Abstract
One-carbon metabolism is a network of biological reactions that plays critical role in DNA methylation and DNA synthesis, and in turn, facilitates the cross-talk between genetic and epigenetic processes. Genetic polymorphisms and supplies of cofactors (e.g. folate, vitamins B) involved in this pathway have been shown to influence cancer risk and even survival. In this review, we summarized the epidemiological evidence for one-carbon metabolism, from both genetics and lifestyle aspects, in relation to breast cancer risk. We also discussed this pathway in relation to breast cancer survival and the modulation of one-carbon polymorphism in chemotherapy. Emerging evidence on modulation of DNA methylation by one-carbon metabolism suggests that disruption of epigenome might have been the underlying mechanism. More results are expected and will be translated to guidance to the general population for disease prevention as well as to clinicians for treatment and management of the disease.
Keywords: one-carbon; breast cancer; folate, genetic polymorphism; DNA methylation, epidemiology
Introduction
Breast cancer is a manifestation of abnormal genetic as well as epigenetic changes (Russo et al., 1998). Global hypomethylation, accompanied by promoter hypermethylation, is a common feature of tumor cells (Esteller, 2008). Global hypomethylation may induce chromosomal instability, reactivate transposons, promote loss of imprinting, and activate proto-oncogenes. Yet, reduced methylation may also protect against C to T mutations (Ulrich et al., 2005). Promoter hypermethylation, in contrast, is associated with the inactivation of genes in virtually all pathways protective of carcinogenesis (e.g. DNA repair, cell cycle control, inflammatory/stress response, detoxification, apoptosis, etc.) (Baylin et al.,1998; Herman, 1999; Widschwendter and Jones, 2002). One-carbon metabolism, also referred as folate-mediated one-carbon metabolism, can impact both genetic and epigenetic pro-carcinogenic processes because of its critical role in both DNA methylation and DNA synthesis (Stern et al., 2000). A low methyl supply can induce DNA global hypomethylation as well as deficient conversion of dUMP to dTMP leading to uracil misincorporation into DNA (Christman et al., 1993). The repair activity by uracil glycosylases can lead to DNA strand breaks, resulting in enhanced mutagenesis and apoptosis (Baylin et al., 2001). Because of the essential roles in these critical processes, one-carbon metabolism not only is capable of influencing the pathogenesis of breast cancers, but also it has been the target pathway for chemotherapy as well as chemoprevention of these diseases. In this review, we summarized the existing epidemiological evidence on the associations between breast cancer and genetic polymorphisms of one-carbon metabolizing genes as well as one-carbon related nutrients (i.e., folate).
Methyl-diet intake and breast cancer risk: epidemiologic evidence
The universal methyl donor in the body is S-adenosylmethionine (SAM) that donates its labile methyl group to more than 80 biological methylation reactions, including the methylation of DNA, RNA and protein. To generate the universal methyl donor SAM in the cell, homocysteine is methylated to methionine via the one-carbon pathway by transferring a methyl group from 5-methyl-tetrahydofolate or betaine. The major external source of methyl groups comes from folate, methionine, and choline in diet (Niculescu, 2002). It has been shown that folate depletion alone is a sufficient perturbing force to diminish the methyl pool (Miller et al., 1994). Other vitamins B, such as vitamins B2, B6 and B12, are also key cofactors for one-carbon metabolism. Alcohol is a folate antagonist; excess consumption of alcohol impairs folate absorption by inhibiting expression of the reduced folate carrier and decreasing the hepatic uptake and renal conservation of circulating folate (Halsted et al., 2002). Associations of folate intake and breast cancer risk have been extensively studied. However, studies on methionine, choline and betaine are limited (Cho et al., 2007; Xu et al., 2008a). In this review, we will focus on epidemiologic evidence of folate consumption and breast cancer.
Association studies using both cohort and case-control study designs have been conducted and subsequently pooled and meta-analyses combining data of individual studies have also been performed. While meta-analyses combine the published results of summary effects such as relative risk (RR) or odds ratio (OR), pooled analyses combine individual-level data that permit a full examination of effect modification within the data. It is important to note that these pooled or meta-analyses are performed retrospectively and subject to inherited limitations such as study heterogeneity and publication bias. Nevertheless, these analyses offer increased power to detect associations, especially as the number of included studies increases.
In epidemiologic studies, folate as an exposure of interest is often assessed from food frequency questionnaires (FFQ) or circulating biomarkers (i.e., plasma folate or red blood cell folate level). Folate intake can be referred to as dietary intake (from food) or total intake (from food and supplements). Folate naturally found in foods are predominantly in the form of 5-methyl-tetrahydrofolate (THF); meanwhile the fully unreduced (e.g. folic acid) and partially reduced forms (e.g. dihydrofolate (DHF)) are also found (Combs, 1992). In contrast, folate in supplements and food fortification is the synthetic form, folic acid, which needs to be reduced before it can participate in cellular reactions (Machlin, 1991). So the term “total folate intake” was used by adding up the folate in diet and in supplements (synthetic folic acid). This approach has limitation because folic acid has 1.7-fold greater bioavailability. Using the crude approach of simply summing up could result in misclassification and bias risk estimates toward null effects. Other approach, such as “Dietary Folate Equivalents (DFE)”, was recommended by the Institute of Medicine of USA (Sauberlich et al., 1987; Hannon-Fletcher et al., 2004), which allows for a combination of dietary and synthetic folic acid into a variable that accounts for this differential bioavailability.
Although a number of cohort and case-control studies have suggested an inverse association between folate status and the risk of breast cancer, these results are far from conclusive. In a meta-analysis summarizing studies published between 1966 and 2006 (Larsson et al., 2007), folate intake (both dietary and total) with 200 μg/day increments was not associated with the risk of breast cancer in 8 prospective studies; however, an inverse association with dietary folate was observed in 13 case-control studies (OR: 0.80; 95% CI: 0.72–0.89). Data from several cohort studies, e.g. Nurses’ Health Study, the Canadian National Breast Screening Study and the Iowa Women’s Health Study, also indicate that adequate folate intake could attenuate the elevated risk associated with moderate alcohol consumption (Zhang et al., 1999; Rohan et al., 2000; Sellers et al., 2001). In addition, there was an indication of inverse associations between blood folate concentrations and breast cancer risk, especially in case-control studies, although these associations failed to reach statistical significance (shown in Table 1).
Table 1.
Summary of meta- and pooled analyses on folate and breast cancer risk
| Author (ref.) | Folate measurement | No. of studies included | No. of cases | No. of controls* | Summary RR or OR | Comparison |
|---|---|---|---|---|---|---|
| Larsson et al.(2007) | dietary intake | 8 cohort | 8,367 | 302,959 | 0.97(0.88–1.07) | 200 µg/day increments |
| total intake | 6 cohort | 8,165 | 306,209 | 1.01(0.97–1.05) | 200 µg/day increments | |
| dietary intake | 13 case-control | 8,558 | 10,812 | 0.80(0.72–0.89) | 200 µg/day increments | |
| total intake | 3 case-control | 2,184 | 3,233 | 0.93(0.81–1.07) | 200 µg/day increments | |
| blood levels | 3 cohor | 970 | 1,979 | 0.81(0.59–1.10) | high vs. low | |
| blood levels | 2 case-control | 269 | 366 | 0.41(0.15–1.10) | high vs. low | |
| Lewis et al. (2006) | dietary intake | 9 cohort | 11,227 | 331,462 | 0.99(0.98–1.01) | 100 µg/day increments |
| dietary intake | 13 case-control | 8,566 | 10,834 | 0.91(0.87–0.96) | 100 µg/day increments |
For cohort study, the numbers are total participant in the study.
Similar results were observed from another meta-analysis by Lewis et al. (2006). A total of 13 case-control studies and 9 cohort studies were included; some overlapped with the study of Larsson et al. (2007). Summary odds ratios for dietary folate were 0.91 (95% CI: 0.87–0.96) for the case—control studies and 0.99 (95% CI: 0.98–1.01) for the cohort studies with a 100 µg/day increase in folate intake. This study lends additional support that dietary folate (not total folate) may be moderately protective against breast cancer (shown in Table 1).
Results from subsequent cohort studies have been inconsistent or even conflicting. In a French cohort study (1,812 cases among 62,739 postmenopausal women with 9-year follow-up), an inverse association was observed (RR: 0.78; 95% CI: 0.67—0.90) (Lajous et al., 2006). In a report from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (691 cases among 25,400 women with 10-year follow-up), an increased risk of breast cancer was observed in postmenopausal women with folic acid supplemental use ≥400 µg/d (Stolzenberg-Solomon et al., 2006). No apparent association between folate status and risk of breast cancer was observed in the Nurses’ Health Study II, in which 90,663 premenopausal women were followed for 12 years with 1,032 documented breast cancer cases (Cho et al., 2007). In the Malmö Diet and Cancer cohort study (392 cases among 11,699 women with 9.5-year follow-up), a higher folate intake (both dietary and total) was associated with a ~40% lower incidence of postmenopausal breast cancer (Ericson et al., 2007).
Alcohol is known to be a folate antagonist. There is quite consistent evidence that lower folate intakes in combination with high alcohol intakes are associated with an increased risk of breast cancer (Larsson et al., 2007). This synergistic effect lends additional support to the notion that one-carbon metabolism is an important process in breast cancer etiology.
There is emerging evidence that indicates the relationship between one-carbon metabolism and breast cancer is complex and non-linear (Ulrich, 2007). While folate- or one-carbon deficiency is thought to increase breast cancer risk, there is also evidence that very high intakes from diet and supplements may result in elevated risk (Stolzenberg-Solomon et al., 2006). This inverted U-shaped relationship is alarming and requires further evaluation (Ulrich, 2007).
In summary, the association of folate intake and breast cancer risk is complex and interactions exist (i.e., with alcohol intake). Further studies especially well-designed clinical trials and mechanistic investigations are needed to clarify the issue.
Genetic polymorphisms in one-carbon metabolizing genes and associations with breast cancer risk
Accumulating evidence from molecular epidemiologic studies has indicated that functional polymorphisms in one-carbon metabolism can influence risk of cancer independently or jointly with dietary factors (folate, other vitamins B and alcohol intake). This section provides an overview of functional genetic polymorphisms involved in one-carbon metabolism and summarizes the results from epidemiologic studies on breast cancer risk.
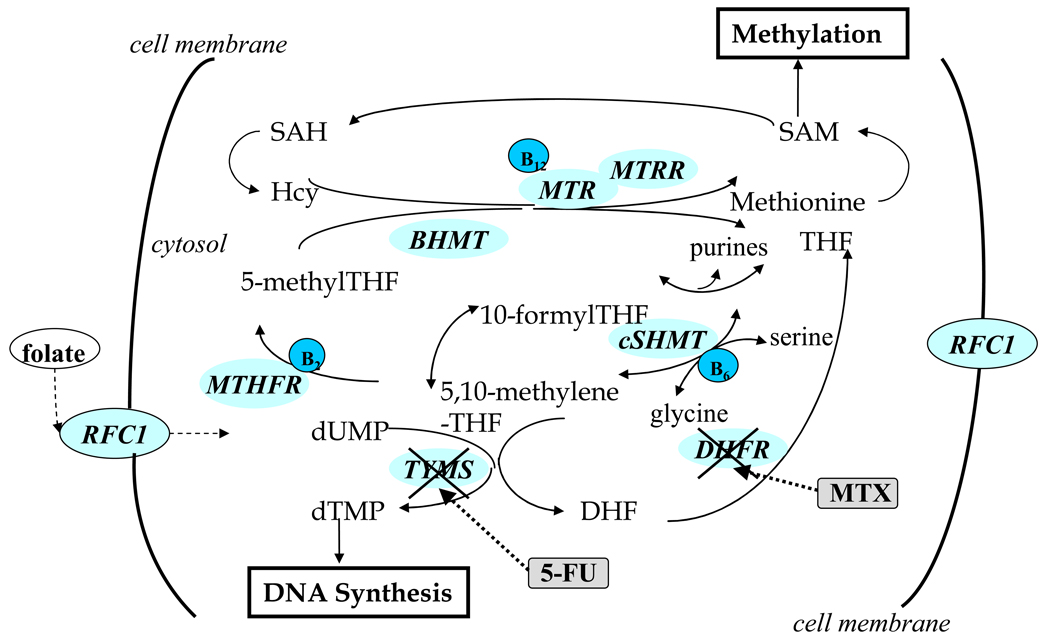
Key enzymes involved in one-carbon metabolism are illustrated in Fig. 1. They include methylenetetrahydrofolate reductase (MTHFR), thymidylate synthase (TS), methionine synthase (MTR), methionine synthase reductase (MTRR), serine hydroxymethyltransferase (SHMT), dihydrofolate reductase (DHFR), betaine-homocysteine methyltransferase (BHMT), cystathionine β-synthase(CBS), methylenetetrahydrofolate dehydrogenase (MTHFD1), reduced folate carrier (RFC1) and transcobalamin II (TCII).
Fig. 1.
Schematic illustration of overview of one-carbon metabolism pathway, linking to methylation reactions and nucleotide synthesis. Hcy, homocysteine; SAM, S-adenosylmethionine; SAH, adenosylhomocysteine; THF, tetrahydrofolate; DHF, dihydrofolate; dUMP, deoxyuridine monophosphate and dUTP, deoxythymidine monophosphate.
An increasing number of genetic polymorphisms have been identified; their information is available on public databases such as the NIH dbSNP database. However, the number of “functional” polymorphisms, i.e., those with confirmed phenotypic changes, such as effects on activity or transcription of the protein, remains limited. Although results on tagging SNPs in one-carbon metabolism have started to emerge, most published studies have been on functional polymorphisms, i.e., those with phenotypic effects, as indicated by biomarker measurements, or have been implicated in studies with disease endpoints (Table 2). These polymorphisms reside in key enzymes of one-carbon matabolism pathway; interruption of their functions may result in aberrant DNA synthesis or methylation (Fig. 1). Several are non-synonymous or reside in regulatory region such as the 5′ or 3′ UTR, and they are generally common in human populations (>5%). Although the biological effects of these polymorphisms are likely to be small or moderate at best, the underlying attributable risk cannot be neglected because of the high prevalence of these polymorphisms in the general population.
Table 2.
Functional polymorphisms involved in one-carbon metabolism
| Genes | Enzyme | Chromosome | Polymorphism | rs number | AA change |
|---|---|---|---|---|---|
| MTHFR | 5,10- methylenetetrahydrofolate reductase |
1p36.3 | 677C > T 1298A > C |
rs1801133 rs1801131 |
Ala222Val Glu429Ala |
| TS | Thymidylate synthase | 18p11.32 | 5’-UTR 28bp tandem repeat; | ||
| 3’-UTR 6bp deletion (1464del6) | |||||
| DHFR | Dihydrofolate reductase | 5q11.2-q13.2 | intron1 -19bp deletion | ||
| MTR(MS) | Methionine synthase | 1q43 | 2756A > G | rs1805087 | Asp919Gly |
| MTRR | Methionine synthase reductase | 5p15.3-p15.2 | 66A > G | rs1801394 | Ile22Met |
| cSHMT | cytosol Serine hydroxymethyltransferase | 17p11.2 | 1420C > T | rs1979277 | Leu474Phe |
| RFC1 | Reduced folate carrier | 21q22.3 | 80G > A | rs1051266 | Arg27His |
| BHMT | Betaine-homocysteine methyltransferase | 5q13.1-q15 | 742G > A | rs3733890 | Arg239Gln |
| CBS | Cystathionine β-synthase | 21q22.3 | 31bp VNTR | ||
| MTHFD1 | methylenetetra-hydrofolate dehydrogenase | 14q24 | 401G > A | rs1950902 | Arg134Lys |
| TCN2 | Transcobalamin II | 22q12.2 | 776C > G | rs1950902 | Pro259Arg |
As illustrated in Fig. 1, functioning of one-carbon metabolism requires several micronutrients, including vitamins B2, B6, and B12, as cofactors of various enzymes. It is important to consider that folate metabolism represents complex and inter-related metabolic reactions with many feedback mechanisms and other regulatory processes that ensure its robustness (Nijhout et al., 2004). Accordingly, one may hypothesize that multiple disturbances within the pathway, or “stress” on the system are needed to result in phenotypic effects. Such “stress” could be present under low intakes of folate or other nutrients, or the presence of genetic polymorphisms with phenotypic changes of the enzymes involved in one-carbon metabolism. Indeed, genetic polymorphisms in MTHFR are most strongly associated with biomarkers, such as homocysteine concentrations, when folate status is low. This provides a rationale for investigating gene-gene and gene-nutrient interactions within this complex system.
5,10-methylenetetrahydrofolate reductase(MTHFR)
MTHFR is a pivotal enzyme in folate and homocysteine metabolism, catalyzing the irreversible reduction of 5,10-methyleneTHF to 5-methyleneTHF. Its critical role in one-carbon metabolism provides strong biological rationale that inherited variability in the enzyme activity may influence cancer risk.
The substrate of the MTHFR enzyme, 5,10-methyleneteTHF, is involved in the conversion of deoxyuridylate monophosphate to deoxythymidylate monophosphate (dTMP for DNA synthesis). Low levels of 5,10-methyleneTHF can result in misincorporation of uracil into DNA, leading to increased rates of point mutations and DNA/chromosome breakage (Blount et al., 1997). A less active form of MTHFR should lead, all other factors being equal, to an accumulation of 5,10-methyleneTHF and less uracil misincorporation, and thus, a presumably lower cancer risk. Recent predictions from a mathematical simulation model of folate metabolism suggest such effects (Luebeck et al., 2008). In contrst, reduced MTHFR activity reduces the level of S-adenosyl-L-methionine, the methyl donor for maintaining DNA methylation patterns, with possible impact on mutation rates and cancer risk (Stern et al., 2000; Davis and Uthus, 2004; Ulrich et al., 2005).
Two SNPs of the MTHFR 677C>T (rs1801133) and 1298A>C (rs1801131) have been most extensively investigated in relation to cancer risk. The 677 variant homozygotes (677TT) have 70% lower enzyme activity compared withwild-type homozygotes (677CC), whereas heterozygotes retain 65% enzyme activity (Frosst et al., 1995). The functionality of the 1298A>C polymorphism is less well established. Individuals who are homozygous for the 1298 variant allele (CC) have about 60% activity compared with subjects carrying the 1298AA genotype in some studies (Weisberg et al., 2001). The compound heterozygotes of the 677C>T and 1298A>C polymorphisms were shown to have similar phenotype as the 677TT genotype (Weisberg et al., 2001); however, this phenotype may be partially explained by the fact that the two SNPs are in linkage disequilibrium (Chen et al., 2005). Nevertheless, the two SNPs have been independently associated with folate and homocysteine concentrations (Bailey and Gregory, 1999; Ulvik et al., 2007).
Associations of MTHFR polymorphisms with risk of breast cancer were examined extensively. Although many individual studies reported significant findings, pooled and meta-analyses revealed no significant associations (Table 3).
Table 3.
Meta-analysis results of MTHFR polymorphisms and breast cancer risk
| Author (ref.) | SNP | No. of studies included | Case/control | Summary OR | P value for heterogeneity |
|---|---|---|---|---|---|
| Lissowaska et al. (2007) | C677T | 21 | 8330/10825 | 0.99(086–1.15) TT vs. CC | 0.02 |
| A1298C | 12 | 6521/8515 | 1.00(085–1.17) CC vs. AA | 0.36 | |
| Macis et al. (2007) | C677T | 18 | 6344/8358 | 1.01(087–1.18) TT vs. CC | |
| Lewis et al. (2006) | C677T | 17 | 6373/8434 | 1.04 (0.94–1.16) TT vs. CC | |
| Zintzaras et al. (2006) | C677T | 18 | 5476/7336 | 1.02(0.95–1.10) T vs. C allele | 0.08 |
| A1298C | 10 | 3768/5276 | 0.97(0.90–1.04) C vs. A allele | 0.21 |
In a meta-analysis by Zintzaras et al. (2006), the overall analysis for the association between the 677 T allele and the breast cancer risk on 18 case-control studies showed no-significant association (OR: 1.02; 95% CI: 0.95–1.10); however, there was indication of heterogeneity among individual studies (P = 0.08). The 677 T allele was not associated with breast cancer risk in Caucasians (nine studies) and in East Asians (four studies) [OR 1.03 (0.93–1.14) and OR 0.96 (0.81–1.15), respectively] or in pre-menopausal (five studies) and postmenopausal (four studies) groups [OR 1.10 (0.94–1.29) and OR 1.06 (0.95–1.18), respectively]. The variant homozygotes (TT vs. CC) produced significant results only for pre-menopausal cases [OR 1.46 (1.05–2.03)]. For the A1298C polymorphism, no association was observed.
Results from a meta-analysis on 17 studies showed no difference in breast cancer risk between MTHFR 677 TT homozygotes and CC homozygotes (OR: 1.04; 95% CI: 0.94–1.16), and there were no interactions between folate intake and MTHFR genotype in relation to breast cancer risk (Lewis et al., 2006).
Macis et al. (2007) conducted another meta-analysis on 17 case-control studies, in which the MTHFR C677T genotype was not associated with breast cancer risk (TT vs. CC+CT OR: 1.01; 95% CI: 0.87—1.18) and (CT+TT vs. CC OR: 1.04; 95% CI; 0.97—1.11). When stratified by menopausal status for seven studies that had such information, MTHFR 677TT genotype was significantly associated with breast cancer in pre-menopausal patients with an OR of 1.42 (95% CI: 1.02—1.98).
Another meta-analysis on 21 studies by Lissowaska et al. (2007) showed no association between MTHFR 677 and breast cancer risk (TT vs. CC: OR: 0.99; 95% CI: 0.86–1.15). Similarly, there was no evidence for an overall association for MTHFR 1298 (OR: 1.00; 95% CI: 0.85–1.17) (Table 3). Results were similar across racial groups and geographical regions. Results from stratified analyses suggested an increased risk of premenopausal breast cancer for women with the homozygous variant genotype (OR: 1.32; 95% CI: 1.00–1.75 for MTHFR C677T and OR: 1.35; 95% CI: 0.85–2.13 for MTHFR A1298C).
A major limitation of meta-analyses and pooled analyses performed on these polymorphisms to date is that they have not been able to take folate-status (or one-carbon status in general) into account. Given the confirmed gene-diet interactions for MTHFR, solitary evaluation of the genetic factor is not very meaningful. In fact, combining results from study populations with divergent nutritional status may negate any effects.
Thymidylate synthase (TS)
A polymorphic 28 bp tandem repeat is located in the 5’-UTR of the TS gene and functions as a cis-acting transcriptional enhancer element (Kaneda et al., 1987). The 3R allele was associated with approximately 2–4 times greater gene expression compared with the 2R allele (Horie et al., 1995; Pullarkat et al., 2001). The association between this polymorphism and breast cancer risk is generally null. Within the 3’UTR of the TS gene, a 6 bp deletion (1494del6) polymorphism was associated with reduced mRNA stability (Ulrich et al., 2000; Mandola et al., 2004). However, the evidence of this polymorphism in relation to cancer risk is sparse and the existing reports generally showed null results.
Methionine synthase (MTR)
The A2756G (Asp919Gly) polymorphism of MTR gene has been proposed to affect plasma homocysteine concentrations (Harmon et al., 1999; Chen et al., 2001). This polymorphism has been investigated by several groups in relation to cancer risk. One group found this SNP was associated with reduced breast cancer risk (Lissowska et al., 2007) while null results were reported in other studies (Justenhoven et al., 2005; Shrubsole et al., 2006; Xu et al., 2007b). Due to the low allele frequency for this variant (~10%), statistical power for investigating gene-diet interactions has been generally insufficient (Goode et al., 2004).
Methionine synthase reductase (MTRR)
An association between the MTRR A66G (Ile22Met) polymorphism and homocysteine concentrations has been reported (Wilson et al., 1999; Gaughan et al., 2001); however, the functional impact of the variant has not been well defined (Jacques et al., 2003). A null association of this polymorphism with breast cancer risk was reported in several studies (Justenhoven et al., 2005; Shrubsole et al., 2006; Lissowska et al., 2007; Xu et al., 2007b).
Dihydrofolate reductase (DHFR)
DHFR converts dihydrofolate into THF and folic acid from supplements needs to be reduced by DHFR before participating in cellular reactions. A 19-base pair (bp) polymorphism in intron 1 of the DHFR gene was identified with potential functionality (Johnson et al., 2004). One study found that although this19-bp deletion polymorphism was not associated with breast cancer risk overall, the deletion allele was associated with increased breast cancer risk among multivitamin users (Xu et al., 2007a). A dose-dependent relationship between DHFR expression and the deletion genotype was observed. Compared to the 19bp +/+ genotype, subjects with the −/− genotype had about 5-fold higher mRNA levels (Xu et al., 2007a).
Other genes
There are many other sporadic reports on other polymorphisms of one-carbon metabolizing genes in relation to cancer risk, yet few researchers have performed systematic investigations. A number of positive findings are included here as examples. Null results were report for the cSHMT C1420T, BHMT G742A, RFC G80A polymorphisms and breast cancer risk in a case-control study (Xu et al., 2007b). The MTHFD1 G401A polymorphism was found to be associated with a significant increased breast cancer risk among postmenopausal women in a case-control study (Stevens et al., 2007).
In summary, a large body of literature on genetic polymorphisms in relation to cancer risk lends support to the concept that genetic variability in one-carbon metabolism may play a critical role in cancer etiology, especially in colorectal and gastric cancer (not discussed here). However, such effect was limited in the context of breast cancer. Most epidemiologic studies on one-carbon polymorphisms have been limited in sample size. Larger and more systematic investigations that provide sufficient statistical power for investigating rarer variants and gene-gene as well as gene-diet interactions are needed. Another method of incorporating the extensive knowledge related to this pathway into the statistical analysis is through the use of mathematical models of one-carbon metabolism (Reed et al., 2006; Ulrich et al., 2006). Initial result from the modeling approach has been promising with model predictions being generally consistent with experimental data (Nijhout HF et al., 2006; Nijhout et al., 2006; Reed et al., 2006; Ulrich et al., 2006). This approach is a powerful tool to better understand gene-gene and gene-diet interactions in the one-carbon metabolism pathway and biological mechanisms linking this pathway to carcinogenesis.
One-carbon metabolism and breast cancer survival
Dietary Methyl Intake and Survival
There are a few studies on one-carbon metabolism and breast survival. These studies usually used the dietary information collected at the baseline interview, which reflects intake patterns for the period prior to and at diagnosis. It is possible that patients change their lifestyle after a diagnosis of cancer (Demark-Wahnefried et al., 2000; McBride et al., 2000). Consequently, results should be interpreted with caution because there are studies which have indicated that after breast cancer diagnosis, women are motivated to change their diet (Salminen, 2000; Maunsell et al., 2002). For example, supplement use among breast cancer patients is high and frequently increases after diagnosis (Velicer and Ulrich, 2008).
There is some evidence that high folate intake is associated with better survival, although the evidence is spare. A report from a study of ~500 postmenopausal breast cancer cases showed that high folate intake also associated with reduced all-cause mortality over 80 months’ follow-up period (McEligot et al., 2006). Results from the 1969 Busselton (Western Australia) Health survey showed that decreased red blood-cell folate was significantly associated with breast cancer morbidity (Rossi et al., 2006). Results from the Iowa Women’s Health Study showed that through a 14-year follow-up period, folate intake was not associated with survival among breast cancer cases treated with chemotherapy (Sellers et al., 2002). In our own study, we found no associations between folate intake and all-cause or breast cancer specific mortality; in contrast, higher vitamins B1 and B3 intakes were associated with better survival (Xu et al., 2008b).
One-carbon polymorphisms and survival
Many studies have been conducted to examine the effects of one-carbon polymorphisms on breast cancer risk, but reports of effects on subsequent survival are relatively sparse. In our own study of the Long Island Breast Cancer Study Project, we found that the MTHFR 677 T allele carriers and BHMT 742 A allele carriers have reduced all-cause mortality (Xu et al., 2008b). Results from the Shanghai Breast Cancer study showed that MTHFR genotypes were not associated with all-cause mortality, but the 677TT genotype was associated with poor survival among those with late stage disease (Shrubsole et al., 2005). Results from a small cohort containing a mixture of Caucasian showed that MTHFR1298 C allele was associated with worse survival compared with the AA genotype (Martin et al., 2006).
Modulation of chemotherapy by one-carbon metabolism
Adjuvant chemotherapy of early breast cancer significantly improves disease free and overall survival (Hanahan and Weinberg, 2000). One major class of chemotherapeutic drugs for the treatment of cancers is characterized as anti-metabolite/anti-folate agents. These are designed to impair nucleotide synthesis and to disrupt normal cellular metabolism; consequently, they inhibit cell growth or induce cell death. These agents that target the one-carbon metabolic pathway can inhibit several intracellular folate metabolizing enzymes. For example, 5-fluorouracil (5-FU), one of the most widely used chemotherapeutic drugs, inhibits TS; methotrexate (MTX), the mainstay for the treatment of acute lymphoblastic leukemia, targets primarily DHFR (Chabner, 2001). Both agents are able to induce cell cycle arrest and apoptosis by inhibiting the cell's ability to synthesize DNA and RNA. Combination chemotherapy with cyclophosphamide, MTX, and 5-FU (CMF) is the treatment of choice for the majority of patients with node-negative breast cancer as trials have shown its efficacy and long-term safety (Clarke, 2000) and this regimen is the form of adjuvant chemotherapy most commonly used worldwide. Resistance to anticancer therapy is one the most critical problems in breast cancer treatment, and it has been estimated that 90% of all cancer deaths are attributable to drug resistance (Young, 1990). Altered one-carbon metabolism may impact the efficacy of such treatment targeting one-carbon pathway. A report on a small series of breast cancer patients showed that the occurrence of severe myelotoxicity after CMF was elevated among individuals with the MTHFR 677TT genotype (Toffoli et al., 2000). However, this genotype was protective against raltitrex-associated toxicity in a phase I trial of the irinotecan/raltitrex combination for treatment of cancer (Stevenson et al., 2001). Another study showed that the MTHFR 677T variant increased chemosensitivity of breast cancer cells to 5-FU, but decreased chemosensitivity to MTX (Sohn et al., 2004). These results provide evidence that the MTHFR C677T polymorphism could affect the concentration and intracellular distribution of folates and changes the growth and chemosensitivity of breast cancer cells. Over-expression of another one-carbon gene, TS, which is associated with the TS*3R allele, increases resistance to fluoropyrimidine derivatives and results in poor clinical outcomes in many investigations, although data specifically on breast cancer are limited (Maring et al., 2005). These polymorphisms may be a useful pharmacogenetic determinant for providing rational and effectively tailored chemotherapy.
One-carbon metabolism and DNA methylation
As discussed previously, observational studies provided strong evidence for the role that one-carbon metabolism plays in breast cancer. Given the fact that one-carbon metabolism provides the essential cofactor in the production of primary methyl donors for DNA methylation, interruption of the pathway could in principle affect the DNA methylation process. This leads to the hypothesis that one-carbon metabolism (dietary methyl content and/or polymorphisms of one-carbon metabolizing genes) influences DNA methylation, and thus, impacts breast cancer etiology and progression.
There is evidence that dietary methyl donors are capable of modulating methylation patterns. In two separate studies on healthy postmenopausal women, folate depletion resulted in lymphocyte DNA hypomethylation, which was reversed following folate repletion (Jacob et al., 1998; Rampersaud et al., 2000). Another study on women with abnormal PAP smear results reported that both cervical tissue folate and serum folate levels were significantly correlated with DNA methylation levels (Fowler et al., 1998). A study on folate and alcohol intake in relation to methylation in colorectal cancer lends strong support that diet is capable of influencing carcinogenesis through epigenetic pathway (van Engeland et al., 2003) by showing that prevalence of promoter hypermethylation of a panel of genes was higher in colorectal tumors derived from patients with low folate/high alcohol intake compared to those with high folate/low alcohol intake.
Furthermore, evidence also exits that functional polymorphisms in one-carbon metabolizing genes could modify DNA methylation status (Stern et al., 2000; Kawakami et al., 2003; Castro et al., 2004). One example is a population study of gastric cancer that showed higher genomic DNA hypomethylation in people with the MTHFR677TT genotype compared to those with the MTHF677CC genotype. Another example is that tumors from patients with MTR 2756GG genotype showed promoter hypermethylation in a large panel of tumor suppressor genes including p16 and BRCA1 (Paz et al., 2002).
Epigenetic alterations in DNA are heritable but do not alter nucleotide sequence. Consequently, unlike genetic changes, epigenetic modifications are potentially reversible (Baylin et al., 2001). Modulating DNA methylation by nutrition may be a fascinating new field of gene and nutrient interaction. Therefore, it is of considerable interest to identify the factors that determine the patterns of methylation, not only to provide evidence for the mechanisms of several pathological conditions, but also to identify at-risk populations in which to conduct appropriate diet-based intervention.
Summary
The associations between one-carbon metabolism and breast cancers have been extensively studied and the associations are complex. These existing results from epidemiological studies help us to understand the contribution of one-carbon metabolism in breast cancer etiology, prognosis, treatment and prevention. The new-emerging epigenetic investigations of the breast cancer could strengthen our knowledge about whether DNA methylation status could be used as prognostic marker as well as whether one-carbon metabolism influences pathogenesis of breast cancer through epigenetic mechanisms. Since epigenetic alterations are reversible and occur early in cancer development, they are promising targets for cancer prevention, i.e., through appropriate diet-based intervention. We are expecting more results that may be translated to guidance to the general population for disease prevention as well as to clinicians for treatment and management of the disease.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (No. CA109753) and the Department of Defense of USA (No. BC031746 and training award W81XWH-06-1-0298).
REFERENCES
- Institute of Medicine. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. Washington, DC: National Academies Press; 1998. [PubMed] [Google Scholar]
- Bailey LB, Gregory JF., 3rd Polymorphisms of methylenetetrahydrofolate reductase and other enzymes: Metabolic significance, risks and impact on folate requirement. J. Nutr. 1999;129:919–922. doi: 10.1093/jn/129.5.919. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. P. Natl. Acad. Sci. USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R, Rivera I, Ravasco P, Camilo ME, Jakobs C, Blom HJ, de Almeida IT. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C-->T and 1298A-->C mutations are associated with DNA hypomethylation. J. Med. Genet. 2004;41:454–458. doi: 10.1136/jmg.2003.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabner B. Cancer Chemotherapy and Biotherapy: Principles and practice. 3rd ed. Philadelphia, PA: Lippincott-Raven Publishers; 2001. [Google Scholar]
- Chen J, Stampfer MJ, Ma J, Selhub J, Malinow MR, Hennekens CH, Hunter DJ. Influence of a methionine synthase (D919G) polymorphism on plasma homocysteine and folate levels and relation to risk of myocardial infarction. Atherosclerosis. 2001;154:667–672. doi: 10.1016/s0021-9150(00)00469-x. [DOI] [PubMed] [Google Scholar]
- Chen J, Gammon MD, Chan W, Palomeque C, Wetmur JG, Kabat GC, Teitelbaum SL, Britton JA, Terry MB, Neugut AI, Santella RM. One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res. 2005;65:1606–1614. doi: 10.1158/0008-5472.CAN-04-2630. [DOI] [PubMed] [Google Scholar]
- Cho E, Holmes M, Hankinson SE, Willett WC. Nutrients Involved in One-Carbon Metabolism and Risk of Breast Cancer among Premenopausal Women. Cancer Epidemiol Biomarkers Prev. 2007;16:2787–2790. doi: 10.1158/1055-9965.EPI-07-0683. [DOI] [PubMed] [Google Scholar]
- Christman JK, Sheikhnejad G, Dizik M, Abileah S, Wainfan E. Reversibility of changes in nucleic acid methylation and gene expression induced in rat liver by severe dietary methyl deficiency. Carcinogenesis. 1993;14:551–557. doi: 10.1093/carcin/14.4.551. [DOI] [PubMed] [Google Scholar]
- Clarke R. Prognostic and predictive factors. Disease of the Breast. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2000. pp. 489–514. [Google Scholar]
- Combs GF. The vitamins: Fundamental aspects in nutrition and health. San Diego, Calif.: Academic Press; 1992. [Google Scholar]
- Davis CD, Uthus EO. DNA Methylation, Cancer Susceptibility, and Nutrient Interactions. Experimental Biology and Medicine. 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- Demark-Wahnefried W, Peterson B, McBride C, Lipkus I, Clipp E. Current health behaviors and readiness to pursue life-style changes among men and women diagnosed with early stage prostate and breast carcinomas. Cancer. 2000;88:674–684. [PubMed] [Google Scholar]
- Ericson U, Sonestedt E, Gullberg B, Olsson H, Wirfalt E. High folate intake is associated with lower breast cancer incidence in postmenopausal women in the Malmo Diet and Cancer cohort. Am J Clin Nutr. 2007;86:434–443. doi: 10.1093/ajcn/86.2.434. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in Cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fowler BM, Giuliano AR, Piyathilake C, Nour M, Hatch K. Hypomethylation in cervical tissue: is there a correlation with folate status? Cancer Epidemiol. Biomarkers Prev. 1998;7:901–906. [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Gaughan DJ, Kluijtmans LA, Barbaux S, McMaster D, Young IS, Yarnell JW, Evans A, Whitehead AS. The methionine synthase reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. 2001;157:451–456. doi: 10.1016/s0021-9150(00)00739-5. [DOI] [PubMed] [Google Scholar]
- Goode EL, Potter JD, Bigler J, Ulrich CM. Methionine Synthase D919G Polymorphism, Folate Metabolism, and Colorectal Adenoma Risk. Cancer Epidemiol Biomarkers Prev. 2004;13:157–162. doi: 10.1158/1055-9965.epi-03-0097. [DOI] [PubMed] [Google Scholar]
- Halsted CH, Villanueva JA, Devlin AM, Chandler CJ. Metabolic Interactions of Alcohol and Folate. J. Nutr. 2002;132:2367S–2372S. doi: 10.1093/jn/132.8.2367S. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hannon-Fletcher MP, Armstrong NC, Scott JM, Pentieva K, Bradbury I, Ward M, Strain JJ, Dunn AA, Molloy AM, Kerr MA, McNulty H. Determining bioavailability of food folates in a controlled intervention study. Am. J. Clin. Nutr. 2004;80:911–918. doi: 10.1093/ajcn/80.4.911. [DOI] [PubMed] [Google Scholar]
- Harmon DL, Shields DC, Woodside JV, McMaster D, Yarnell JW, Young IS, Peng K, Shane B, Evans AE, Whitehead AS. Methionine synthase D919G polymorphism is a significant but modest determinant of circulating homocysteine concentrations. Genet. Epidemiol. 1999;17:298–309. doi: 10.1002/(SICI)1098-2272(199911)17:4<298::AID-GEPI5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin. Cancer Biol. 1999;9:359–367. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5'-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct. Funct. 1995;20:191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J. Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- Jacques PF, Bostom AG, Selhub J, Rich S, Ellison RC, Eckfeldt JH, Gravel RA, Rozen R National Heart, Lung and Blood Institute, National Institutes of Health. Effects of polymorphisms of methionine synthase and methionine synthase reductase on total plasma homocysteine in the NHLBI Family Heart Study. Atherosclerosis. 2003;166:49–55. doi: 10.1016/s0021-9150(02)00204-6. [DOI] [PubMed] [Google Scholar]
- Johnson WG, Stenroos ES, Spychala JR, Chatkupt S, Ming SX, Buyske S. New 19 bp deletion polymorphism in intron-1 of dihydrofolate reductase (DHFR): a risk factor for spina bifida acting in mothers during pregnancy? Am. J. Med. Genet. A. 2004;124:339–345. doi: 10.1002/ajmg.a.20505. [DOI] [PubMed] [Google Scholar]
- Justenhoven C, Hamann U, Pierl CB, Rabstein S, Pesch B, Harth V, Baisch C, Vollmert C, Illig T, Bruning T, Ko Y, Brauch H. One-carbon metabolism and breast cancer risk: no association of MTHFR, MTR, and TYMS polymorphisms in the GENICA study from Germany. Cancer Epidemiol. Biomarkers Prev. 2005;14:3015–3018. doi: 10.1158/1055-9965.EPI-05-0592. [DOI] [PubMed] [Google Scholar]
- Kaneda S, Takeishi K, Ayusawa D, Shimizu K, Seno T, Altman S. Role in translation of a triple tandemly repeated sequence in the 5'-untranslated region of human thymidylate synthase mRNA. Nuclear Acids Res. 1987;15:1259–1270. doi: 10.1093/nar/15.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Watanabe G, Iacopetta B. The folate pool in colorectal cancers is associated with DNA hypermethylation and with a polymorphism in methylenetetrahydrofolate reductase. Clin. Cancer Res. 2003;9:5860–5865. [PubMed] [Google Scholar]
- Lajous M, Romieu I, Sabia S, Boutron-Ruault MC, Clavel-Chapelon F. Folate, vitamin B12 and postmenopausal breast cancer in a prospective study of French women. Cancer Causes Control. 2006;17:1209–1213. doi: 10.1007/s10552-006-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson SC, Giovannucci E, Wolk A. Folate and risk of breast cancer: A meta-analysis. J. Natl. Cancer Inst. 2007;99:64–76. doi: 10.1093/jnci/djk006. [DOI] [PubMed] [Google Scholar]
- Lewis SJ, Harbord RM, Harris R, Smith GD. Meta-analyses of observational and genetic association studies of folate intakes or levels and breast cancer risk. J. Natl. Cancer Inst. 2006;98:1607–1622. doi: 10.1093/jnci/djj440. [DOI] [PubMed] [Google Scholar]
- Lissowska J, Gaudet MM, Brinton LA, Chanock SJ, Peplonska B, Welch R, Zatonski W, Szeszenia-Dabrowska N, Park S, Sherman M, Garcia-Closas M. Genetic polymorphisms in the one-carbon metabolism pathway and breast cancer risk: a population-based case-control study and meta-analyses. Int. J. Cancer. 2007;120:2696–2703. doi: 10.1002/ijc.22604. [DOI] [PubMed] [Google Scholar]
- Luebeck EG, Moolgavkar SH, Liu AY, Boynton A, Ulrich CM. Does Folic Acid Supplementation Prevent or Promote Colorectal Cancer? Results from Model-Based Predictions. Cancer Epidemiol. Biomarkers Prev. 2008;17:1360–1367. doi: 10.1158/1055-9965.EPI-07-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin LJ. Handbook of vitamins. 2nd ed., rev. and expanded. ed. New York: M. Dekker; 1991. [Google Scholar]
- Macis D, Maisonneuve P, Johansson H, Bonanni B, Botteri E, Iodice S, Santillo B, Penco S, Gucciardo G, D'Aiuto G, Rosselli, Del Turco M, Amadori M, Costa A, Decensi A. Methylenetetrahydrofolate reductase (MTHFR) and breast cancer risk: a nested-case-control study and a pooled meta-analysis. Breast Cancer Res. Treat. 2007;106:263–271. doi: 10.1007/s10549-006-9491-6. [DOI] [PubMed] [Google Scholar]
- Mandola MV, Stoehlmacher J, Zhang W, Groshen S, Yu MC, Iqbal S, Lenz HJ, Ladner RD. A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics. 2004;14:319–327. doi: 10.1097/00008571-200405000-00007. [DOI] [PubMed] [Google Scholar]
- Maring JG, Groen HJ, Wachters FM, Uges DR, de Vries EG. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5:226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- Martin DN, Boersma BJ, Howe TM, Goodman JE, Mechanic LE, Chanock SJ, Ambs S. Association of MTHFR gene polymorphisms with breast cancer survival. BMC Cancer. 2006;6:257. doi: 10.1186/1471-2407-6-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunsell E, Drolet M, Brisson J, Robert J, Deschenes L. Dietary change after breast cancer: extent, predictors, and relation with psychological distress. J. Clin. Oncol. 2002;20:1017–1025. doi: 10.1200/JCO.2002.20.4.1017. [DOI] [PubMed] [Google Scholar]
- McBride CM, Clipp E, Peterson BL, Lipkus IM, Demark-Wahnefried W. Psychological impact of diagnosis and risk reduction among cancer survivors. Psychooncology. 2000;9:418–427. doi: 10.1002/1099-1611(200009/10)9:5<418::aid-pon474>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- McEligot AJ, Largent J, Ziogas A, Peel D, Anton-Culver H. Dietary fat, fiber, vegetable, and micronutrients are associated with overall survival in postmenopausal women diagnosed with breast cancer. Nutr. Cancer. 2006;55:132–140. doi: 10.1207/s15327914nc5502_3. [DOI] [PubMed] [Google Scholar]
- Miller JW, Nadeau MR, Smith J, Smith D, Selhub J. Folate-deficiency-induced homocysteinaemia in rats: disruption of S-adenosylmethionine's co-ordinate regulation of homocysteine metabolism. Biochem. J. 1994;298(Pt 2):415–419. doi: 10.1042/bj2980415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J. Nutr. 2002;132:2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Reed MC, Anderson DF, Mattingly JC, James SJ, Ulrich CM. Long-range allosteric interactions between the folate and methionine cycles stabilize DNA methylation reaction rate. 2006;1:81–87. doi: 10.4161/epi.1.2.2677. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Reed M, Lam S, Shane B, Gregory J, Ulrich C. In silico experimentation with a model of hepatic mitochondrial folate metabolism. 2006;3:40. doi: 10.1186/1742-4682-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout HF, Reed MC, Budu P, Ulrich CM. A Mathematical Model of the Folate Cycle: NEW INSIGHTS INTO FOLATE HOMEOSTASIS. J. Biol. Chem. 2004;279:55008–55016. doi: 10.1074/jbc.M410818200. [DOI] [PubMed] [Google Scholar]
- Paz MF, Avila S, Fraga MF, Pollan M, Capella G, Peinado MA, Sanchez-Cespedes M, Herman JG, Esteller M. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res. 2002;62:4519–4524. [PubMed] [Google Scholar]
- Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong YP, Ingles SA, Sherrod A, Warren R, Tsao-Wei D, Groshen S, Lenz HJ. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am. J. Clin. Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- Reed MC, Nijhout HF, Neuhouser ML, Gregory JF, III, Shane B, James SJ, Boynton A, Ulrich CM. A Mathematical Model Gives Insights into Nutritional and Genetic Aspects of Folate-Mediated One-Carbon Metabolism. J. Nutr. 2006;136:2653–2661. doi: 10.1093/jn/136.10.2653. [DOI] [PubMed] [Google Scholar]
- Rohan TE, Jain MG, Howe GR, Miller AB. Dietary folate consumption and breast cancer risk. J. Natl. Cancer Inst. 2000;92:266–269. doi: 10.1093/jnci/92.3.266. [DOI] [PubMed] [Google Scholar]
- Rossi E, Hung J, Beilby JP, Knuiman MW, Divitini ML, Bartholomew H. Folate levels and cancer morbidity and mortality: prospective cohort study from Busselton, Western Australia. Ann. Epidemiol. 2006;16:206–212. doi: 10.1016/j.annepidem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Russo J, Yang X, Hu YF, Bove BA, Huang Y, Silva ID, Tahin Q, Wu Y, Higgy N, Zekri A, Russo IH. Biological and molecular basis of human breast cancer. Front. Biosci. 1998;3:D944–D960. doi: 10.2741/a335. [DOI] [PubMed] [Google Scholar]
- Salminen EK. Does breast cancer change patients' dietary habits? Eur. J. Clin. Nutr. 2000;54:844–848. doi: 10.1038/sj.ejcn.1601103. [DOI] [PubMed] [Google Scholar]
- Sauberlich HE, Kretsch MJ, Skala JH, Johnson HL, Taylor PC. Folate requirement and metabolism in nonpregnant women. Am. J. Clin. Nutr. 1987;46:1016–1028. doi: 10.1093/ajcn/46.6.1016. [DOI] [PubMed] [Google Scholar]
- Sellers TA, Kushi LH, Cerhan JR, Vierkant RA, Gapstur SM, Vachon CM, Olson JE, Therneau TM, Folsom AR. Dietary folate intake, alcohol, and risk of breast cancer in a prospective study of postmenopausal women. Epidemiology. 2001;12:420–428. doi: 10.1097/00001648-200107000-00012. [DOI] [PubMed] [Google Scholar]
- Sellers TA, Alberts SR, Vierkant RA, Grabrick DM, Cerhan JR, Vachon CM, Olson JE, Kushi LH, Potter JD. High-folate diets and breast cancer survival in a prospective cohort study. Nutr. Cancer. 2002;44:139–144. doi: 10.1207/S15327914NC4402_03. [DOI] [PubMed] [Google Scholar]
- Shrubsole MJ, Shu XO, Ruan ZX, Cai Q, Cai H, Niu Q, Gao YT, Zheng W. MTHFR genotypes and breast cancer survival after surgery and chemotherapy: a report from the Shanghai Breast Cancer Study. Breast Cancer Res. Treat. 2005;91:73–79. doi: 10.1007/s10549-004-7265-6. [DOI] [PubMed] [Google Scholar]
- Shrubsole MJ, Gao Y, Ca iQ, Shu XO, Dai Q, Jin F, Zheng W. MTR and MTRR Polymorphisms, Dietary Intake, and Breast Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2006;15:586–588. doi: 10.1158/1055-9965.EPI-05-0576. [DOI] [PubMed] [Google Scholar]
- Sohn K, Croxford R, Yates Z, Lucock M, Kim Y. Effect of the Methylenetetrahydrofolate Reductase C677T Polymorphism on Chemosensitivity of Colon and Breast Cancer Cells to 5-Fluorouracil and Methotrexate. J. Natl. Cancer Inst. 2004;96:134–144. doi: 10.1093/jnci/djh015. [DOI] [PubMed] [Google Scholar]
- Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol. Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- Stevens VL, McCullough ML, Pavluck AL, Talbot JT, Feigelson HS, Thun MJ, Calle EE. Association of Polymorphisms in One-Carbon Metabolism Genes and Postmenopausal Breast Cancer Incidence. Cancer Epidemiol Biomarkers Prev. 2007;16:1140–1147. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- Stevenson JP, Redlinger M, Kluijtmans LA, Sun W, Algazy K, Giantonio B, Haller DG, Hardy C, Whitehead AS, O'Dwyer PJ. Phase I clinical and pharmacogenetic trial of irinotecan and raltitrexed administered every 21 days to patients with cancer. J. Clin. Oncol. 2001;19:4081–4087. doi: 10.1200/JCO.2001.19.20.4081. [DOI] [PubMed] [Google Scholar]
- Stolzenberg-Solomon RZ, Chang SC, Leitzmann MF, Johnson KA, Johnson C, Buys SS, Hoover RN, Ziegler RG. Folate intake, alcohol use, and postmenopausal breast cancer risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am. J. Clin. Nutr. 2006;83:895–904. doi: 10.1093/ajcn/83.4.895. [DOI] [PubMed] [Google Scholar]
- Toffoli G, Veronesi A, Boiocchi M, Crivellari D. MTHFR gene polymorphism and severe toxicity during adjuvant treatment of early breast cancer with cyclophosphamide, methotrexate, and fluorouracil (CMF) Ann. Oncol. 2000;11:373–374. doi: 10.1023/a:1008337900349. [DOI] [PubMed] [Google Scholar]
- Ulrich CM, Curtin K, Samowitz W, Bigler J, Potter JD, Caan B, Slattery ML. MTHFR Variants Reduce the Risk of G:C->A:T Transition Mutations within the p53 Tumor Suppressor Gene in Colon Tumors. J. Nutr. 2005;135:2462–2467. doi: 10.1093/jn/135.10.2462. [DOI] [PubMed] [Google Scholar]
- Ulrich CM, Bigler J, Velicer CM, Greene EA, Farin FM, Potter JD. Searching Expressed Sequence Tag Databases: Discovery and Confirmation of a Common Polymorphism in the Thymidylate Synthase Gene. Cancer Epidemiol Biomarkers Prev. 2000;9:1381–1385. [PubMed] [Google Scholar]
- Ulrich CM, Nijhout HF, Reed MC. Mathematical Modeling: Epidemiology Meets Systems Biology. Cancer Epidemiol Biomarkers Prev. 2006;15:827–829. doi: 10.1158/1055-9965.EPI-06-0252. [DOI] [PubMed] [Google Scholar]
- Ulrich CM. Folate and cancer prevention: a closer look at a complex picture. Am J Clin Nutr. 2007;86:271–273. doi: 10.1093/ajcn/86.2.271. [DOI] [PubMed] [Google Scholar]
- Ulvik A, Ueland PM, Fredriksen A, Meyer K, Vollset SE, Hoff G, Schneede J. Functional inference of the methylenetetrahydrofolate reductase 677C > T and 1298A > C polymorphisms from a large-scale epidemiological study. Hum. Genet. 2007;121:57–64. doi: 10.1007/s00439-006-0290-2. [DOI] [PubMed] [Google Scholar]
- van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruine AP, Goldbohm RA, van den Brandt PA, Baylin SB, de Goeij AF, Herman JG. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–3137. [PubMed] [Google Scholar]
- Velicer CM, Ulrich CM. Vitamin and mineral supplement use among US adults after cancer diagnosis: a systematic review. J. Clin. Oncol. 2008;26:665–673. doi: 10.1200/JCO.2007.13.5905. [DOI] [PubMed] [Google Scholar]
- Weisberg IS, Jacques PF, Selhub J, Bostom AG, Chen Z, Curtis ER, Eckfeldt JH, Rozen R. The 1298A-->C polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis. 2001;156:409–415. doi: 10.1016/s0021-9150(00)00671-7. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- Wilson A, Platt R, Wu Q, Leclerc D, Christensen B, Yang H, Gravel RA, Rozen R. A common variant in methionine synthase reductase combined with low cobalamin (vitamin B12) increases risk for spina bifida. Mol. Genet. Metab. 1999;67:317–323. doi: 10.1006/mgme.1999.2879. [DOI] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Wetmur JG, Rao M, Gaudet MM, Teitelbaum SL, Britton JA, Neugut AI, Santella RM, Chen J. A functional 19-base pair deletion polymorphism of dihydrofolate reductase (DHFR) and risk of breast cancer in multivitamin users. Am. J. Clin. Nutr. 2007a;85:1098–1102. doi: 10.1093/ajcn/85.4.1098. [DOI] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Zhang H, Wetmur JG, Rao M, Teitelbaum SL, Britton JA, Neugut AI, Santella RM, Chen J. Polymorphisms of One-carbon Metabolizing Genes and Risk of Breast Cancer in a Population-based Study. Carcinogenesis. 2007b;28:1504–1509. doi: 10.1093/carcin/bgm061. [DOI] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Zeisel SH, Lee YL, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. Choline metabolism and risk of breast cancer in a population-based study. FASEB J. 2008a;22:2045–2052. doi: 10.1096/fj.07-101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Wetmur JG, Bradshaw PT, Teitelbaum SL, Neugut AI, Santella RM, Chen J. B-Vitamin Intake, One-Carbon Metabolism, and Survival in a Population-Based Study of Women with Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2008b;17:2109–2116. doi: 10.1158/1055-9965.EPI-07-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RC. Mechanisms to improve chemotherapy effectiveness. Cancer. 1990;65:815–822. doi: 10.1002/1097-0142(19900201)65:3+<815::aid-cncr2820651329>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hunter DJ, Hankinson SE, Giovannucci EL, Rosner BA, Colditz GA, Speizer FE, Willett WC. A Prospective Study of Folate Intake and the Risk of Breast Cancer. JAMA. 1999;281:1632–1637. doi: 10.1001/jama.281.17.1632. [DOI] [PubMed] [Google Scholar]
- Zintzaras E. Methylenetetrahydrofolate reductase gene and susceptibility to breast cancer: a meta-analysis. Clin. Genet. 2006;69:327–336. doi: 10.1111/j.1399-0004.2006.00605.x. [DOI] [PubMed] [Google Scholar]