Abstract
Genetic mapping provides a powerful approach to identify genes and biological processes underlying any trait influenced by inheritance, including human diseases. We discuss the intellectual foundations of genetic mapping of Mendelian and complex traits in humans, examine lessons emerging from linkage analysis of Mendelian diseases and genome-wide association studies of common diseases, and discuss questions and challenges that lie ahead.
By the early 1900s, geneticists understood that Mendel’s laws of inheritance underlie the transmission of genes in diploid organisms. They noted that some traits are inherited according to Mendel’s ratios, as a result of alterations in single genes, and they developed methods to map the genes responsible. They also recognized that most naturally occurring trait variation, while showing strong correlation among relatives, involves the action of multiple genes and nongenetic factors.
Although it was clear that these insights applied to humans as much as to fruit flies, it took most of the century to turn these concepts into practical tools for discovering genes contributing to human diseases. Starting in the 1980s, the use of naturally occurring DNA variation as markers to trace inheritance in families led to the discovery of thousands of genes for rare Mendelian diseases. Despite great hopes, the approach proved unsuccessful for common forms of human diseases—such as diabetes, heart disease, and cancer—that show complex inheritance in the general population.
Over the past year, a new approach to genetic mapping has yielded the first general progress toward mapping loci that influence susceptibility to common human diseases. Still, most of the genes and mutations underlying these findings remain to be defined, let alone understood, and it remains unclear how much of the heritability of common disease they explain. Below, we discuss the intellectual foundations of genetic mapping, examine emerging lessons, and discuss questions and challenges that lie ahead.
Genetic Mapping by Linkage and Association
Genetic mapping is the localization of genes underlying phenotypes on the basis of correlation with DNA variation, without the need for prior hypotheses about biological function. The simplest form, called linkage analysis, was conceived by Sturtevant for fruit flies in 1913 (1). Linkage analysis involves crosses between parents that vary at a Mendelian trait and at many polymorphic variants (“markers”); because of meiotic recombination, any marker showing correlated segregation (“linkage”) with the trait must lie nearby in the genome.
In the 1970s, the ability to clone and sequence DNA made it possible to tie genetic linkage maps in model organisms to the underlying DNA sequence, and thereby to molecularly clone the genes responsible for any Mendelian trait solely on the basis of their genomic position (2, 3). Such studies typically involved three steps: (i) identifying the locus responsible through a genome-wide search; (ii) sequencing the region in cases and controls to define causal mutation(s); and (iii) studying the molecular and cellular functions of the genes discovered. So-called “positional cloning” became a mainstay of experimental genetics, identifying pathways that are crucial in development and physiology.
Linkage analysis in humans
For most of the 20th century, genome-wide linkage mapping was impractical in humans: Family sizes are small crosses are not by design, and there were too few classical genetic markers to systematically trace inheritance. Progress in identifying the genes contributing to human traits was initially limited to studies of biological candidates such as blood-type antigens (4) and hemoglobin β protein in sickle-cell anemia (5).
In 1980, Botstein and colleagues, building on their use of DNA polymorphisms to study linkage in yeast (6) and the finding of DNA polymorphism at the globin locus in humans (7, 8), proposed the use of naturally occurring DNA sequence polymorphisms as generic markers to create a human genetic map and systematically trace the transmission of chromosomal regions in families (9). The feasibility of genetic mapping in humans was soon demonstrated with the localization of Huntington disease in 1983 (10). A rudimentary genetic linkage map with ~400 DNA markers was generated by 1987 (11) and was fleshed out to ~5000 markers by 1996 (12). Physical maps providing access to linked chromosomal regions were developed by 1995 (13). With these tools, positional cloning became possible in humans, and the number of disorders tied to a specific gene grew from ~100 in the late 1980s to >2200 today (14).
Several lessons emerged from studies of Mendelian disease genes: (i) The “candidate gene” approach was woefully inadequate; most disease genes were completely unsuspected on the basis of previous knowledge, (ii) Disease-causing mutations often cause major changes in encoded proteins, (iii) Loci typically harbor many disease-causing alleles, mostly rare in the population, (iv) Mendelian diseases often revealed great complexity, such as locus heterogeneity, incomplete penetrance, and variable expressivity.
Geneticists were eager to apply genetic mapping to common diseases, which also show familial clustering. Mendelian subtypes of common diseases [such as breast cancer (15), hypertension (16), and diabetes (17)] were elucidated, but mutations in these genes explained few cases in the population. In common forms of common disease, risk to relatives is lower than in Mendelian cases, and linkage studies with excellent power to detect a single causal gene yielded equivocal results.
These features were consistent with, but did not prove, a polygenic model. The idea that commonly varying traits might be polygenic in nature was offered by East in 1910 (18). By 1920, linkage mapping was used to identify multiple unlinked factors influencing truncate wings in Drosophila (19), and Fisher had developed a mathematical framework for relating Mendelian factors and quantitative traits (20). In the late 1980s, linkage mapping of complex traits was made feasible for experimental organisms through the use of genetic mapping in large crosses (21). But there was little success in humans.
Genetic association in populations
A possible path forward emerged from population genetics and genomics. Instead of mapping disease genes by tracing transmission in families, one might localize them through association studies—that is, comparisons of frequencies of genetic variants among affected and unaffected individuals.
Genetic association studies were not a new idea. In the 1950s, such studies revealed correlations between blood-group antigens and peptic ulcer disease (4); in the 1960s and 1970s, common variation at the human leukocyte antigen (HLA) locus was associated with autoimmune and infectious diseases (22); and in the 1980s, apolipoprotein E was implicated in the etiology of Alzheimer’s disease (23). Still, only about a dozen extensively reproduced associations of common variants (outside the HLA locus) were identified in the 20th century (24).
A central problem was that association studies of candidate genes were a shot in the dark: They were limited to specific variants in biological candidate genes, each with a tiny a priori probability of being disease-causing. Moreover, association studies were susceptible to false positives due to population structure, because there was no way to assess differences in the genetic background of cases and controls. Although many claims of associations were published, the statistical support tended to be weak and few were subsequently replicated (25).
In the mid-1990s, a systematic genome-wide approach to association studies was proposed (26–28): to develop a catalog of common human genetic variants and test the variants for association to disease risk. The focus on common variants as a mapping tool was a matter of practicality, grounded in population genetics. The human population has recently grown exponentially from a small size. As predicted by classical theory (29), humans have limited genetic variation: The heterozygosity rate for single-nucleotide polymorphisms (SNPs) is ~1 in 1000 bases (30–32). Moreover, perhaps 90% of heterozygous sites in each individual are common variants, typically shared among continental populations (33).
If most genetic variation in an individual is common, then why are mutations responsible for Mendelian diseases typically rare? One answer is natural selection: Mutations that cause strongly deleterious phenotypes—as most Mendelian diseases appear to be—are lost to purifying selection. But if deleterious mutations are typically rare, how could common variants play a role in disease? Common diseases often have late onset, with modest or no obvious impact on reproductive fitness. Mildly deleterious alleles can rise to moderate frequency, particularly in populations that have undergone recent expansion (34). Moreover, some alleles that were advantageous or neutral during human evolution might now confer susceptibility to disease because of changes in living conditions accompanying civilization. Finally, disease-causing alleles could be maintained at high frequency if they were under balancing selection, with disease burden offset by a beneficial phenotype (as in sickle-cell disease and malaria resistance).
These lines of reasoning led to the so-called “common disease–common variant” (CD-CV) hypothesis: the proposal that common polymorphisms (classically defined as having a minor allele frequency of >1%) might contribute to susceptibility to common diseases (26–28). If so, genome-wide association studies (GWASs) of common variants might be used to map loci contributing to common diseases. The concept was not that all causal mutations at these genes should be common (to the contrary, a full spectrum of alleles is expected), only that some common variants exist and could be used to pinpoint loci for detailed study.
It took a decade to develop the tools and methods required to test the CD-CV hypothesis: (i) catalogs of millions of common variants in the human population, (ii) techniques to genotype these variants in studies with thousands of patients, and (iii) an analytical framework to distinguish true associations from noise and artifacts.
Cataloging SNPs and linkage disequilibrium
Pilot projects in the late 1990s showed that it was possible to identify thousands of SNPs and to perform highly multiplexed genotyping by means of DNA microarrays (35). A public-private partnership, the SNP Consortium, built an initial map of 1.4 million SNPs (32); this has grown to more than 10 million SNPs (36) and is estimated to contain 80% of all SNPs with frequencies of >10% (37).
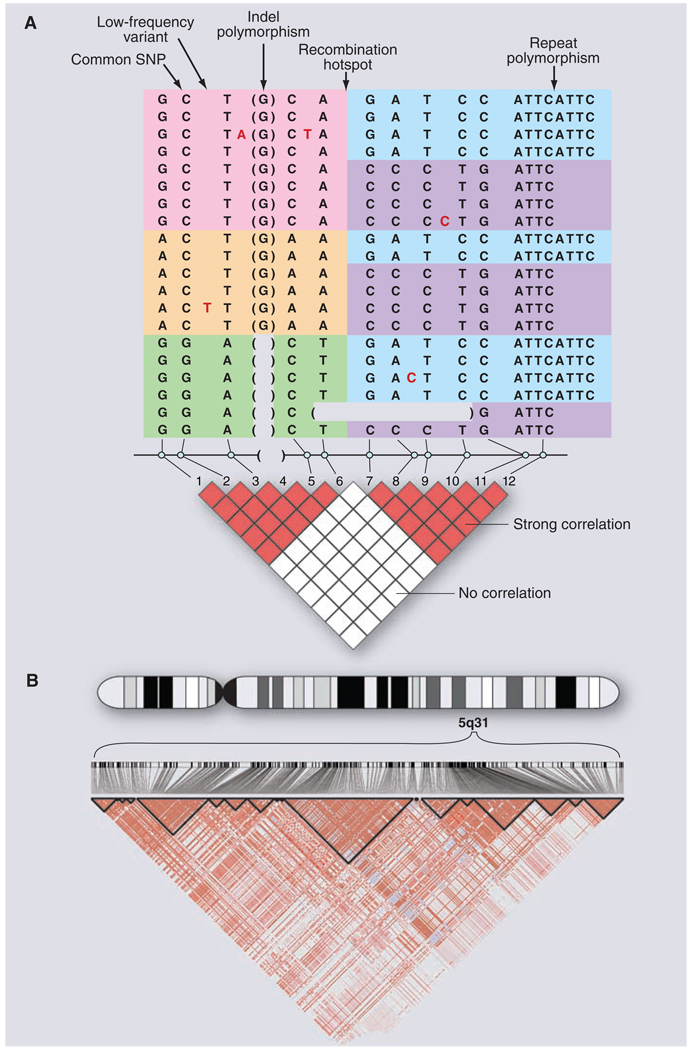
As the SNP catalog grew, a critical question loomed: Would GWASs require directly testing each of the ~10 million common variants for association to disease? That is, if only 5% of variants were tested, would 95% of associations be missed? Or could a subset serve as reliable proxies for their neighbors? Experience from Mendelian diseases suggested that substantial efficiencies might be possible. Each disease-causing mutation arises on a particular copy of the human genome and bears a specific set of common alleles in cis at nearby loci, termed a haplotype. Because the recombination rate is low [~1 crossover per 100 megabases (Mb) per generation], disease alleles in the population typically show association with nearby marker alleles for many generations, a phenomenon termed linkage disequilibrium (LD) (Fig. 1).
Fig. 1. DNA sequence variation in the human genome.
(A) Common and rare genetic variation in 10 individuals, carrying 20 distinct copies of the human genome. The amount of variation shown here is typical for a 5-kb stretch of genome and is centered on a strong recombination hotspot. The 12 common variations include 10 SNPs, an insertion-deletion polymorphism (indel), and a tetranucleotide repeat polymorphism. The six common polymorphisms on the left side are strongly correlated. Although these six polymorphisms could theoretically occur in 26 possible patterns, only three patterns are observed (indicated by pink, orange, and green). These patterns are called haplotypes. Similarly, the six common polymorphisms on the right side are strongly correlated and reside on only two haplotypes (indicated by blue and purple). The haplotypes occur because there has not been much genetic recombination between the sites. By contrast, there is little correlation between the two groups of polymorphisms, because a hotspot of genetic recombination lies between them. The pairwise correlation between the common sites is shown by the red and white boxes below, with red indicating strong correlation and white indicating weak correlation. In addition to the common polymorphisms, lower-frequency polymorphisms also occur in the human genome. Five rare SNPs are shown, with the variant nucleotide marked in red and the reference nucleotide not shown. In addition, on the second to last chromosome, a larger deletion variant is observed that removes several kilobases of DNA. Such larger deletion or duplication events (i.e., CNVs) may be common and segregate as other DNA variants. (B) Small regions such as in (A) are often embedded in genomic regions with much greater extents of LD. The diagram shows actual data from the International HapMap Project, showing 420 genetic variants in a region of 500 kb on human chromosome 5q31. Positions of the variants and the pairwise correlations are shown below. Blocks of strong correlation are indicated by the black outlines. Longer-range patterns are often more complex than shown in (A) because weaker recombination hotspots may reduce, but not completely eliminate, marker-to-marker correlation.
Early studies had demonstrated LD of nearby polymorphisms at the globin locus (38), which proved useful in tracking sickle-cell mutation. In the mid-1980s, it was proposed that a genome-wide search might be performed in genetically isolated populations, scanning the genome for a haplotype shared among unrelated patients carrying the same founder mutation (39). Such “LD mapping” in essence treated the entire population as a very large and very old extended family. This method soon proved useful in fine-mapping the founder Δ508 mutation in the transmembrane conductance regulator CFTR as a cause of cystic fibrosis (40) and in screening the entire genome in isolated populations such as Finland (41).
The key question was whether the same approach could be used more generally to study common alleles in large human populations, where recombination had more time to whittle down haplotypes. A simulation study suggested that LD might typically be too short to be useful with a SNP every 5 kb (500,000 SNPs across the genome) providing very weak LD (average correlation r2 = 0.1) (42). Studies of individual loci showed great heterogeneity in local LD (43).
As denser genetic maps became available, a clear picture emerged. Nearby variants were observed to form a block-like structure consisting of regions characterized by little evidence for historical recombination and limited haplo-type diversity (44, 45). Within such regions, which soon proved general (46), genotypes of common SNPs could be inferred from knowledge of only a few empirically determined tag SNPs (45–47). These patterns were shaped by hot and cold spots of recombination in the human genome (48–50), as well as historical population bottlenecks (51).
The International HapMap Project was launched in 2002, with the goal of characterizing SNP frequencies and local LD patterns across the human genome in 270 samples from Europe, Asia, and West Africa. The project genotyped ~1 million SNPs by 2005 (37) and more than 3 million by 2007 (52). Sequence data collected by the project confirmed that the vast majority of common SNPs are strongly correlated to one or more nearby proxies: 500,000 SNPs provide excellent power to test >90% of common SNP variation in out-of-Africa populations, with roughly twice that number required in African populations (37).
Massively parallel genotyping
SNP genotyping was initially performed one SNP at a time, at a cost of ~$1 per measurement. Multiplex genotyping of hundreds of SNPs on DNA microarrays was demonstrated in 1998 (35), and capacity per array grew from 10,000 to 100,000 SNPs in 2002 to 500,000 to 1 million SNPs in 2007. In parallel, cost fell to $0.001 per genotype, or less than $1000 per sample for a whole-genome analysis. By 2006, several technologies could simultaneously genotype hundreds of thousands of SNPs at >99% completeness and >99% accuracy.
Copy-number variation
SNPs are only one type of genetic variation (Fig. 1). Using microarray technology, two groups in 2004 observed that individual copies of the human genome contain large regions (tens to hundreds of kilobases in size) that are deleted, duplicated, or inverted relative to the reference sequence (53, 54). Structural variants had been previously associated with developmental disorders and were often assumed to be pathogenic; the presence of so many segregating copy-number variations (CNVs) in the general population was surprising. The generality of CNVs was soon established (55–59). Many CNVs display tight LD with nearby SNPs (56, 57) and thus can be proxied by nearby SNPs in GWASs. Others occur in regions that are difficult to follow with SNPs, are highly mutable, or are rare (58, 59). Hybrid genotyping platforms have recently been developed to genotype SNPs and CNVs simultaneously (60).
Statistical analysis
Recognizing causal loci amid a genome’s worth of random fluctuation required advances in statistical design, analysis, and interpretation. The risk of false negatives was illustrated by a study of type 2 diabetes (T2D) and the Pro12 → Ala polymorphism in peroxisome proliferator-activated receptor γ. Whereas an initial positive report (61) had not been confirmed in four modest-sized replication studies, larger studies produced strong and consistent evidence of increased risk by a factor of 1.2 (62, 63). The negative studies were actually consistent with the level of increased risk, but simply lacked adequate power to detect it.
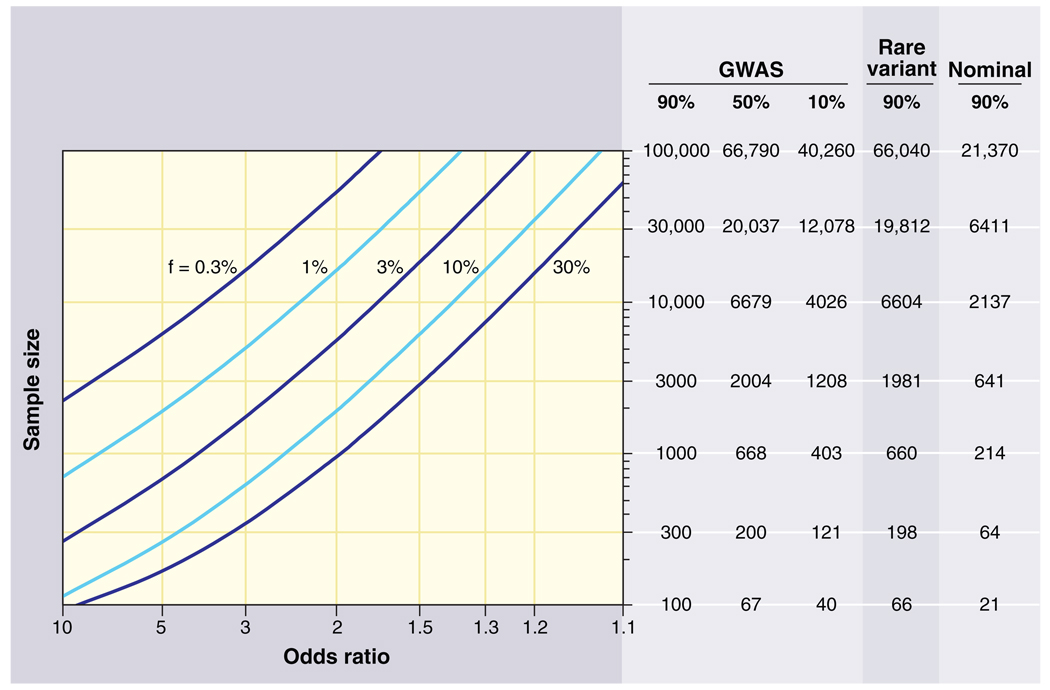
Conversely, stringent thresholds for statistical significance are needed to avoid false positives due to multiple hypothesis testing. Simulations indicated that a dense genome-wide scan of common variants involves the equivalent of ~1 million independent hypotheses (64). A significance level of P = 5 × 10−8 thus represents a finding expected by chance once in 20 GWASs. Large sample sizes would be needed to reach such a stringent threshold (Fig. 2).
Fig. 2.
Sample sizes required for genetic association studies. The graphs show the total number N of samples (consisting of N/2 cases and N/2 controls) required to map a genetic variant as a function of the increased risk due to the disease-causing allele (x axis) and the frequency of the disease-causing allele (various curves). The required sample size is shown in the table on the right for various different kinds of association studies. The first three columns pertain to GWASs using common variants across the entire genome; the columns correspond to different levels of statistical power to achieve a significant result at P < 10−8. The fourth column pertains to a search for rare variants where the frequency listed is the collective frequency of rare variants in controls, and the odds ratio is the excess in cases as compared to controls. Sample sizes assume correction for a genome-wide search of ~20,000 protein-coding genes in the genome (aiming to achieve P < 10−5 with one test performed per gene). The fifth column pertains to a test of a single hypothesis (e.g., testing association with a single SNP). For example, in a GWAS, 1000 samples provide 90% statistical power to detect a 30% allele with a factor of 2 effect. In a genome-wide search via exon sequencing, 660 samples provide 90% power to detect a gene in which rare variants have aggregate population frequency 1% and convey a factor of ~8 increase in risk. Note that the sample size to test essentially all common SNPs in the human genome is only 5 times the sample size to test a single SNP.
Systematic biases could also cause false positives. Differences in ancestry between cases and controls would yield spurious associations (65), suggesting the need for family-based controls (66). It was later recognized that genome-wide studies provide their own internal control: Mismatched ancestry is readily detectable because it produces frequency differences at thousands of SNPs, which could not all reflect causal associations. Methods were developed to detect and adjust for such biases (67–69) as well as unexpected relatedness between subjects. Technical artifacts, which are particularly problematic if cases and controls are not genotyped in parallel (70), were overcome by improved genotyping methods, quality control, and stringent filtering. To maximize efficiency and power, several groups developed methods of selecting tag SNPs (47, 71–73) from empirical LD data and using them to impute genotypes at other SNPs not genotyped in clinical samples (74) on the basis of LD relationships in the HapMap.
Genome-Wide Associations: Lessons
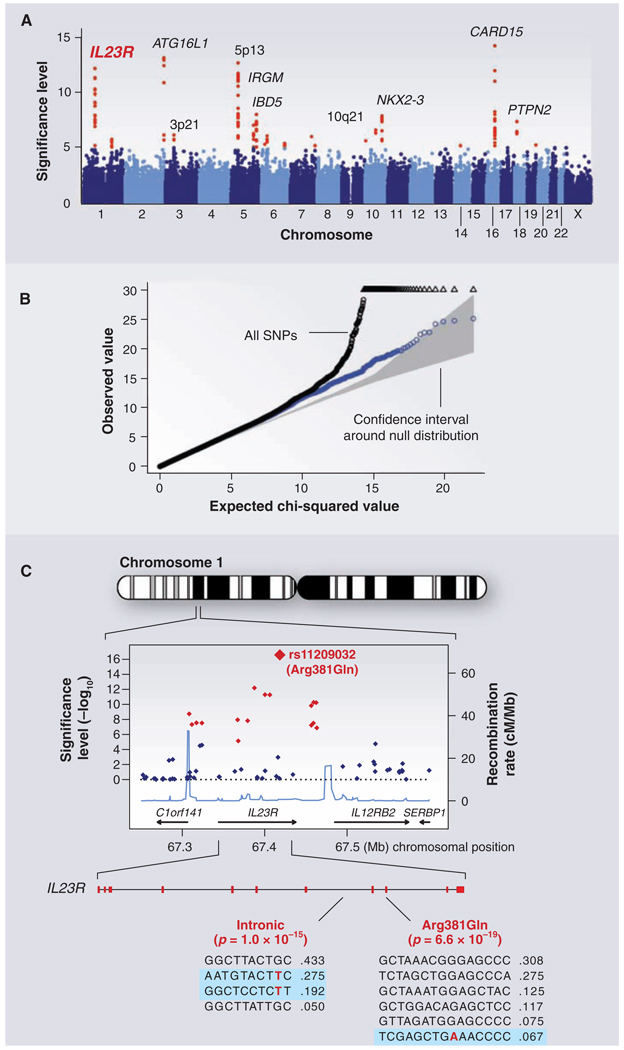
By early 2006, the tools were in place and studies were under way in many laboratories to resolve the hotly debated issue (75, 76) of whether genetic mapping of common SNPs would shed light on common disease. Since then, scores of publications have reported the localization of common SNPs associated with a wide range of common diseases and clinical conditions (age-related macular degeneration, type 1 and type 2 diabetes, obesity, inflammatory bowel disease, prostate cancer, breast cancer, colorectal cancer, rheumatoid arthritis, systemic lupus erythematosus, celiac disease, multiple sclerosis, atrial fibrillation, coronary disease, glaucoma, gallstones, asthma, and restless leg syndrome) as well as various individual traits (height, hair color, eye color, freckles, and HIV viral set point). Figure 3 illustrates data from a paradigmatic genome-wide association study of Crohn’s disease performed by the Wellcome Trust Case Control Consortium.
Fig. 3.
GWAS for Crohn's disease. The panels show data from the study of Crohn's disease by the Wellcome Trust Case Control Consortium. (A) Significance level (P value on log10 scale) for each of the 500,000 SNPs tested across the genome. SNP locations reflect their positions across the 23 human chromosomes. SNPs with significance levels exceeding 10−5 (corresponding to 5 on the y axis) are colored red; the remaining SNPs are in blue. Ten regions with multiple significant SNPs are shown, labeled by their location or by the likely disease-related gene (e.g., IL23R on chromosome 1). (B) The fact that the SNPs in red are extreme outliers is made clear from a so-called Q-Q plot. A Q-Q plot is made as follows: The SNPs are ordered (from 1 to n) according to their observed P values; observed and expected P values are plotted for each SNP. Under the null distribution, the expected P value for the ith SNP is i/n. If there are no significant associations, the Q-Q plot will lie along the 45° line; the gray region corresponds to a 95% confidence region around this null expectation. Black points correspond to all 500,000 SNPs studied that passed strict quality control; they diverge strongly from the null expectation. Blue points reflect the P values that remain when the SNPs in the 10 most significant regions are removed; there is still some excess of significant P values, indicating the presence of additional loci of more modest effect. (C) Close-up of the region around the IL23R locus on chromosome 1. The first part shows the significance levels for SNPs in a region of ~400 kb, with colors as in (A). The highest significance level occurs at a SNP in the coding region of the IL23R gene (causing an Arg381 → Gln change). The light blue curve shows the inferred local rate of recombination across the region. There are two clear hotspots of recombination, with SNPs lying between these hotspots being strongly correlated in a few haplotypes. The second part shows that the IL23R locus harbors at least two independent, highly significant disease-associated alleles. The first site is the Arg381 → Gln polymorphism, which has a single disease-associated haplotype (shaded in blue) with frequency of 6.7%. The second site is in the intron between exons 7 and 8; it tags two disease-associated haplotypes with frequencies of 27.5% and 19.2%.
Various lessons have already emerged about genetic mapping by GWAS:
1) GWASs work. Before 2006, only about two dozen reproducible associations outside the HLA locus had been discovered (25). By early 2008, more than 150 relationships were identified between common SNPs and disease traits (table S1). In most diseases studied, GWASs have revealed multiple independent loci, although some traits have not yet yielded associations that meet stringent thresholds (e.g., hypertension). It is not clear whether this reflects inadequate sample size, phenotypic definition, or a different genetic architecture.
2) Effect sizes for common variants are typically modest. In a few cases, common variants with effects of a factor of ≥2 per allele have been found: APOE4 in Alzheimer’s disease (23), CFH in age-related macular degeneration (77–79), and LOXL1 in exfoliative glaucoma (80). In the vast majority of cases, however, the estimated effects are much smaller—mostly increases in risk by a factor of 1.1 to 1.5 per associated allele.
3) The power to detect associations has been low. Given the effect sizes now known to exist, and the need to exceed stringent statistical thresholds, the first wave of GWASs provided low power to discover disease-causing loci (81, 82). For example, achieving 90% power to detect an allele with 20% frequency and a factor of 1.2 effect at a statistical significance of 10−8 requires 8600 samples (Fig. 2). Thus, although it is unlikely that common alleles of large effect have been missed, GWASs of hundreds to several thousand cases have necessarily identified only a fraction of the loci that can be found with larger sample sizes. This prediction has been empirically confirmed in T2D (83), serum lipids (84, 85), Crohn’s disease (86), and height (87–90). Across these four traits and diseases, individual GWASs together documented 29 associations. Increasing the power by pooling the samples to perform meta-analysis and replication genotyping has increased this yield to more than 100 replicated loci for these four conditions.
4) Association signals have identified small regions for study but have not yet identified causal genes and mutations. Genetic mapping is a double-edged sword Local correlation of genetic variants facilitates the initial identification of a region but makes it difficult to distinguish causal mutation(s). Luckily, whereas family-based linkage methods typically yield regions of 2 to 10 Mb in span, GWASs typically yield more manageable regions of 10 to 100 kb.
These regions have yet to be scrutinized by fine-mapping and resequencing to identify the specific gene and variants responsible. Even when a locus is identified by SNP association, the causal mutation itself need not be a SNP. For example, the IRGM gene was associated with Crohn’s disease on the basis of GWAS. Subsequent study suggests that the causal mutation is a deletion upstream of the promoter affecting tissue-specific expression (91).
5) A single locus can contain multiple independent common risk variants. Intensive study has already identified seven independent alleles at 8q24 for prostate cancer (92), three at complement factor H (CFH) for age-related macular degeneration (93, 94), three at IRF5 for systemic lupus erythematosus (95), and two at IL23R for Crohn’s disease (96). Multiple distinct alleles with different frequencies and risk ratios may well be the rule.
6) A single locus can harbor both common variants of weak effect and rare variants of large effect. In recent GWASs, studies of common SNPs enabled the identification of 19 loci as influencing low- or high-density lipoprotein (LDL, HDL) or triglycerides (84, 85). Nine of these 19 were already known to carry rare Mendelian mutations with large effects, such as the loci for the LDL receptor (LDLR) and familial hypercholesterolemia (FH). Similarly, the genes encoding Kir6.2, WFS1, and TCF2 are all known to cause Mendelian syndromes including T2D, as well as common SNPs with modest effects.
7) Because allele frequencies vary across human populations, the relative roles of common susceptibility genes can vary among ethnic groups. One example is the association of prostate cancer at 8q24: SNPs in the region play a role in all ethnic groups, but the contribution is greater in African Americans. This is not because the risk alleles yet found confer greater susceptibility in African Americans, but because they occur at higher frequencies (92), contributing to the higher incidence among African American men than among men of European ancestry.
Lessons have also emerged about the functions and phenotypic associations of genes related to common diseases:
1) A subset of associations involve genes previously related to the disease. Of 19 loci meeting genome-wide significance in a recent GWAS of LDL, HDL, or triglyceride levels, 12 contained genes with known functions in lipid biology (84, 85). The gene for 3-hydroxy-3-methyl glutaryl–coenzyme A reductase (HMGCR), encoding the rate-limiting enzyme in cholesterol biosynthesis and the target of statin medications, was found by GWAS to carry common genetic variation influencing LDL levels (84,85). Similarly, SNPs in the β-cell zinc transporter encoded by SLC30A8 were associated with risk of T2D (97).
2) Most associations do not involve previous candidate genes. In some cases, GWAS results immediately suggest new biological hypotheses— for example, the role of complement factor H in age-related macular degeneration (77–79), FGFR2 in breast cancer (98), and CDKN2A and CDKN2B in T2D (99–101). In many other cases, such as LOC387715/HTRAl with age-related macular degeneration (102), nearby genes have no known function.
3) Many associations implicate non-protein-coding regions. Although some associated non-coding SNPs may ultimately prove attributable to LD with nearby coding mutations, many are sufficiently far from nearby exons to make this outcome unlikely. Examples include the region at 8q24 associated with prostate, breast, and colon cancer, 300 kb from the nearest gene (103, 104), and the region at 9q21 associated with myocardial infarction and T2D, 150 kb from the nearest genes encoding CDKN2A and CDKN2B (99–101, 105–107).
A role for noncoding sequence in disease risk is not surprising: Comparative genome analysis has shown that 5% of the human genome is evolutionarily conserved and thus functional; less than one-third of this 5% consists of genes that encode proteins (108). Noncoding mutations with roles in disease susceptibility will likely open new doors to understanding genome biology and gene regulation. Regulatory variation also suggests different therapeutic strategies: Modulating levels of gene expression may prove more tractable than replacing a fully defective protein or turning off a gain-of-function allele.
4) Some regions contain expected associations across diseases and traits. Crohn’s disease, psoriasis, and ankylosing spondylitis have long been recognized to share clinical features; the association of the same common polymorphisms in IL23R in all three diseases points to a shared molecular cause (96, 109, 110). SNPs in STAT4 (signal transducer and activator of transcription 4) are associated with rheumatoid arthritis and systemic lupus, two diseases that share clinical features. Multiple variants associated with T2D are associated with insulin secretion defects in nondiabetic individuals (101, 111–116), highlighting the role of β-cell failure in the pathogenesis of T2D.
5) Some regions reveal surprising associations. For example, unexpected connections have emerged among T2D, inflammatory diseases (two loci), and cancer (four loci). A single intron of CDKAL1 was found to contain a SNP associated with T2D and insulin secretion defects (99–101, 116), and another with Crohn’s disease and psoriasis (117). A coding variant in glucokinase regulatory protein is associated with triglyceride levels and fasting glucose (101) but also with C reactive protein levels (118, 119) and Crohn’s disease (86). A SNP in TCF2 is associated with protection from T2D, as expected on the basis of Mendelian mutations at the same gene (120). Unexpectedly, the same association signal turned up in a GWAS for prostate cancer (121). Similarly, JAZF1 was identified as containing SNPs associated with T2D (83) and prostate cancer (122), and TCF7L2 with T2D (123) and colon cancer (124, 125).
From Common SNPs to the Full Allelic Spectrum
The current HapMap provides reliable proxies for the vast majority of SNPs at frequencies above 5%, but its coverage declines rapidly for lower-frequency alleles (37). Such lower-frequency alleles may be particularly important: Alleles with strong deleterious effects are constrained by natural selection from becoming too common. We divide these alleles into two conceptually distinct classes:
1) Common variants with frequencies below 5%. By “common,” we refer to variants that occur at sufficient frequency to be cataloged in studies of the general population and measured (directly, or indirectly through LD) in association studies. In practice, this class may include allele frequencies in the range of 0.5% and above. A GWAS of 2000 cases and 2000 controls provides good power for a 1% allele causing a factor of 4 increase in risk (even at P < 10−8) (Fig. 2).
The value of lower-frequency common variants is illustrated by PCSK9 (proprotein convelíase subtilisin/kexin type 9). The gene encoding PCSK9 contains very rare mutations causing autosomal dominant hypercholesterolemia (discovered by linkage analysis), as well as high-frequency common variants with modest effects. The former are too rare and the latter too weak to enable effective clinical study of PCSK9 with respect to coronary artery disease risk. Hobbs and Cohen sequenced the gene (126, 127) and identified low-frequency common variants (0.5 to 1%), which allowed epidemiological research documenting a protective effect on myocardial infarction (128).
2) Rare variants. Most Mendelian diseases involve rare mutations that are essentially never observed in the general population. Rare mutations likely also play an important role in common diseases. Because they are numerous and individually rare, it is not possible to create a complete catalog in the general population. Instead, they must be identified by sequencing in cases and controls in each study. Moreover, because each variant is too rare to prove statistical evidence of association, the mutations must be aggregated as a class to compare the overall frequency of cases versus controls.
A few examples are known through candidate gene studies. Rare nonsynonymous mutations in MC4R are found in patients with extreme early-onset obesity (129). Rare nonsynonymous mutations in ABCA1 are more common in patients with extremely low HDL than in those with high HDL (130). An excess of rare mutations in renal salt-handling genes has been associated with lower blood pressure and protection against hypertension (131).
The sample size required to perform a genome-wide search based on coding mutations depends on the background frequency (μ) of mutations that confer disease risk and the level (ω) of increased risk for each such mutation. ABCA1 is a favorable case because μ and ω are high (the gene has an unusually large coding region of ~7 kb, and mutations confer a factor of ~6 increase in risk). Achieving genome-wide significance will likely require resequencing studies of thousands of cases and controls, similar to GWASs (Fig. 2).
GWASs of rare variants are already under way for large structural variants through the use of micro-array analysis. A recent GWAS of autism revealed that a highly penetrant, recurrent microdeletion and microduplication of a 593-kb region in 16pll.2 explains 1% of cases (132). Moreover, several recent studies report that patients with autism and schizophrenia may have an excess of rare deletions across the genome relative to unaffected controls (133,134). Although these studies did not identify specific loci (none of the novel loci were observed more than once), they suggest that the universe of rare structural changes contributing to each disease may be as large and diverse as that of common SNPs.
The Genetic Architecture of Common Disease
Variants so far identified by GWASs together explain only a small fraction of the overall inherited risk of each disease (for example, ~10% of the variance for Crohn’s and ~5% for T2D). Where is the remaining genetic variance to be found? There are several answers:
1) At disease loci already identified by GWAS, the locus-attributable risk will often be higher than currently estimated. This is because marker SNPs used in GWASs will typically be imperfect proxies for the actual causal mutation that led to the association signal. The causal gene will often contain additional mutations not tagged by the initial marker SNPs, both common and rare. Determining the contribution of each gene will require intensive studies of variants at each locus.
2) Many more disease loci remain to be identified by GWAS. As noted above, GWASs to date have had low statistical power and thus necessarily missed many loci with common variants of similar and smaller effects. The first studies did not have proxies for common structural variants and have failed to capture lower-frequency common variants (0.5 to 5%). Moreover, the vast majority of studies have been performed only in samples of European ancestry. Larger, more comprehensive, and more diverse GWASs will reveal many more loci.
3) Some disease loci will contain only rare variants. Such loci (if not already found by Mendelian genetics) cannot be identified by study of common variants alone. They will require systematic resequencing of all genes in large samples (Fig. 2).
4) Current estimates of the variance explained are based on simplifying assumptions. Because the genotype-phenotype correlation has yet to be well characterized, the estimates assume that the variants interact in a simple additive manner. Yet gene-gene and gene-environment interactions play important roles in disease risk. Although searches have not yet found much evidence for epistasis [e.g., (93, 94, 135)], this may simply reflect limited power to assess the many possible modes of interaction, including pairwise interactions and threshold effects. Once patterns of association and interaction are understood, effects of specific gene and environmental exposures on each phenotype may be larger.
For these reasons, it is premature to make inferences about the overall genetic architecture of common disease. Only by systematically exploring each of these directions over the coming years will a general picture emerge—with the likely outcome being that different diseases will each be characterized by a different balance of allele frequencies, interactions, and types. Although the proportion of genetic variance explained is certain to grow in the coming years, it is unlikely to approach 100% because of practical limitations, such as the difficulty of detecting common variants with extremely small effects, genes harboring rare variants at very low frequency, and complex interactions among genes and with the environment.
Disease Risk Versus Disease Mechanism
The primary value of genetic mapping is not risk prediction, but providing novel insights about mechanisms of disease. Knowledge of disease pathways (not limited to the causal genes and mutations) can suggest strategies for prevention, diagnosis, and therapy From this perspective, the frequency of a genetic variant is not related to the magnitude of its effect, nor to the potential clinical value that may be obtained.
The classic example is Brown and Goldstein’s studies of FH, which affects ~0.2% of the population and accounts for a tiny fraction of the heritability of LDL and myocardial infarction. Studies of FH led to the discovery of the LDL receptor and supported the development of HMGCR inhibitors (statins) for lowering LDL, the use of which is not limited to FH carriers.
More recently, GWASs have shown that common genetic variation in LDLR and HMGCR influences LDL levels (84, 85). Although SNPs in HMGCR have only a small effect (~5%) on LDL levels, drugs targeting the encoded protein decrease LDL levels by a much greater extent (~30%). This is because the effect of an inherited variant is limited by natural selection and pleiotropy, whereas the effect of a drug treatment is not.
The Path Ahead
Given the long-standing success of genetic mapping in providing new insights into biology and disease etiology, and the recent proof that systematic association studies can identify novel loci, our aim should be nothing less than identifying all pathways at which genetic variation contributes to common diseases. We sketch key steps in achieving this goal.
Expanding clinical studies
Current studies are underpowered for the types of SNP alleles that we now know exist, and available evidence indicates that increasing sample size will yield substantial returns. A study of 1000 cases and 1000 controls provides only 1% power to detect a 20% variant that increases risk by a factor of 1.3, but a study of 5000 cases and 5000 controls provides 98% power (Fig. 2). Moreover, early data on rare single-nucleotide (130, 131) and structural variants (133, 134, 136) indicate that similarly large samples will be needed to achieve the levels of statistical significance required to detect rare events in a genome-wide search.
Nearly all GWASs to date have been performed in populations of European ancestry. Even if a variant has the same effect in all ancestry groups, it may be more readily detected in one population simply because it happens to have higher frequency. Genetic effects will likely vary across groups because of modification by environment and behavior, which may vary more across groups than does genotype.
Many important diseases remain to be studied by GWAS. Disease-related intermediate traits can also offer substantial insight, particularly in conjunction with clinical endpoints. For example, newly described variants on chromosome 1 (near SORTI) are associated both with levels of LDL cholesterol (84, 85) and with risk of myocardial infarction (106); this provides not only increased statistical confidence, but also a biomarker for gene function and pathophysiological insight. Genetic variants that influence gene expression [e.g., (137)] hold promise for elucidating regulatory pathways. Mapping of modifiers of Mendelian mutations—for example, genes that influence the age of onset in carriers of BRCA1 and BRCA2 mutations—may suggest ways to reverse high risk due to mutations.
Correlations between genetic variants and phenotypes are limited by the accuracy with which each is measured. The ability to measure genotype now far exceeds our ability to measure phenotype. Continuous ambulatory monitoring, imaging methods, and comprehenive (“-omic”) approaches to biological samples all have promise in improving the accuracy of phenotype measurement.
Environmental exposures play a larger role in human phenotypic variation than does genetic variation, but environmental exposures are fundamentally more difficult to measure. DNA is stable throughout life, with a single physical chemistry that enables generic approaches to measurement. Environmental exposures are heterogeneous and may be fleeting. Improved methods for measuring environmental exposures, perhaps based on epigenetic marks they leave, are sorely needed.
Expanding the range of genetic variation
The lowest-hanging fruit will be to resequence loci that have been definitively implicated in disease by Mendelian genetics or by GWAS. Because the prior probability of a true association is higher, such regions will be the best setting to develop methods for understanding the statistical significance and biological importance of rare mutations. Initially, resequencing of coding exons will be easiest to interpret. Rare coding mutations with large effect will be especially valuable, because physiological studies of mutation carriers can help illuminate the biological basis of the disease, and because coding mutations of large effect are more straightforwardly transferred to cellular and animal models for mechanistic studies.
Extending GWASs to include structural variants and lower-frequency common variants will require comprehensive catalogs of genomic variation, as well as characterization of LD relationships. With new massively parallel sequencing technologies, an accurate map of all 1% alleles (both single-nucleotide and structural) should be achievable. A “1000 Genomes Project” was recently launched toward this end (138).
Some loci may harbor neither common variants nor rare structural variants, and thus will be missed by array and LD-based approaches. Discovering such genes will require sequencing in thousands of cases and controls. Initial studies will likely focus on exons, where functional mutations are enriched to the greatest extent. Highly parallel methods to capture hundreds of thousands of exons, and other targets of interest, are under development (139).
Multiple instances of de novo coding mutations at a locus (by comparing affected individuals with parents) could provide particularly powerful association information, because the human mutation rate is so low (in the range of 10−8). But identifying de novo mutations without being overwhelmed by false positives will require extraordinary sequencing accuracy (far better than finished genome sequence). Because such studies will be expensive at first, priority should go to disorders with high heritability, where there is an unmet medical need, and for which other approaches have met with limited success. Psychiatric disorders might represent one such target.
Eventually, it will become practical to resequence entire genomes from thousands of cases and controls. The problem of interpretation will be much harder for noncoding functional elements, because it is unclear either how to aggregate elements to achieve a large enough target size, or to develop ways to recognize function-altering changes.
Routine genome sequencing of deeply phenotyped cohorts will fundamentally change the nature of genetic mapping: from the current serial process (in which initial localization by linkage or GWAS is followed by scrutiny of DNA variation and phenotypes) to a joint estimation procedure combining variation information of all types, frequencies, and phenotypes to discover and characterize genotype-phenotype correlations. New statistical methods will be required to combine evidence from rare and common alleles at a locus and across multiple loci, phenotypes, and nongenetic exposures. A particular challenge will be to identify mutations in regions without known function or evolutionary conservation.
There may be inherent limits to our ability to relate phenotypic variation and genotypic variation. To the extent that disease is influenced by tiny effects at hundreds of loci or highly heterogeneous rare mutations, it may be impractical to assemble sufficiently large samples to give a complete accounting.
Implications for Biology, Medicine, and Society
Genetic mapping is only a first step toward biological understanding and clinical application. Useful tools will include maps of evolutionary conservation (108) and chromatin state (140), as well as databases of cell-state signatures, such as genome-wide expression patterns, that may integrate aspects of cell biology under resting and provoked conditions (141). Creation of disease models, both in human cell culture and nonhuman animals, will be key. Physiological studies in patients classified by genotype may inform disease processes and lead to useful nongenetic bio-markers. Given the limits of human clinical research, rare alleles of strong effect may be more useful than common alleles of weak effect.
The high failure rate of clinical trials testifies to the limited predictive value of current approaches. By focusing attention on genes and processes, human genetics has the potential to yield productive targets and predictive animal models. In clinical trials, the ability to stratify patients by genotype or biological pathway may reveal differences in therapeutic response. Genetics may also increase the efficiency of outcome trials by focusing on patients at higher-than-average risk.
The extent to which genetic information will figure in “personalized medicine” will depend on whether predictive accuracy beyond conventional measures can be attained, and whether there are interventions whose effectiveness is improved by knowledge of a genetic test. Knowledge of a common variant that increases T2D risk by 20% may eventually lead to new understanding and therapeutic strategies, but whether an increase in absolute risk (from 8% to 10%) is useful for patients remains to be seen. Although it is tempting to think that knowledge of individual risk might promote greater adherence to a healthy lifestyle, human behavior is complex and risk estimates are challenging to interpret. Even where genotype can predict response to a drug with a narrow therapeutic window, it cannot be assumed that genetic testing will necessarily lead to improved clinical outcomes.
Our understanding of complex disease will be in constant flux over the coming years. The pace of discovery, while scientifically exhilarating, poses daunting challenges. Direct-to-consumer marketing of genetic information is already under way. It will be a challenge for the public to understand the difference between relative and absolute risk, and to figure in their thinking the larger component of genetic and environmental factors not yet captured by today’s technologies. Rigorous assessment of health benefit and cost are needed, including costs of testing and treatment that may flow from an altered sense of risk. As genetic information is shown to be useful, equitable access will be critical.
Finally, we must ensure that the promise of research on genetic factors in complex disease does not encourage a mistaken sense of genetic determinism. This is especially important for behavioral traits, which are especially prone to misinterpretation and misguided policy. We must constantly remind the public—and ourselves— that although genes play a role (and can lead us to new biological insight), our traits are powerfully shaped by the environment, and the solutions to important problems will often lie outside our genes.
Footnotes
Information about obtaining reprints of this article or about obtaining permission to reproduce this article in whole or in part can be found at: http://www.sciencemag.org/about/permissions.dtl
Supporting Online Material
www.sciencemag.org/cgi/content/full/322/5903/881/DCl Table S1
References
- 1.Sturtevant A. J. Exp. Zool. 1913;14:43. [Google Scholar]
- 2.Clarke L, Carbon J. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2173. doi: 10.1073/pnas.77.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender W, et al. Science. 1983;221:23. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 4.Aird I, Bentall HH, Mehigan JA, Roberts JA. Br. Med. J. 1954;2:315. doi: 10.1136/bmj.2.4883.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingram VM. Nature. 1956;178:792. doi: 10.1038/178792a0. [DOI] [PubMed] [Google Scholar]
- 6.Petes TD, Botstein D. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5091. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffreys AJ. Cell. 1979;18:1. doi: 10.1016/0092-8674(79)90348-9. [DOI] [PubMed] [Google Scholar]
- 8.Kan YW, Dozy AM. Proc. Natl. Acad. Sci. U.S.A. 1978;75:5631. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botstein D, White RL, Skolnick M, Davis RW. Am. J. Hum. Genet. 1980;32:314. [PMC free article] [PubMed] [Google Scholar]
- 10.Gusella JF, et al. Nature. 1983;306:234. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 11.Donis-Keller H, et al. Cell. 1987;51:319. doi: 10.1016/0092-8674(87)90158-9. [DOI] [PubMed] [Google Scholar]
- 12.Dib C, et al. Nature. 1996;380:152. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 13.Hudson TJ, et al. Science. 1995;270:1945. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 14.Online Mendelian Inheritance in Man. ( www.ncbi.nlm.nih.gov/sites/entrez?db=omim)
- 15.Welcsh PL, King MC. Hum. Mol. Genet. 2001;10:705. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 16.Lifton RP. Harvey Lect. 2004;100:71. [PubMed] [Google Scholar]
- 17.Bell GI, Polonsky KS. Nature. 2001;414:788. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- 18.East E. Am. Nat. 1910;44:65. [Google Scholar]
- 19.Altenburg E, Muller HJ. Genetics. 1920;5:1. doi: 10.1093/genetics/5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RA. Trans. R. Soc. Edinburgh. 1918;52:399. [Google Scholar]
- 21.Paterson AH, et al. Nature. 1988;335:721. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- 22.Klein J, Sato A. N. Engl. J. Med. 2000;343:782. doi: 10.1056/NEJM200009143431106. [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter WJ, Roses AD. Annu. Rev. Neurosci. 1996;19:53. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- 24.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. Genet. Med. 2002;4:45. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn NJ. Nat. Genet. 2003;33:177. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 26.Collins FS, Guyer MS, Chakravarti A. Science. 1997;278:1580. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 27.Lander ES. Science. 1996;274:536. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 28.Risch N, Merikangas K. Science. 1996;273:1516. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M, Ota T. Genetics. 1973;75:199. doi: 10.1093/genetics/75.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris H. Proc. R. Soc. London Ser. B. 1966;164:298. doi: 10.1098/rspb.1966.0032. [DOI] [PubMed] [Google Scholar]
- 31.Li WH, Sadler LA. Genetics. 1991;129:513. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachidanandam R, et al. Nature. 2001;409:928. [Google Scholar]
- 33.Lewontin R. In: Evolutionary Biology 6. Dobzhansky T, Hecht MK, Steere WC, editors. New York: Appleton-Century-Crofts; 1972. pp. 391–398. [Google Scholar]
- 34.Reich DE, Lander ES. Trends Genet. 2001;17:502. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang DG, et al. Science. 1998;280:1077. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 36.Entrez SNP. ( www.ncbi.nlm.nih.gov/sites/entrez?db=snp)
- 37.International HapMap Consortium. Nature. 2005;437:1299. [Google Scholar]
- 38.Antonarakis SE, Boehm CD, Giardina PJ, Kazazian HH., Jr Proc. Natl. Acad. Sci. U.S.A. 1982;79:137. doi: 10.1073/pnas.79.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lander ES, Botstein D. Cold Spring Harb. Symp. Quant. Biol. 1986;51:49. doi: 10.1101/sqb.1986.051.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Kerem B, et al. Science. 1989;245:1073. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 41.Hastbacka J, et al. Nat. Genet. 1992;2:204. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 42.Kruglyak L. Nat. Genet. 1999;22:139. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 43.Ardlie KG, Kruglyak L, Seielstad M. Nat Rev. Genet. 2002;3:299. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 44.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. Nat. Genet. 2001;29:229. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 45.Patil N, et al. Science. 2001;294:1719. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 46.Gabriel SB, et al. Science. 2002;296:2225. doi: 10.1126/science.1069424. published online 23 May 2002. [DOI] [PubMed] [Google Scholar]
- 47.Johnson GC, et al. Nat. Genet. 2001;29:233. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 48.Reich DE, et al. Nat. Genet. 2002;32:135. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 49.Crawford DC, et al. Nat. Genet. 2004;36:700. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 50.T. McVean GA, et al. Science. 2004;304:581. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 51.Tishkoff SA, Verrelli BC. Annu. Rev. Genomics Hum. Genet. 2003;4:293. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap Consortium. Nature. 2007;449:851. [Google Scholar]
- 53.Sebat J, et al. Science. 2004;305:525. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 54.Iafrate AJ, et al. Nat. Genet. 2004;36:949. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 55.Tuzun E, et al. Nat. Genet. 2005;37:727. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 56.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Nat. Genet. 2006;38:82. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 57.McCarroll SA, et al. Nat. Genet. 2006;38:86. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 58.Locke DP, et al. Am. J. Hum. Genet. 2006;79:275. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redon R, et al. Nature. 2006;444:444. [Google Scholar]
- 60.McCarroll SA, et al. Nat. Genet. 2008;40:1166. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 61.Deeb SS, et al. Nat. Genet. 1998;20:284. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 62.Altshuler D, et al. Nat. Genet. 2000;26:76. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 63.Florez JC, Hirschhorn JN, Altshuler D. Annu. Rev. Genomics Hum. Genet. 2003;4:257. doi: 10.1146/annurev.genom.4.070802.110436. [DOI] [PubMed] [Google Scholar]
- 64.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Genet. Epidemiol. 2008;32:381. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 65.Knowler WC, Williams RC, Pettitt DJ, Steinberg AG. Am. J. Hum. Genet. 1988;43:520. [PMC free article] [PubMed] [Google Scholar]
- 66.Spielman RS, McGinnis RE, Ewens WJ. Am. J. Hum. Genet. 1993;52:506. [PMC free article] [PubMed] [Google Scholar]
- 67.Devlin B, Roeder K. Biometrics. 1999;55:997. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 68.Pritchard JK, Rosenberg NA. Am. J. Hum. Genet. 1999;65:220. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AL, et al. Nat. Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 70.Clayton DG, et al. Nat. Genet. 2005;37:1243. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 71.Chapman JM, Cooper JD, Todd JA, Clayton DG. Hum. Hered. 2003;56:18. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 72.Hinds DA, et al. Science. 2005;307:1072. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 73.de Bakker PIW, et al. Nat. Genet. 2005;37:1217. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 74.Marchini J, Howie B, Myers S, McVean G, Donnelly P. Nat. Genet. 2007;39:906. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 75.Weiss KM, Terwilliger JD. Nat. Genet. 2000;26:151. doi: 10.1038/79866. [DOI] [PubMed] [Google Scholar]
- 76.Couzin J. Science. 2002;296:1391. doi: 10.1126/science.296.5572.1391. [DOI] [PubMed] [Google Scholar]
- 77.Klein RJ, et al. Science. 2005;308:385. published online 10 March 2005. [Google Scholar]
- 78.Edwards AO, et al. Science. 2005;308:421. published online 10 March 2005. [Google Scholar]
- 79.Haines JL, et al. Science. 2005;308:419. published online 10 March 2005. [Google Scholar]
- 80.Thorleifsson G, et al. Science. 2007;317:1397. doi: 10.1126/science.1146554. published online 9 August 2007. [DOI] [PubMed] [Google Scholar]
- 81.Wellcome Trust Case Control Consortium. Nature. 2007;447:661. [Google Scholar]
- 82.Altshuler D, Daly M. Nat. Genet. 2007;39:813. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 83.Zeggini E, et al. Nat. Genet. 2008;40:638. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kathiresan S, et al. Nat. Genet. 2008;40:189. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Willer CJ, et al. Nat. Genet. 2008;40:161. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barrett JC, et al. Nat. Genet. 2008;40:955. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lettre G, et al. Nat. Genet. 2008;40:584. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weedon MN, et al. Nat. Genet. 2008;40:575. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanna S, et al. Nat. Genet. 2008;40:198. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gudbjartsson DF, et al. Nat. Genet. 2008;40:609. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 91.McCarroll SA, et al. Nat. Genet. 2008;40:1107. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haiman CA, et al. Nat. Genet. 2007;39:638. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maller J, et al. Nat. Genet. 2006;38:1055. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 94.Li M, et al. Nat. Genet. 2006;38:1049. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Graham RR, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6758. [Google Scholar]
- 96.Duerr RH, et al. Science. 2006;314:1461. doi: 10.1126/science.1135245. published online 26 October 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sladek R, et al. Nature. 2007;445:881. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 98.Easton DF, et al. Nature. 2007;447:1087. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeggini E, et al. Science. 2007;316:1336. doi: 10.1126/science.1142364. published online 25 April 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scott LJ, et al. Science. 2007;316:1341. doi: 10.1126/science.1142382. published online 25 April 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Diabetes Genetics Initiative of Broad Institute of Harvard MIT, Lund University, Novartis Institutes for BioMedical Research. Science. 2007;316:1331. published online 26 April 2007. [Google Scholar]
- 102.Rivera A, et al. Hum. Mol. Genet. 2005;14:3227. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 103.Amundadottir LT, et al. Nat. Genet. 2006;38:652. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 104.Freedman ML, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14068. [Google Scholar]
- 105.McPherson R, et al. Science. 2007;316:1488. doi: 10.1126/science.1142447. published online 2 May 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Samani NJ, et al. N. Engl. J. Med. 2007;357:443. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Helgadottir A, et al. Science. 2007;316:1491. doi: 10.1126/science.1142842. published online 2 May 2007. [DOI] [PubMed] [Google Scholar]
- 108.Waterston RH, et al. Nature. 2002;420:520. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 109.Burton PR, et al. Nat. Genet. 2007;39:1329. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cargill M, et al. Am. J. Hum. Genet. 2007;80:273. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Florez JC, et al. N. Engl. J. Med. 2006;355:241. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grarup N, et al. Diabetes. 2007;56:3105. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 113.Pascoe L, et al. Diabetes. 2007;56:3101. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 114.Saxena R, et al. Diabetes. 2006;55:2890. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 115.Staiger H, et al. PLoS One. 2007;2:e832. doi: 10.1371/journal.pone.0000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Steinthorsdottir V, et al. Nat. Genet. 2007;39:770. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 117.Wolf N, et al. J. Med. Genet. 2008;45:114. doi: 10.1136/jmg.2007.053595. [DOI] [PubMed] [Google Scholar]
- 118.Reiner AP, et al. Am. J. Hum. Genet. 2008;82:1193. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ridker PM, et al. Am. J. Hum. Genet. 2008;82:1185. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Winckler W, et al. Diabetes. 2007;56:685. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 121.Gudmundsson J, et al. Nat. Genet. 2007;39:977. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 122.Thomas G, et al. Nat. Genet. 2008;40:310. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 123.Grant SFA, et al. Nat. Genet. 2006;38:320. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 124.Hazra A, et al. Cancer Causes Control. 2008;19:975. doi: 10.1007/s10552-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Folsom AR, et al. Diabetes Care. 2008;31:905. doi: 10.2337/dc07-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kotowski IK, et al. Am. J. Hum. Genet. 2006;78:410. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cohen J, et al. Nat. Genet. 2005;37:161. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 128.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. N. Engl. J. Med. 2006;354:1264. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 129.Hirschhorn JN, Altshuler D. J. Clin. Endocrinol. Metab. 2002;87:4438. doi: 10.1210/jc.2002-021329. [DOI] [PubMed] [Google Scholar]
- 130.Cohen JC, et al. Science. 2004;305:869. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 131.Ji W, et al. Nat. Genet. 2008;40:592. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weiss LA, et al. N. Engl. J. Med. 2008;358:667. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 133.Walsh T, et al. Science. 2008;320:539. doi: 10.1126/science.1155174. published online 27 March 2008. [DOI] [PubMed] [Google Scholar]
- 134.Sebat J, et al. Science. 2007;316:445. doi: 10.1126/science.1138659. published online 14 March 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rioux JD, et al. Nat. Genet. 2007;39:596. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weiss LA, et al. N. Engl. J. Med. 2008;358:667. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 137.Emilsson V, et al. Nature. 2008;452:423. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 138.1000 Genomes. ( www.1000genomes.org)
- 139.Albert TJ, et al. Nat. Methods. 2007;4:903. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 140.Mikkelsen TS, et al. Nature. 2007;448:553. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lamb J, et al. Science. 2006;313:1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]