysteine scanning of β3 showed that the only activating mutation in the juxtamembrane/cytoplasmic domain is K716C (Fig. 5A). Furthermore, this mutation is more activating than the single other activating Cys mutation identified, G708C in the TM domain. These results confirm the important structural role of Lys-716 in the αIIbβ3 interface. β3 Lys-716 was also mutated to Ser, Arg, Glu, Leu, and Pro. All substitutions were activating except for Arg (Fig. 5B), the only other sidechain which could form a similar hydrogen bond to an αIIb backbone carbonyl oxygen, and is found at this position in integrin β subunits (Fig. S10). Mutation showed that in contrast to Arg, polar (Ser, Glu) and aliphatic (Leu) sidechains could not substitute for Lys-716. These findings lend credence to the hydrogen bond between the β3 Lys-716 sidechain and the αIIb backbone.
Arg-995 of the αIIb GFFKR motif is nearby both β3 Asp-723 and Glu-726 (Fig. 4E), consistent with the proposal based on mutational studies of a salt bridge between Arg-995 and Asp-723 (Hughes et al., 1996). However, the sidechains of Arg-995 and Asp-723 sample a large conformational space, and are within hydrogen-bonding distance in only 30% of the cluster 1 structural ensemble and 16% of low energy structures (Table S2).
We have consistently observed little effect of mutation of β3 Asp-723, the putative salt-bridge partner of αIIb Arg-995 in either 293T transfectants (Fig. 5A, B) or transient CHO-K1 transfectants (Fig. 5C). Since Asp-723 mutants have been found to be active in stable CHO transfectants (Hughes et al., 1996), this may reflect differences in level of cell surface density or in assay sensitivity.
Structure of the full-length αIIbβ3 integrin
Models of the entire receptor were generated by assembling the structures of the TM+cytoplasmic domains with the ectodomain crystal structures (Zhu et al., 2008). Redox buffer disulfide restraints were used in the 6-residue αIIb and 4-residue β3 linkers between these segments (Fig. 2A,B and Table S3B). Restraints were validated based on crosslinking between nearby residues defined in αIIbβ3 and αVβ3 crystal structures; because of flexibility in crystal structures of the β3 tail domain (Zhu et al., 2008), restraints were loosened relative to the TM region (Fig. S12). The linkers were modeled while simultaneously optimizing the rigid-body orientation of the ectodomain with respect to the membrane. Two long, disordered loops at the membrane-proximal base of the αIIb calf-2 domain were included as flexible loops in models because they may impact orientation on the cell surface. The lowest energy models exhibit a diversity of ectodomain orientations (Fig. 5A). However, the range of orientations was more restricted than would be imposed solely by the physical barrier of the membrane bilayer. These results show that the TM and linker regions do not completely restrict the orientation of the ectodomain, which is still able to explore a substantial amount of conformational space. This is an important finding, because it shows that models of signal transmission between the integrin ecto and TM domains have to be compatible with substantial flexibility at the interfaces between these domains.
Discussion
Structures for intact receptors with two associating TM domains
There are currently no crystal or NMR structures of intact signaling receptors with a single transmembrane domain per monomer. NMR studies to date have been on small fragments of subunits that either constitutively associate (Call et al., 2006; MacKenzie et al., 1997; Smith et al., 2001) or undergo regulated association but are devoid of the regulating extracellular domains (Bocharov et al., 2008a; Bocharov et al., 2008b). Even the two fragment structures that nominally represent regulated receptors may be special cases, because the EphA1 receptor dimerizes in SDS, and ErbB2 physiologically only forms heterodimers, not homodimers. NMR experiments on fragments are capable of yielding more detailed structural information, but not of defining for regulated receptors whether the conformations that are experimentally captured are physiologically relevant, and whether they correspond to “on” or “off” states. The method developed here is complementary, because it characterizes structures in the presence of extracellular and cytoplasmic domains that regulate transmembrane association in intact receptors on the cell surface. The method is applicable to a large class of signaling receptors that have a single transmembrane domain per monomer or subunit and undergo regulated dimerization.
Our method combines sparse experimental data (disulfide restraints) with energy terms in Rosetta analogously to how experimental and energy terms are used in NMR and crystal model building, except that there are more energy terms in Rosetta. In crystal and NMR structures, energy terms for bond angles and distances and van der Waals repulsion are generally used; additional energy terms may be used in lower resolution structures. In lower resolution crystal and NMR structures, there is less experimental data, and the energy terms receive more weight. We may similarly think of the Membrane Rosetta models made here as low resolution structures because the weight of the energy terms relative to the experimental data is high.
Our GPA ensemble is as close to the solid state NMR structure as the solution state NMR ensemble is to the solid-state structure. The GPA Disulfide/Rosetta structure suggests that the GPA TM domain NMR structures determined in dodecylphosphocholine detergent (MacKenzie et al., 1997) and dimyristoylphosphatidylcholine or 1-palmitoyl-2-oleoylphosphatidylcholine multilamellar dispersions (Smith et al., 2001) are similar to the GPA TM domain structure on the cell surface.
Methods for validating structures with sparse experimental data such as reported here are an important area for further development (Das and Baker, 2008). However, analogous to cross-validation in NMR, or omission of data from refinement of crystal structures (Rfree) (Brunger et al., 1993), we have used restraint omission to attempt to evaluate the accuracy of our integrin structure. The estimated RMSD Cα accuracy is 1 Å for the TM portion of the integrin and 0.8 Å for GPA, which is comparable to the RMSD between our GPA membrane structure and the NMR structure. The lack of restraint violation by mutationally important residues that were omitted from the structure refinement provides further support for our integrin structure.
Previous experience with Rosetta has shown that structural features are most accurately modeled when shared by the majority of low energy structures, and when alternative low energy structures can also be ruled out by experimental data (Das and Baker, 2008). Consequently, we have focused not on a single structure, but on the overall characteristics of the 10% lowest energy structures. We have only emphasized conclusions when they were validated by a majority of low energy structures, and when alternatives could be ruled out. The most critical example of this is the GFFKR motif. We obtained restraints between the αIIb and β3 TM helices preceding GFFKR, and between the helical JM portion of β3 and the K and R residues of GFFKR and several following residues. Thus, low energy structures of the GFF moiety had to be found by Rosetta that were consistent with these restraints. The structure of the GFF moiety illustrates one of the strengths of Rosetta, which uses a large library of fragments from high-resolution structures. The most common C-cap motif for α-helices involves a Gly in left handed helical conformation (Richardson and Richardson, 1989), and the backbone hydrogen bonds in this motif include the first F in GFFKR (Fig. 4E). Thus, this conformation was readily found, and 91% of low energy structures were built including this motif. The other 9% of structures continued in right-handed α-helical conformation through the G, and do not have GFF in αIIb and K716 in β3 in the interface. These structures can be ruled out, because mutational studies show that GFF in αIIb and K716 in β3 have a critical role in αIIbβ3 association. Interestingly, the only cluster lacking the turn at the G showed an unnatural, continuously curved TM β3 α-helix, as a consequence of satisfying disulfide restraints between αIIb and β3 residues on either side of the GFF motif (Fig. S7B), presenting yet another criterion for discarding this model. We therefore conclude that both the TM and JM portions of our αIIbβ3 membrane structure are strongly supported.
The integrin TM interface
Our study definitively identifies the interface between the αIIb and β3 TM α-helices. Previous disulfide crosslinking of the first 9 residues of the TM domains established only the approximate orientation between their helices, and did not include the second G of the GXXXG motif (Luo et al., 2004). Based on that data, approximate orientations between the αIIb and β3 TM helices (Lau et al., 2008a) or detailed models (Gottschalk, 2005) have subsequently been presented. As an example of the lack of sufficiency of the previous data, the detailed model has an Cα RMSD of 2.7 Å over 44 TM residues, and does not follow the ridge-in-groove packing we describe in the inner half of the membrane (Gottschalk, 2005). Subsequent to (Luo et al., 2004), another group used a different approach, and produced two models that superimpose on our structure over 44 TM residues with Cα RMSD of 6.8 and 8.0 Å (Partridge et al., 2005). Thus, there has previously been no consensus on the orientation between the αIIb and β3 TM domains.
Compared to previous homodimer TM structures, the integrin TM heterodimer structure illustrates the important principle that ridge in groove packing in heterodimers is not constrained by symmetry, enabling packing of the ridge at intermediate positions in the groove not allowed in symmetric homodimers (vertically in Fig. 4G and H). This finding is likely to reflect general differences between homodimeric and heterodimeric TM domain association. Many single-span receptors, such as the EGFR/ErbB family, can associate either as homo or heterodimers. ErbB family heterodimers have an important role in signaling, and their TM domains associate more stably than the corresponding homodimers (Duneau et al., 2007). It will be interesting to compare homo and heterodimers within a single receptor family to determine whether differences in packing modes between homo and heterodimers regulates the stability or signaling of TM domains.
While association between GPA TM domains is constitutive and occurs even in SDS, association between the integrin TM domains is regulated and labile to detergents (Li et al., 2001; MacKenzie et al., 1997). The finding here that integrin ectodomain fusion directly to the GPA TM/cytoplasmic domains in the αIIbβ3/GPA 71–131 chimera results in resistance to activation is compatible with the greater stability of the GPA dimer. This stability is not related to surface area buried in the interface, which with a 1.4 Å radius probe is 780 Å2 for GPA, compared to 1050 Å2 for the TM domain portion of integrin and 1300 Å2 when the membrane-embedded JM portion is included. The β-branched residues, Ile, Thr, and Val, that only have one available rotamer in α-helices, are present in the GPA TM domain interface. This minimizes loss of entropy when these residues are transferred from the lipid environment to a protein interface (Liu et al., 2003; MacKenzie et al., 1997). By contrast, integrins have a substantial number of interfacial Leu and Met residues, for which the conformational entropy loss upon TM helix-helix association will be much greater. Furthermore, hydrogen-bonded Thr residues stabilize the GPA but not integrin interface (MacKenzie et al., 1997; Smith et al., 2001).
The JM interface and comparison of associated and dissociated integrin subunits
The juxtamembrane αIIbβ3 structure is most unusual. The αIIb α-helix is terminated by a turn at the Gly of GFFKR motif that brings its Phe residues into the interface with the β3 subunit and partially embeds them in the hydrophobic portion of the membrane bilayer. Furthermore, a Lys residue immediately following the β3 TM hydrophobic segment also participates in the interface with αIIb. These key structural findings are all supported by mutational studies. Our study demonstrates the importance of juxtamembrane segments in dimerization. Previous homodimeric NMR structures of associating TM domains (Bocharov et al., 2008a; Bocharov et al., 2008b; Call et al., 2006; MacKenzie et al., 1997; Smith et al., 2001) show different packing modes between two TM α-helices, but do not contain additional segments that contribute to packing within the membrane. JM amphipathic segments intervene between TM domains and intracellular signaling domains in many receptors, including the epidermal growth factor receptor, and thus may be of key importance in signal transduction (McLaughlin et al., 2005). The demonstration here that the integrin α subunit GFFKR motif is an amphipathic, JM segment that directly participates in the association between the α and β TM domains raises the intriguing possibility that amphipathic JM segments in other receptors may participate in dimerization interactions between their TM domains.
Careful studies by Ulmer et al. have described the NMR structures of isolated αIIb and β3 TM/cytoplasmic domain fragments in bicelles (Lau et al., 2008a; Lau et al., 2008b). In contrast to findings that similar αIIb and β3 constructs formed homomultimers in detergents (Li et al., 2001), Ulmer and coworkers found that these subunits were monomeric in the more bilayer-like bicelle environment. Comparisons between the isolated peptide and membrane complex structures show a remarkable similarity in the αIIb structures in the GFFKR motif (Fig. 6B–D); it should be noted that no information from the monomer structures was used in the calculation of the αIIbβ3 complex structure. The backbone conformation is identical through the GFFKR motif to Lys-994, which is the last structured residue in the bicelle αIIb structure (Lau et al., 2008a). The same hydrogen bonds that support the turn at Gly-991, between the backbone O atom of Met-987 and N atoms of Gly-991 and Phe-992, are found in both structures, and in general, in the C-cap motif with Gly in left-handed helical conformation (Richardson and Richardson, 1989). Moreover, Phe-993, which is central in the αIIbβ3 complex structure, and has a well-defined rotamer in the αIIb structure, has the same rotamer in the complex structure (Fig. 6B–D).
Figure 6. Structure in the membrane and comparison to isolated αIIb and β3 TM domains and β3 talin complex.
A. Low energy orientations of the αIIbβ3 ectodomain on the cell surface. Four favorable cell surface orientations of intact αIIbβ3 are shown, superimposed on the TM domains. The ligand-binding αIIbβ-propeller and β3 I domains are shown in magenta, and ligand-binding I domain Mg2+ ion as an orange sphere. The outer bounds of the hydrophobic, interface, and polar regions of the membrane are shown as black, red, and green lines, respectively. B–D. The NMR bicelle structures of β3 (Lau et al., 2008b) (B) and αIIb (Lau et al., 2008a) (D) are shown in the same orientation as the subunits of the αIIbβ3 complex (C). E. Superposition on the αIIbβ3 complex of the NMR structure of β3 integrin cytoplasmic tail fragment in complex with talin F3 domain (Wegener et al., 2007). The integrin heterodimer structure is in green. The talin F3 domain is in cyan and the β3 cytoplasmic tail fragment of the NMR structure is in red. The side chains of the two phenylalanine residues (F727 and F730) are shown as sticks.
The isolated αIIb and β3 TM domain structures provide the basis for comparisons to our complex structure. There is no precedent for association between TM monomers, one of which has a significant juxtamembrane segment that associates with the α-helical TM segment. It was unknown whether the αIIb juxtamembrane segment would alter conformation after association. Indeed, there was good reason to expect that the conformation of the αIIb GFFKR segment would alter after association with β3, because as pointed out (Lau et al., 2008a), residues in this motif were found to be in α-helical conformation in an αIIbβ3 complex NMR structure containing the entire GFFKR motif and two preceding residues (Vinogradova et al., 2002). Thus, it is surprising that the only significant difference between the structured portions of the peptide and complex structures is a difference in tilt between the TM and JM segments. This tilt brings Phe-993 in monomeric αIIb 2 Å closer to Val-984 to pack against it in van der Waals contact (Fig. 6D). In the αIIbβ3 complex, β3 Leu-712 contacts αIIb Val-984, and there is not room for Phe-993 to come as close (Fig. 6C). Thus, some conformational adjustments must occur between the associated and dissociated states of the αIIb subunit.
Another interesting juxtamembrane feature is the sidechain-backbone intersubunit hydrogen bond donated by the β3 Lys-716 sidechain. A caveat is that this hydrogen bond appears in only 65% of structures in the ensemble. However, the presence of such a bond is strongly supported by the ability of Arg but not other amino acids to substitute for Lys. This bond is to the αIIb backbone near the end of the GFFKR motif, and thus stabilizes both αβ association and GFFKR conformation. In our structure, Lys-716 is by far the most important β3 residue for interaction with αIIb in the JM region, and it is fitting that β3 Lys-716 and the αIIb GFFKR motif, which interact with one another, are the sites where mutations are most disruptive to αβ interaction. Arg and Lys are abundant at the boundary between TM and cytoplasmic domains where they determine the polarity of membrane insertion by the positive-inside rule (von Heijne, 1992); thus, while Lys and Arg are of general importance, a specific role for β3 Lys-716 was unanticipated.
Comparison to previous complex structures
Surprisingly, our results disagree with a prominent NMR structure of the αIIbβ3 JM/cytoplasmic complex (Vinogradova et al., 2002). There is no similarity whatsoever in the interfaces between the NMR complex structure in aqueous media and our cell surface complex structure (Fig. S13A). The previous comprehensive study showed that mutations that activate integrins disrupted the αIIbβ3 complex, and that the talin head domain bound to the β3 JM/cytoplasmic fragment, and disrupted its complex with the αIIb JM/cytoplasmic fragment (Vinogradova et al., 2002). Other NMR studies have yielded conflicting results. Refinement by one group did not converge on a single NMR structure, but on two different structures (Weljie et al., 2002), neither of which is in agreement with that of (Vinogradova et al., 2002) or described here (Fig. S13B,C). Furthermore, the same αIIb JM/cytoplasmic fragments that associated in aqueous media were found not to associate in the presence of dodecylphosphocholine (Vinogradova et al., 2004). Moreover, a lack of complex formation has been reported even with proximity enforced by fusion of αIIb and β3 JM/cytoplasmic fragments to coiled-coils (Ulmer et al., 2001).
In retrospect, perhaps it is not surprising that none of the NMR structures resemble the Disulfide/Rosetta structure (Fig. S13). The two Phe residues of the GFFKR motif pack in the hydrophobic lipid environment, and in an interface against the sidechains of the α and β TM helices, all of which are missing in the discrepant structures. The lack of appropriate structure of the juxtamembrane/cytoplasmic segments when isolated from the TM segments, and the lack of association between the α and β TM segments in intact cells when the GFFKR motif is deleted (Luo et al., 2004), demonstrate cooperativity between these segments for folding and assembly, consistent with their intimate interaction in the Disulfide/Rosetta structure.
Integrin structure on the cell surface
Using a crystal structure of the αIIbβ3 extracellular domain (Zhu et al., 2008), the αIIbβ3 membrane structure, and disulfide restraints on the short extracellular linkers, we characterized the structure of intact αIIbβ3 on the cell surface. Multiple low-energy orientations between the extracellular and transmembrane domains were found. This finding suggests that the orientation of integrins on the cell surface is dynamic rather than fixed, and that the 6-residue αIIb and 4-residue β3 linkers between the extracellular and TM domains are flexible. The lack of a fixed structure for this region is consistent with marked variation in the sequence and length of these linkers, particularly among integrin subunits. For example, the αV subunit, which also associates with β3, has an 8-residue QPAPMPVP linker compared to the 6-residue EERAIP linker in αIIb.
In contrast to our results, a cryoEM structure of native αIIbβ3 in octylglucoside detergent suggested a defined, extended orientation between the extracellular and TM domains (Adair and Yeager, 2002). However, the cryoEM extracellular domain structure does not agree with that determined in crystals or negative stain (Zhu et al., 2008) and it has been suggested that the cryoEM structure may have averaged over bent and extended integrin conformations (Luo et al., 2007).
Our finding that limited but not extensive flexibility between the extracellular and TM domain is compatible with signal transduction has important implications. Flexibility and variation in the ectodomain-TM domain linkers are compatible with activation by large-scale movements such as separation in the plane of the membrane between the TM domains (Luo et al., 2004), but not with small-scale twisting or pistoning motions with which the linkers could comply. On the other hand, extreme flexibility in the linkers decouples extracellular domain activation from the TM domains, as shown here by the αIIbβ3/GPA 60–131 chimera and previously by insertion of artificial linkers (Xiong et al., 2003). Decoupling likely arises because long flexible linkers enable integrin ectodomain extension and activation while the TM domains remain associated.
Inside-out activation of integrins
Ligand binding by integrins can be activated by binding of the actin cytoskeleton to the β cytoplasmic domain. Two classes of cytoskeleton-associated proteins mediate this linkage, talin and kindlins (Moser et al., 2008; Wegener and Campbell, 2008). Talin and kindlin bind through their FERM domains to NPXY motifs at β3 residues 744–747 and 756–759, respectively. Talin also binds to more membrane-proximal β3 regions, including Phe-727 and Phe-730 (Wegener et al., 2007). The Disulfide/Rosetta structure shows that these residues are in the β3 α-helix and face the αIIb cytoplasmic tail. Superposition of the talin β3 and αIIbβ3 complexes demonstrates that talin clashes with αIIb residues that interact with the β3 α-helix, and immediately follow the GFFKR motif, i.e. αIIb residues N996-P998 (Fig. 6E). However, the position of the segment with talin-binding residues Phe-727 and Phe-730 is not well defined, since some clusters show two helices in the β3 cytoplasmic domain separated by a turn near Glu-726 (Fig. S7A and B)). Furthermore, the talin complex structure shows a loss of helicity in β3 N-terminal to His-722, and isolated β3 in bicelles loses α-helicity after residue His-722 (Lau et al., 2008b). Thus, it is plausible either that talin binding to β3 would perturb its association with αIIb, or that flexibility in the β3 α-helix between its sites of association with αIIb and talin would eliminate clashes in a talin/αIIbβ3 complex. Since the evidence that talin interferes with association between αIIb and β3 comes from an NMR study on a complex that our results suggest is non-physiologic, the mechanism by which talin activates integrins requires further structural study.
Once integrins bind through talins or kindlins to actin filaments, lateral force exerted by the cytoskeleton will favor an extended integrin conformation with the hybrid domain in β swung out in the direction of the pulling force in the open conformation with high affinity for ligand (Zhu et al., 2008). This mechanism can explain activation by talin as well as by kindlins, which bind to an NPXY motif that is distal from the αIIb/β3 interface (Moser et al., 2008). Lateral force would tilt the dissociated β3 TM domain in the plane of the membrane. The long continuous α-helix seen in the dissociated β3 TM domain is well suited to tilting (Lau et al., 2008b).
We have described the structure of an intact receptor containing two TM domains on the cell surface. The results shed important light on the structural basis for transmembrane signaling through integrins, demonstrate that the principles differ for association between heterodimeric and homodimeric TM domains, and demonstrate that JM domains can have an important role in TM domain association. The method can be extended to many other important classes of cell surface receptors with two associating TM domains, and yield information on how regulated changes in the association state of the TM domains transmits signals between the extracellular and intracellular environments.
Methods
Disulfide crosslinking and immunoprecipitation
293T transfectants were pretreated with 15 μg/ml of 2-BP for 1 hour, labeled with [35S] cysteine/methionine (10mCi/ml, PerkinElmer Life Science) for 1.5 h, and chased for 17 h. Cells were detached and kept intact or broken by freeze-thaw, treated on ice with 200 μM CuSO4/1000 μM o-phenanthroline for 10 min., with 10 mM N-ethylmaleimide for 10 min, and lysed in 2% Triton X-100. Lysate supernatants were immunoprecipitated with anti-β3 mAb AP3 and protein G agarose, and subjected to nonreducing 7.5% SDS-PAGE. Radioactivity was quantitated with a PhosphorImager. Crosslinking efficiency was quantitated as disulfide-linked heterodimer as % of heterodimer plus αIIb and β3 monomers. For constitutively crosslinked residues, crosslinking was also measured after 1 h in 5 mM cysteamine/1 mM cystamine redox buffer and after DTT treatment followed by Cu-phenanthroline. Details are in the Supplement.
Membrane structure generation
The energy terms used in Membrane Rosetta (Barth et al., 2007; Barth et al., 2009) include a Lennard-Jones potential, a backbone torsional term, a knowledge-based pairwise interaction term between amino acid residues, a penalty term for the disulfide restraints that is proportional to distance beyond the upper bound, an implicit Lazaridis-Karplus solvation term for the aqueous and hydrophobic environments (with the hydrophobic potential based on experimental transfer free energies from water to cyclohexane), and an orientation-dependent hbonding term. Models are first built with centroids for sidechains, and then refined in all-atom mode. The all atom mode membrane environment has an inner, slab-shaped, 24-Å thick isotropic, hydrophobic phase representing the lipid alkyl groups, outer isotropic phases representing the headgroup and membrane-external regions, and in between, two 10-Å thick, anisotropic phases representing the interface region where alkyl, acyl, and glycerol groups are located. The centroid membrane environment is similar, but replaces each anisotropic slab with three isotropic slabs. The solvation and hbond potentials differ in the hydrophobic and aqueous environments, and are interpolated in the interface region. The hbond potential includes weak CαH-O bonds and bifurcated hbonds. The position of models in the membrane slab is varied to find the lowest energy embedding.
Structures were built in several stages. First, twelve-residue ideal helices were docked. Then, all but two residues of each helix were discarded, and chain-growing with fragment assembly was used to build longer structures that could in principle have any secondary structure. A total of two (GPA) or three (integrin) chain growing stages were used, alternating in N to C and C to N direction, and previously built residues were discarded in each stage, so that no remnant from the first one (GPA) or two stages (integrin) were present in the final structures (Fig. S6). Approximately 10,000 models were generated in each stage. Disulfide restraints were used as part of the total energy function in each Monte Carlo step within each chain-growing stage. Furthermore, between each stage, the 100 models used as seeds for the next stage were selected by the independent criteria of low restraint violation and low energy (in this case, energy did not include the restraint violation penalty). The stages are summarized in more detail in Fig. S6 and Supplementary Materials. Structures have been deposited in the protein databank as codes ……
Modeling the intact integrin in the membrane bilayer
To provide starting TM structures, one additional stage of chain-growing was performed from the TM regions to grow the linker regions, using redox buffer disulfide restraints for the linker region, and distance restraints for the two C-terminal residues present in αIIb and β3 in the crystal structure (Zhu et al., 2008). After joining these TM structures to the crystal structure, the two linkers were simultaneously remodeled with Membrane Rosetta using the linker disulfide restraints to find low energy orientations with respect to the membrane (Supplementary Materials). Two long, membrane-proximal loops in the calf-2 domain that are disordered in the crystal structure were included in the models.
Ligand binding assay
Ligand mimetic IgM PAC-1 (Becton Dickinson, San Jose, CA) and FITC-labeled human fibrinogen (Enzyme Research Laboratories, South Bend, IN) binding to transfected cells was as described (Luo et al., 2004).
Supplementary Material
Acknowledgments
We thank Junichi Takagi and Guo-hui Li for early help with the project. Supported by NIH grant HL-48675and HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adair BD, Yeager M. Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci USA. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P, Wallner BDB. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci USA. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, Artemenko EO, Efremov RG, Arseniev AS. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J Biol Chem. 2008a doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008b;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Clore GM, Gronenborn AM, Saffrich R, Nilges M. Assessing the quality of solution nuclear magnetic resonance structures by complete cross-validation. Science (New York, NY) 1993;261:328–331. doi: 10.1126/science.8332897. [DOI] [PubMed] [Google Scholar]
- Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Levitt M, Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Duneau JP, Vegh AP, Sturgis JN. A dimerization hierarchy in the transmembrane domains of the HER receptor family. Biochemistry. 2007;46:2010–2019. doi: 10.1021/bi061436f. [DOI] [PubMed] [Google Scholar]
- Gottschalk KE. A coiled-coil structure of the αIIbβ3 integrin transmembrane and cytoplasmic domains in its resting state. Structure. 2005;13:703–712. doi: 10.1016/j.str.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Diaz-Gonzalez F, Leong L, Wu C, McDonald JA, Shattil SJ, Ginsberg MH. Breaking the integrin hinge. J Biol Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Yang X, Kolesnikova TV, Hemler ME. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem J. 2004;377:407–417. doi: 10.1042/BJ20031037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Dua V, Ulmer TS. Structure of the integrin alphaIIb transmembrane segment. J Biol Chem. 2008a;283:16162–16168. doi: 10.1074/jbc.M801748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Partridge AW, Ginsberg MH, Ulmer TS. Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry. 2008b;47:4008–4016. doi: 10.1021/bi800107a. [DOI] [PubMed] [Google Scholar]
- Li R, Babu CR, Lear JD, Wand AJ, Bennett JS, Degrado WF. Oligomerization of the integrin aIIbb3: Roles of the transmembrane and cytoplasmic domains. Proc Natl Acad Sci USA. 2001;98:12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Metcalf DG, Gorelik R, Li R, Mitra N, Nanda V, Law PB, Lear JD, Degrado WF, Bennett JS. A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci USA. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Crocker E, Siminovitch DJ, Smith SO. Role of side-chain conformational entropy in transmembrane helix dimerization of glycophorin A. Biophys J. 2003;84:1263–1271. doi: 10.1016/S0006-3495(03)74941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Imm. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci USA. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science (New York, NY. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nature medicine. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- O’Toole TE, Katagiri Y, Faull RJ, Peter K, Tamura R, Quaranta V, Loftus JC, Shattil SJ, Ginsberg MH. Integrin cytoplasmic domains mediate inside-out signal transduction. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain packing stabilizes integrin aIIbb3 in the low affinity state. J Biol Chem. 2005;280:7294–7200. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Principles and Patterns of Protein Conformation. In: Fasman GD, editor. Prediction of Protein Structure and the Principles of Protein Conformation. New York: Plenum Press; 1989. pp. 1–98. [Google Scholar]
- Smith SO, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Yaspan B, Ginsberg MH, Campbell ID. NMR analysis of structure and dynamics of the cytosolic tails of integrin alpha IIb beta 3 in aqueous solution. Biochemistry. 2001;40:7498–7508. doi: 10.1021/bi010338l. [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Vaynberg J, Kong X, Haas TA, Plow EF, Qin J. Membrane-mediated structural transitions at the cytoplasmic face during integrin activation. Proc Natl Acad Sci USA. 2004;101:4094–4099. doi: 10.1073/pnas.0400742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. A structural mechanism of integrin αIIbβ3 “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (review) Mol Membr Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Weljie AM, Hwang PM, Vogel HJ. Solution structures of the cytoplasmic tail complex from platelet αIIb- and β3-subunits. Proc Natl Acad Sci USA. 2002;99:5878–5883. doi: 10.1073/pnas.092515799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YM, Chen J, Zhang L. Modulation of CD11b/CD18 adhesive activity by its extracellular, membrane-proximal regions. J Immunol. 2003;171:1042–1050. doi: 10.4049/jimmunol.171.2.1042. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a Complete Integrin Ectodomain in a Physiologic Resting State and Activation and Deactivation by Applied Forces. Mol Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.