Abstract
X chromosome inactivation (XCI) is the mammalian mechanism that compensates for the difference in gene dosage between XX females and XY males. Genetic and epigenetic regulatory mechanisms induce transcriptional silencing of one X chromosome in female cells. In mouse embryos, XCI is initiated at the preimplantation stage following early whole-genome activation. It is widely thought that human embryos do not employ XCI prior to implantation. Here, we show that female preimplantation embryos have a progressive accumulation of XIST RNA on one of the two X chromosomes, starting around the 8-cell stage. XIST RNA accumulates at the morula and blastocyst stages and is associated with transcriptional silencing of the XIST-coated chromosomal region. These findings indicate that XCI is initiated in female human preimplantation-stage embryos and suggest that preimplantation dosage compensation is evolutionarily conserved in placental mammals.
Introduction
Mammalian XX females equalize gene dosage relative to XY males by the inactivation of one of their X chromosomes in each cell. The X chromosome inactivation (XCI) center contains several genetic elements essential for the transcription initiation of long noncoding RNAs that are involved in XCI. Initiation of XCI requires the cis accumulation of a nontranslated mouse Xist RNA or human XIST RNA (Xist/XIST RNA) (MIM 314670) that coats the X chromosome.1–3 This is followed by various epigenetic changes on the future inactive X (Xi) chromosome that contribute to chromosome silencing.4 In somatic cells, the Xi chromosome is visible as a region of dense chromatin called the Barr body.5
There are two different forms of XCI: random XCI and imprinted XCI. Random XCI of either the maternal or the paternal X chromosome takes place in all somatic cell lineages of eutherian mammals, starting around gastrulation. Random XCI has no specific preference for inactivation of one of the parental X chromosomes.6–8 In contrast, imprinted XCI results in preferential inactivation of the paternal X chromosome and occurs in female marsupials and mouse placental tissues.9–11 Although expression of Xist RNA and a preferential expression of Xist from the paternal allele has long been observed in preimplantation mouse embryos, the prevailing view has been that actual inactivation of the X chromosome, and thus dosage compensation, begins only after differentiation of the placental precursor cells.12–14 In recent years, however, it has become apparent that XCI of the paternal X chromosome is already present from the 4-cell stage onward in all cells of preimplantation mouse embryos.15–17 Imprinted XCI in the mouse persists until the blastocyst stage and continues in the trophectoderm and the primitive endoderm.10,11,18 However, the inactive paternal X chromosome is reactivated in the inner cell mass (ICM) that forms the embryo proper15–17 and is followed by random XCI in somatic cell lineages.5,16,19 It is still unclear exactly how the earlier imprinted XCI in cleavage-stage embryos, trophectoderm cells, and primitive endoderm cells is programmed by the parental germline. Evidence exists for a mark on the maternal X chromosome that allows it to remain active.20,21 On the other hand, there is also evidence for a preference of Xist-mediated inactivation of the paternal X chromosome.12,15,22 However, these two mechanisms need not be mutually exclusive.
Data regarding the mechanism of human XCI are not easy to obtain, because of restrictions on the use of human embryos and the generation of human embryonic stem (ES) cell lines. Only a minority of human ES cell lines have two active X chromosomes in their undifferentiated state and will start a process of random XCI upon differentiation, similar to mouse ICM cells and ES cell lines.16,23–25 The majority of the undifferentiated human ES cell lines so far examined have already inactivated one X chromosome, evident from the single XIST cloud in 20%–70% of the cells and the accumulation of specific chromatin modifications.24–27 One study showed that differentiation of a human ES cell line resulted in either random XCI or preferential inactivation of a single allele, depending on the differentiated cell type. Only trophoblast cells showed a preferential inactivation of a single X chromosome, similar to mouse trophoblast tissue.23 Although the parental origin of the X chromosome could not be identified in this ES cell line, which was generated from anonymously donated embryos, it does suggest that a form of preferential XCI, such as imprinted XCI, may exist during the first stages of human trophoblast development, similar to mouse extra-embryonic tissues.10,11,18 Studies of human placental tissue have shown a variety of patterns of XCI. Some reports describe preferential expression from the maternal X chromosome, similar to mice, suggesting conservation of imprinted XCI.28–31 However, other papers report a random XCI or an XCI moderately skewed in favor of an inactive paternal X chromosome.32–36 If imprinted XCI occurs in humans, it is possible that human trophoblasts gradually lose their imprint and perform random XCI at later stages, as has been demonstrated in vitro.37,38 This may result in an XCI pattern skewed toward an inactivated paternal X chromosome, which would explain the mixed results observed in the analysis of placentas.28–36
Defects in dosage compensation prior to implantation of the embryo lead to abnormal development in a majority of the embryos and early lethality, as demonstrated by analysis of parthenogenetic mouse embryos that have two maternal genomes.13,39 Similarly, female mutant embryos that inherit a paternal X chromosome with a deletion of the Xist gene are not able to inactivate this X chromosome, resulting in two active X chromosomes and early lethality.40,41
Previous studies using PCR analysis of human preimplantation embryos detected XIST expression both in female and in male embryos.42,43 Because female-specific XIST expression was not detected, it was concluded that XIST RNA was not functional at this stage of development and that dosage compensation was not initiated in human preimplantation embryos.42,43 However, in mice, a brief “pinpoint” expression of Xist from both the paternal and the maternal X chromosome was later reported in male and female preimplantation embryos.17,19,44,45 The initial Xist expression on the maternal X chromosome subsequently disappears while the Xist expression from the paternal X chromosome accumulates in female preimplantation embryos to coat the future Xi chromosome.17,44–46 Similar pinpoint signals are also observed during the onset of random XCI in male and female ES cells.17,19,46 Thus, the XIST expression previously reported in male human embryos could be attributable to a brief window of expression of XIST from the paternal X chromosome and does not exclude XCI in female embryos.
We therefore decided to reinvestigate XCI in human preimplantation embryos at the single-cell level to analyze XIST RNA localization and the transcriptional and epigenetic features of XCI.
Material and Methods
Collection of Surplus Embryos and Cryopreserved Embryos Donated for Research
Ovarian stimulation, oocyte retrieval, and in vitro fertilization (IVF) and/or intracytoplasmic sperm injection (ICSI) procedures were performed as described previously.47 This study was approved by the Dutch Central Committee on Research Involving Human Subjects (CCMO, NL11448) and the local ethics review committee of the Erasmus Medical Center Hospital (MEC 2007-130). Written consent was obtained from the couples for confirmation that the surplus or cryopreserved embryos could be used for research purposes.
RNA FISH, DNA FISH, and Immunofluorescence
Immunofluorescence followed by RNA and/or DNA fluorescence in situ hybridization (FISH) was performed as described previously,17 with some modifications. The zona pellucida of fresh or thawed cryopreserved embryos was removed with 0.05% pronase (Sigma Aldrich) in calcium- and magnesium-free medium (G-PGD, Vitrolife, Kungsbacka, Sweden). Embryos of the 8-cell and morula stage were incubated in calcium- and magnesium-free medium so that single cells could be obtained. Single cells and blastocysts were washed in phosphate-buffered saline (PBS) and fixed on slides in 1% paraformaldehyde (Sigma Aldrich) containing 0.5% Triton X-100 for 0.5–1 hr. Slides were washed with PBS and stored in 70% ethanol at −20°C. Cumulus cells and amniocytes were fixed similarly. Primary antibodies used for immunofluorescence were H3K27Me3 (Abcam, 1:50), macroH2A.1 (Upstate, 1:100), and H3K9ac (Upstate, 1:100). Secondary antibodies (Invitrogen), used at 1:250 dilutions, were as follows: goat anti-rabbit Alexa 594, goat anti-mouse Alexa 488, and goat anti-mouse Pacific Blue.
For the detection of XIST RNA, a 16.4 kb plasmid covering the complete RNA sequence of the XIST gene48 was used on nondenatured cells. Cot1 RNA detection was performed with the use of labeled Cot1 DNA (Invitrogen) as a probe. For the detection of chromosomes X, Y, and 15, the following DNA probes were used: X centromere (pBAMX5), Y chromosome heterochromatin (RPN1305X), and chromosome 15 satellite III region f (D15Z1). The Y probe occasionally produces a diffuse signal because of the highly polymorphic heterochromatin region.49 Overlapping CHIC1 fosmid clones (G248P389032C3 and G248P86549C3) were located with the UCSC genome browser (UC Santa Cruz) and obtained from BACPAC Resources (Oakland). RNA and DNA FISH probes were labeled by nick translation with fluorochromes Alexa 594 and 555 (Molecular Probes, Invitrogen, Leiden, The Netherlands), diethylaminocoumarin-5-UTP (NEN Life Science Products, Boston, MA, USA), or Bio-16-dUTP (Roche). Probes were validated in cultured lymphocytes of a normal XY male. A significant distance between the signals of the X centromere probe and XIST RNA was often observed, because XIST is located 80 Mb distal to the centromere. XIST expression was detected as a pinpoint signal, a small cloud, or a full cloud. The distinction between the three was usually easy to detect. A pinpoint signal was equivalent in size and intensity to a locus-specific DNA FISH signal, a small cloud was 10–20 times the size of a pinpoint signal, and a full cloud was 100 or more times the size of a pinpoint signal.
Slides were examined with a Zeiss Axioplan 2 epifluorescence microscope equipped with appropriate filters (Chroma, Rockingham, VT, USA). Images were captured with the ISIS FISH Imaging System (MetaSystems, Altlussheim, Germany), and background correction was applied via Adobe Photoshop CS2 when necessary. For each embryo, the positions of all nuclei were mapped in detail, which allowed an accurate analysis of each nucleus. Chromosomally chaotic embryos (ascertained on the basis of chromosome X, Y, and 15 analysis) were excluded from the analysis, and mosaic embryos were included only if less than 50% of the cells were aneuploid for X and Y.
Results
Donated cryopreserved and surplus embryos from in vitro fertilization (IVF) treatments were dissociated and fixed at the 8-cell, morula, and blastocyst stages. RNA FISH with a human XIST probe was performed for the detection of the Xi chromosome in single cells, followed by DNA FISH with chromosome X-, Y-, and 15-specific probes for the identification of the sex and diploid status of the embryos (see Figure S1, available online).
XIST Expression in Early Human Embryos
Only a fraction of the blastomeres from male embryos showed XIST RNA signals. These signals were small “pinpoint”-like signals reminiscent of the expression of unstable XIST RNA.44,46 This form of XIST expression was observed mostly at the morula stage, and the pinpoints never accumulated to a “cloud”-like signal in blastocysts, indicating the absence of XCI in male preimplantation embryos (Figures 1A–1C and Table 1). In contrast, female embryos showed a different XIST-staining pattern: the majority of cells had pinpoint signals for XIST RNA at the 8-cell stage. The XIST signal gradually accumulated to a full cloud on one of the X chromosomes at the late morula and blastocyst stages in female embryos (Figures 1D–1F). Distinct patterns of XIST expression were observed for different developmental stages (Table 1): In 8-cell-stage embryos, 65% of the blastomeres displayed one pinpoint signal of XIST, 4% of the blastomeres had a small cloud of XIST, and 19% showed two pinpoints of XIST RNA. The remaining 12% of blastomeres had no expression of XIST. In morulas, XIST expression had progressively accumulated, resulting in 49% of the cells displaying a single XIST cloud, suggesting that XCI was initiated in these cells, and 19% of the cells had a pinpoint of XIST RNA. The percentage of cells with two pinpoints was reduced from 19% at the 8-cell stage to 2% in morulas. The rest of the blastomeres had no XIST, two small clouds of XIST, or a pinpoint together with a cloud signal. Different cells of the same embryo regularly showed variable levels of XIST and distinct XIST expression patterns (Figure S2), similar to previous observations in mouse embryos.13,44,50,51
Figure 1.
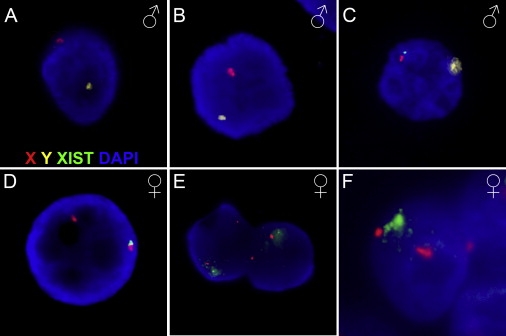
XIST Expression in Male and Female Human Preimplantation Embryos
RNA and DNA FISH staining with probes detecting XIST RNA (green), the X (red) and Y (yellow) chromosome, and DAPI counterstain. Human male embryos (A–C) do not generally show XIST signals at the 8-cell stage (A) or at the morula stage (B). A minority of male cells at the morula stage show a pinpoint of XIST staining (C). Female embryos (D–F) show an XIST pinpoint in the majority of embryos at the 8-cell stage (D). Two cells at the morula stage each show a beginning cloud of XIST on one of their two X chromosomes (E). At the blastocyst stage, this has further accumulated to a full cloud on one of the two X chromosomes (F). A third diffuse red signal is an X chromosome from an adjacent cell that is in a different focal plane.
Table 1.
XIST Patterns in Human Embryos at Different Developmental Stages
| Embryo Stage | No. of ♀ Embryos (No. of Cells) |
XIST Pattern in ♀ Embryos |
No. of ♂ Embryos (No. of Cells) |
XIST Pattern in ♂ Embryos |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8-cell | 5 (26) | 12 | 65 | 19 | - | 4 | - | 6 (30) | 94 | 6 |
| Morula | 13 (89) | 19 | 19 | 2 | 6 | 49 | 4 | 9 (91) | 86 | 14 |
| Blastocyst | 6 (>100) | 5 | - | - | - | 90 | 5 | 5 (98) | 93 | 7 |
Percentages of analyzable cells that have the indicated pattern of XIST signals. Signals in blastomeres were scored as negative (−), pinpoint signal (•), small cloud (•), or full cloud (•).
Blastocysts did not disaggregate well during the fixation procedure and were therefore examined as intact embryos. Nuclei that could be visualized showed a full XIST cloud in 90% of the cells (Figure 1 and Table 1). The remainder of cells either had no XIST signal or had two small cloud signals. In summary, whereas female embryos at the 8-cell stage showed a clear single pinpoint or small cloud of XIST that accumulated to a proper XIST cloud at the late morula and blastocyst stage, male embryos showed only brief expression of XIST in a minority of the cells. Thus, a clear difference in the timing, duration, and level of XIST expression between male and female human embryos suggests the occurrence of XCI in female preimplantation embryos.
Absence of Transcriptional Activity on the XIST-Coated Chromosomal Region
We analyzed the transcriptional activity together with XIST staining, because XIST RNA accumulation on the X chromosome itself does not automatically imply an inactivated status of the X chromosome. Transcriptional activity was investigated by Cot1 RNA FISH staining, which highlights areas of ongoing hnRNA transcription; trancriptionally silent nuclear compartments, such as an Xi chromosome, are devoid of Cot1 RNA.15,17 Human embryos were stained by RNA and DNA FISH with probes for Cot1 RNA, XIST RNA, chromosome 15, and the X and Y chromosomes. Cumulus cells of human follicle complexes were used as positive controls in each experiment (Figures S3A–S3C). Cells of human blastocysts showed XIST RNA clouds corresponding to areas where Cot1 RNA (Figures 2A–2F) was excluded. Figures 2D–2F show a representative cell with an XIST-coated X in a Cot1-depleted region and the second X chromosome in a Cot1-positive area. The Cot1 exclusion coincided with the XIST signal in 89% of the cells (n = 47). Comparable percentages of Cot1 exclusion from Xist/XIST-positive areas have been observed in mouse embryos and differentiating human ES cells.15,22,24 The initiation of transcriptional silencing was confirmed with RNA FISH detection of nascent transcripts of the X-linked CHIC1 gene. Blastocyst cells showed a single spot of CHIC1 expression close to one X centromere and an XIST cloud on the other Xi chromosome (Figures 2G–2J). Biallelic expression was not observed in blastocysts and was not assayed in earlier stages. Thus, the X chromosome that is at least partially coated with XIST RNA in the human embryo is in a transcriptionally silent area, demonstrating that XCI and dosage compensation commences in preimplantation female human embryos.
Figure 2.
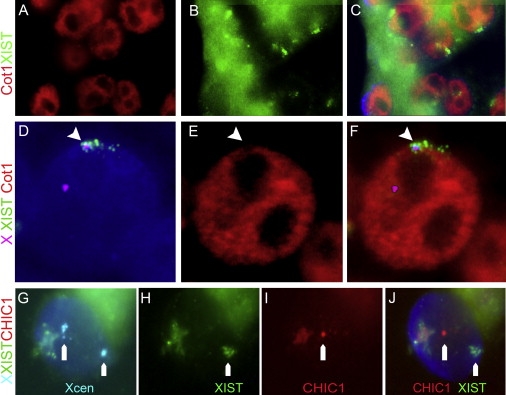
Transcriptional Changes on the Inactive X Chromosome
(A–F) Cot1 exclusion around XIST. (A–C) Cells of a female blastocyst embryo, stained for Cot1 RNA (red in A) and XIST RNA (green in B) showing depleted regions of Cot-1 RNA around the XIST signals indicating the position of the Xi chromosome (merged in C). (D–F) Representative cell of a female blastocyst with staining for the X centromeres and XIST RNA (D; Xcen in magenta, XIST in green) together with Cot1 (red in E). Transcription of Cot1 RNA was absent in a region that overlaps with XIST RNA staining (F), whereas the active X without XIST staining overlaps with a Cot1-positive region.
(G–J) Female blastocyst cell with two X centromeres (cyan in G) has a single XIST cloud on one X chromosome (green in H) and monoallelic expression of CHIC1 on the other X chromosome (red in I, merged in J). A dust spot is visible in all colors and is therefore nonspecific staining.
Chromatin Conformational Changes on the Inactive X Chromosome
To further explore XCI in human preimplantation embryos, we investigated histone modifications that are established hallmarks of a silenced X chromosome. Specific accumulation of one or more histone modifications forms macrochromatin bodies that indicate the position of the Xi chromosome, as shown for XCI in mouse preimplantation embryos and in differentiating mouse and human ES cells.1,4,16,17,25,52–54
Identification of the chromatin state of X chromosomes was carried out with antibodies that detect hypoacetylation of lysine 9 on histone H3 (H3K9ac), accumulation of trimethylation of lysine 27 on H3 (H3K27Me3), and the enrichment of the histone variant macroH2A,1,4,16,17,52,53 followed by DNA FISH for the gender identification of the embryo. As a control, we used cumulus cells in which the Xi chromosome can be identified as a chromatin-dense Barr body or amniocytes in which immunostaining together with XIST detection is possible. Human cumulus cells showed DAPI-dense Barr bodies that were positively stained for macroH2A and H3K27Me3, marks associated with inactive chromatin (Figures S3D–S3G). Amniocytes showed the same accumulation of chromatin markers overlaying the XIST signal (Figures S3H–S3K). In addition, staining with an antibody against H3K9 acetylation, an active chromatin mark, showed exclusion from Barr bodies in cumulus cells or from XIST-stained regions in amniocytes (Figures S3L–S3S). These findings confirm the specificity of the antibodies used to indicate a macrochromatin body as the Xi. Staining of cells from preimplantation blastocysts with the use of the same antibodies showed identical results; i.e., double staining showed a single region where accumulated H3K27Me3 formed an exact overlay with the region of H3K9 hypoacetylation (Figures 3A–3C), indicating the presence of an Xi. Furthermore, macroH2A enrichment and accumulation of H3K27Me3 (Figures 3D–3F) colocalized exactly in blastocyst cells. Up to 30% of the analyzable cells in blastocysts showed a double immunostaining of chromatin marks that are specific for XCI (either H3K27Me3 together with macroH2A or H3K27Me3 in a depleted region of H3K9ac). The other 70% of the cells had no visible accumulation (or depletion, in the case of H3K9ac) of either antibody, and single accumulations were rarely found (<5%). In contrast, male embryos did not show accumulation of H3K27Me3 or macroH2a, and no specific exclusion of H3K9ac was observed (data not shown).
Figure 3.
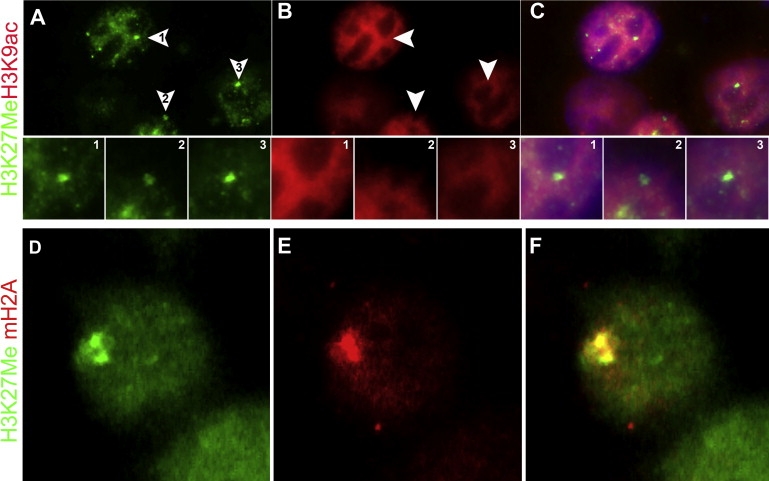
Epigenetic Changes on the Inactive X Chromosome
(A–C) Three adjacent blastocyst cells show a single nuclear domain with H3K27Me3 hypermethylation (arrowheads in J and enlarged panels 1–3), and staining for H3K9 acetylation (B, 1–3) shows an H3K9ac-depleted region overlaying the H3K27Me3 accumulation (C, 1–3), indicating the position of the Xi chromosome.
(D–F) Representative blastocyst cell shows a single nuclear domain with H3K27 hypermethylation (green, D) and enrichment for macroH2A (red, E) with a clear overlap (yellow, F), analogous to the signal around an Xi chromosome.
Taken altogether, these observations show that once XIST RNA coats the X chromosome in human embryos, epigenetic changes that are known to lead to XCI are induced, similar to that which has been observed in mouse preimplantation embryos and ES cells.4,55,56
Discussion
In contrast to previous suggestions that XCI may not be present in the human preimplantation embryo,4,42,43,55,57 our observations of XIST RNA accumulation in female embryos, local accumulation of Xi-specific chromatin modifications, and the absence of active transcription in XIST RNA-coated areas indicate that XCI occurs in human female preimplantation embryos, starting from the 8-cell stage. However, the extent of the coating by XIST and the extent of transcriptional silencing other than that of the CHIC1 gene remain unknown.
Conservation of Timing of X Chromosome Inactivation
The developmental stage at which embryonic genome activation occurs in mammalian species varies considerably: at the 1- to 2-cell stage in mice, at the 4- to 8-cell stage in cows and humans, and at the 8- to 16-cell stage in sheep and rabbits.58,59 The necessity of dosage compensation by XCI is likely to start at the onset of genome activation; thus, around the 2-cell stage in mice and around the 8-cell stage in humans. Initiation of XCI can be detected in mice from the 2-cell stage onward by Xist RNA FISH, showing a small pinpoint of Xist in 67% of female cells, and complete X-associated Xist accumulation occurs by the 8-cell stage.15,17 In human embryos, we observed pinpoint XIST signals in 65% of female 8-cell blastomeres and cis accumulation (cloud-like signals) of XIST in late morulas and in blastocysts (Figure 1 and Table 1). We could not analyze embryos before the 6- to 8-cell stage because such embryos were not available for research. Given that our observations in 6- to 8-cell-stage human embryos show XIST RNA with exactly the same pinpoint signals and at a similar frequency as those in 2-cell-stage mouse embryos (67% versus 65%; Table 1 and 15), it is likely that the 8-cell stage represents the actual onset of XIST expression in humans. The later stage of XCI in humans relative to mice suggests a correlation with the later transition from maternal to embryonic or zygotic gene expression in humans.60
XIST patterns were analyzed only in euploid cells, and because similar results were observed in cells derived form normal or mosaic embryos (Table S1), these data were combined. No consistent pattern of XIST expression was discernible in the sex aneuploid cells (Table S1). The quality of the embryos did not influence the results; we observed similar patterns of XCI in euploid cells of surplus embryos and cryopreserved embryos. It is likely that aneuploidies other than those detected with the chromosome X, Y, or 15 probes were present in the analyzed cells, because human embryos are known to have high aneuploidy rates.61 However, aneuploidies may have only a subtle effect on the skewing of the selected X chromosome.62
Comparison of several characteristics of XCI between mouse and human embryos indicates that although the onset of XIST expression may occur later in humans than in mice, the timeline and order of XIST expression initiation through actual XCI is similar in mice and men (Figure 2 and 63).
Detection of XIST in Male Embryos
In contrast to previous reports on the detection of XIST in human embryos,42,43 we found a clear difference in XIST expression between male and female embryos (Figure 1). The previous data on human male embryos were obtained by nested PCR, which, in contrast to our FISH method, detects very low levels of XIST RNA. The large number of cycles in this method probably masked the difference in XIST levels between male and female embryos, leading to the erroneous conclusion that human embryos do not initiate XCI.42,43 In general, large differences in levels of Xist/XIST expression are found between embryos and between cells of a single embryo in both mice17,50 and humans (Table 1 and Figure S2). PCR detection of RNA expression in pooled embryos or even single embryos can mask these variations,50 and interpretations of these results are therefore unreliable. Thus, although PCR is more sensitive than the FISH method, the ability to localize the transcripts, as in a FISH experiment, is essential for studying the X-inactivation process.
The onset of Xist/XIST expression in male embryos, detected as a pinpoint signal by Xist/XIST RNA FISH, is slightly later than that in female embryos, both in mice and in humans (17,44,45, and Table 1). Although our XIST RNA FISH probe did not overlap with the reported transcribed TSIX sequence (MIM 300181),64 which is transcribed in the opposite orientation and may be involved in downregulation of XIST, we cannot exclude that the transient male and early female pinpoint signals are (in part) TSIX RNA. We therefore tested TSIX probes that had been used previously to detect TSIX in human fetal cells,64 but we failed to obtain specific signals on either embryos or control fetal amniocytes from routine amniocentesis. Single-strand XIST and TSIX RNA probes produced too much background. Thus, whether human TSIX plays a role in the initiation of XCI in humans remains to be investigated.
Similar to those in mouse embryos, the pinpoint signals in human male embryos never accumulated to a complete cloud, indicating that initial XIST and/or TSIX expression does not lead to actual inactivation of the X chromosome in male embryos. (17,44,45, and Table 1).
What Is the Mechanism of X Chromosome Inactivation?
Our data on human embryos show a variety of XIST-staining patterns that are similar to mouse Xist embryo patterns (13,15,17,19,44, Table 1, and Figure S1). Slight discrepancies between published data can be explained by differences in detection sensitivity, because double Xist signals were observed more frequently when larger probes and increased signal amplification were used.13,15,44
Both random XCI and imprinted XCI can result in patterns of XCI that are comparable to the patterns that we have observed in human embryos: Of the different models for random XCI,8 the stochastic model—in which every X chromosome has a certain probability of being inactivated, resulting in a majority of cells with one Xi, as well as cells with no Xi or two Xis—7 best explains our results. Only this model accommodates the variety of XIST expression patterns such as we have observed; other random XCI models view this variation as errors of the XCI mechanism. Imprinted XCI has a preferential expression of the paternal Xist allele but is only manifested in 70%–90% of the cells, whereas the remainder of the cells show no Xist expression. These Xist-negative cells may at a later stage inactivate the correct number of X chromosomes, akin to random XCI.65 The human data could thus be explained by both imprinted and random mechanisms of XCI. The majority of blastomeres from our 8-cell human female embryos showed only one pinpoint XIST signal, suggesting a preference for activation of a single XIST allele that could be indicative of imprinted XCI, similar to that which is observed in mice. The obvious solution would be parental tracing of the expressed XIST gene. Unfortunately, this was not possible with these anonymously donated embryos. Future experiments are necessary in order to determine whether XCI in human embryos is a random process or whether imprinted XCI is fully conserved.
In summary, we find X-associated accumulation of XIST RNA in female cleavage-stage and blastocyst embryos, together with transcriptional silencing and Xi-specific histone modifications. These results indicate that (at least part of) the X chromosome is silenced in human preimplantation embryos. Our findings therefore suggest that X-linked dosage compensation in mammalian preimplantation embryos is evolutionary conserved.
Supplemental Data
Supplemental Data include three figures and one table and can be found with this article online at http://www.ajhg.org/.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
University of California, Santa Cruz (USSC) Genome Browser, http://genome.ucsc.edu/
Acknowledgments
We thank Frank Grosveld and Willy Baarends for valuable comments; Eric Steegers for financial support; Cindy Eleveld, Arletta Bol, Shimaira van Beusekom, Bert Eussen, and Erwin Brosens for technical assistance; the Erasmus MC IVF team for support; and the patients for their generous embryo donation.
References
- 1.Brockdorff N., Ashworth A., Kay G.F., Cooper P., Smith S., McCabe V.M., Norris D.P., Penny G.D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 3.Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 4.Heard E., Disteche C.M. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 5.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 6.Avner P., Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 7.Monkhorst K., Jonkers I., Rentmeester E., Grosveld F., Gribnau J. X inactivation counting and choice is a stochastic process: Evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 8.Starmer J., Magnuson T. A new model for random X chromosome inactivation. Development. 2009;136:1–10. doi: 10.1242/dev.025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graves J.A. Mammals that break the rules: Genetics of marsupials and monotremes. Annu. Rev. Genet. 1996;30:233–260. doi: 10.1146/annurev.genet.30.1.233. [DOI] [PubMed] [Google Scholar]
- 10.Takagi N., Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 11.West J.D., Frels W.I., Papaioannou V.E., Karr J.P., Chapman V.M. Development of interspecific hybrids of Mus. J. Embryol. Exp. Morphol. 1977;41:233–243. [PubMed] [Google Scholar]
- 12.Kay G.F., Barton S.C., Surani M.A., Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 13.Nesterova T.B., Barton S.C., Surani M.A., Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev. Biol. 2001;235:343–350. doi: 10.1006/dbio.2001.0295. [DOI] [PubMed] [Google Scholar]
- 14.Zuccotti M., Boiani M., Ponce R., Guizzardi S., Scandroglio R., Garagna S., Redi C.A. Mouse Xist expression begins at zygotic genome activation and is timed by a zygotic clock. Mol. Reprod. Dev. 2002;61:14–20. doi: 10.1002/mrd.1126. [DOI] [PubMed] [Google Scholar]
- 15.Huynh K.D., Lee J.T. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 16.Mak W., Nesterova T.B., de Napoles M., Appanah R., Yamanaka S., Otte A.P., Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 18.Sugawara O., Takagi N., Sasaki M. Correlation between X-chromosome inactivation and cell differentiation in female preimplantation mouse embryos. Cytogenet. Cell Genet. 1985;39:210–219. doi: 10.1159/000132137. [DOI] [PubMed] [Google Scholar]
- 19.Matsui J., Goto Y., Takagi N. Control of Xist expression for imprinted and random X chromosome inactivation in mice. Hum. Mol. Genet. 2001;10:1393–1401. doi: 10.1093/hmg/10.13.1393. [DOI] [PubMed] [Google Scholar]
- 20.Lyon M.F., Rastan S. Parental source of chromosome imprinting and its relevance for X chromosome inactivation. Differentiation. 1984;26:63–67. doi: 10.1111/j.1432-0436.1984.tb01375.x. [DOI] [PubMed] [Google Scholar]
- 21.Tada T., Obata Y., Tada M., Goto Y., Nakatsuji N., Tan S., Kono T., Takagi N. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development. 2000;127:3101–3105. doi: 10.1242/dev.127.14.3101. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto I., Arnaud D., Le Baccon P., Otte A.P., Disteche C.M., Avner P., Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- 23.Dhara S.K., Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva S.S., Rowntree R.K., Mekhoubad S., Lee J.T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall L.L., Byron M., Butler J., Becker K.A., Nelson A., Amit M., Itskovitz-Eldor J., Stein J., Stein G., Ware C. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J. Cell. Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman L.M., Hall L., Batten J.L., Young H., Pardasani D., Baetge E.E., Lawrence J., Carpenter M.K. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y., Matsuno Y., Fouse S.D., Rao N., Root S., Xu R., Pellegrini M., Riggs A.D., Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl. Acad. Sci. USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison K.B., Warburton D. Preferential X-chromosome activity in human female placental tissues. Cytogenet. Cell Genet. 1986;41:163–168. doi: 10.1159/000132221. [DOI] [PubMed] [Google Scholar]
- 29.Ropers H.H., Wolff G., Hitzeroth H.W. Preferential X inactivation in human placenta membranes: Is the paternal X inactive in early embryonic development of female mammals? Hum. Genet. 1978;43:265–273. doi: 10.1007/BF00278833. [DOI] [PubMed] [Google Scholar]
- 30.Harrison K.B. X-chromosome inactivation in the human cytotrophoblast. Cytogenet. Cell Genet. 1989;52:37–41. doi: 10.1159/000132835. [DOI] [PubMed] [Google Scholar]
- 31.Goto T., Wright E., Monk M. Paternal X-chromosome inactivation in human trophoblastic cells. Mol. Hum. Reprod. 1997;3:77–80. doi: 10.1093/molehr/3.1.77. [DOI] [PubMed] [Google Scholar]
- 32.Migeon B.R., Wolf S.F., Axelman J., Kaslow D.C., Schmidt M. Incomplete X chromosome dosage compensation in chorionic villi of human placenta. Proc. Natl. Acad. Sci. USA. 1985;82:3390–3394. doi: 10.1073/pnas.82.10.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Looijenga L.H., Gillis A.J., Verkerk A.J., van Putten W.L., Oosterhuis J.W. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am. J. Hum. Genet. 1999;64:1445–1452. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bamforth F., Machin G., Innes M. X-chromosome inactivation is mostly random in placental tissues of female monozygotic twins and triplets. Am. J. Med. Genet. 1996;61:209–215. doi: 10.1002/(SICI)1096-8628(19960122)61:3<209::AID-AJMG4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 35.Uehara S., Tamura M., Nata M., Ji G., Yaegashi N., Okamura K., Yajima A. X-chromosome inactivation in the human trophoblast of early pregnancy. J. Hum. Genet. 2000;45:119–126. doi: 10.1007/s100380050197. [DOI] [PubMed] [Google Scholar]
- 36.Zeng S.M., Yankowitz J. X-inactivation patterns in human embryonic and extra-embryonic tissues. Placenta. 2003;24:270–275. doi: 10.1053/plac.2002.0889. [DOI] [PubMed] [Google Scholar]
- 37.Migeon B.R., Schmidt M., Axelman J., Cullen C.R. Complete reactivation of X chromosomes from human chorionic villi with a switch to early DNA replication. Proc. Natl. Acad. Sci. USA. 1986;83:2182–2186. doi: 10.1073/pnas.83.7.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migeon B.R., Axelman J., Jeppesen P. Differential X reactivation in human placental cells: Implications for reversal of X inactivation. Am. J. Hum. Genet. 2005;77:355–364. doi: 10.1086/432815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao C., Takagi N. An extra maternally derived X chromosome is deleterious to early mouse development. Development. 1990;110:969–975. doi: 10.1242/dev.110.3.969. [DOI] [PubMed] [Google Scholar]
- 40.Lee J.T. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 41.Takagi N. Imprinted X-chromosome inactivation: Enlightenment from embryos in vivo. Semin. Cell Dev. Biol. 2003;14:319–329. doi: 10.1016/j.semcdb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 42.Daniels R., Zuccotti M., Kinis T., Serhal P., Monk M. XIST expression in human oocytes and preimplantation embryos. Am. J. Hum. Genet. 1997;61:33–39. doi: 10.1086/513892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray P.F., Winston R.M., Handyside A.H. XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum. Mol. Genet. 1997;6:1323–1327. doi: 10.1093/hmg/6.8.1323. [DOI] [PubMed] [Google Scholar]
- 44.Sheardown S.A., Duthie S.M., Johnston C.M., Newall A.E., Formstone E.J., Arkell R.M., Nesterova T.B., Alghisi G.C., Rastan S., Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 45.Latham K.E., Patel B., Bautista F.D., Hawes S.M. Effects of X chromosome number and parental origin on X-linked gene expression in preimplantation mouse embryos. Biol. Reprod. 2000;63:64–73. doi: 10.1095/biolreprod63.1.64. [DOI] [PubMed] [Google Scholar]
- 46.Panning B., Dausman J., Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 47.Hohmann F.P., Macklon N.S., Fauser B.C. A randomized comparison of two ovarian stimulation protocols with gonadotropin-releasing hormone (GnRH) antagonist cotreatment for in vitro fertilization commencing recombinant follicle-stimulating hormone on cycle day 2 or 5 with the standard long GnRH agonist protocol. J. Clin. Endocrinol. Metab. 2003;88:166–173. doi: 10.1210/jc.2002-020788. [DOI] [PubMed] [Google Scholar]
- 48.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 49.Repping S., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 50.Hartshorn C., Rice J.E., Wangh L.J. Differential pattern of Xist RNA accumulation in single blastomeres isolated from 8 cell stage mouse embryos following laser zona drilling. Mol. Reprod. Dev. 2003;64:41–51. doi: 10.1002/mrd.10223. [DOI] [PubMed] [Google Scholar]
- 51.Hartshorn C., Rice J.E., Wangh L.J. Developmentally-regulated changes of Xist RNA levels in single preimplantation mouse embryos, as revealed by quantitative real-time PCR. Mol. Reprod. Dev. 2002;61:425–436. doi: 10.1002/mrd.10037. [DOI] [PubMed] [Google Scholar]
- 52.Costanzi C., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 53.Chadwick B.P., Willard H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaumeil J., Okamoto I., Guggiari M., Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- 55.Chow J.C., Yen Z., Ziesche S.M., Brown C.J. Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 2005;6:69–92. doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- 56.Latham K.E. X chromosome imprinting and inactivation in preimplantation mammalian embryos. Trends Genet. 2005;21:120–127. doi: 10.1016/j.tig.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Brown C.J., Robinson W.P. XIST expression and X-chromosome inactivation in human preimplantation embryos. Am. J. Hum. Genet. 1997;61:5–8. doi: 10.1086/513914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz G.A., Heyner S. Gene expression in pre-implantation mammalian embryos. Mutat. Res. 1992;296:17–31. doi: 10.1016/0165-1110(92)90029-9. [DOI] [PubMed] [Google Scholar]
- 59.Dobson A.T., Raja R., Abeyta M.J., Taylor T., Shen S., Haqq C., Pera R.A. The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 60.Wells D., Bermudez M.G., Steuerwald N., Thornhill A.R., Walker D.L., Malter H., Delhanty J.D., Cohen J. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum. Reprod. 2005;20:1339–1348. doi: 10.1093/humrep/deh778. [DOI] [PubMed] [Google Scholar]
- 61.Baart E.B., Martini E., van den Berg I., Macklon N.S., Galjaard R.J., Fauser B.C., Van Opstal D. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum. Reprod. 2006;21:223–233. doi: 10.1093/humrep/dei291. [DOI] [PubMed] [Google Scholar]
- 62.Bretherick K., Gair J., Robinson W.P. The association of skewed X chromosome inactivation with aneuploidy in humans. Cytogenet. Genome Res. 2005;111:260–265. doi: 10.1159/000086898. [DOI] [PubMed] [Google Scholar]
- 63.Okamoto I., Heard E. The dynamics of imprinted X inactivation during preimplantation development in mice. Cytogenet. Genome Res. 2006;113:318–324. doi: 10.1159/000090848. [DOI] [PubMed] [Google Scholar]
- 64.Migeon B.R., Lee C.H., Chowdhury A.K., Carpenter H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am. J. Hum. Genet. 2002;71:286–293. doi: 10.1086/341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huynh K.D., Lee J.T. Imprinted X inactivation in eutherians: A model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 2001;13:690–697. doi: 10.1016/s0955-0674(00)00272-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


