Abstract
There are extensive data indicating that some glacial refuge zones of southern Europe (Franco-Cantabria, Balkans, and Ukraine) were major genetic sources for the human recolonization of the continent at the beginning of the Holocene. Intriguingly, there is no genetic evidence that the refuge area located in the Italian Peninsula contributed to this process. Here we show, through phylogeographic analyses of mitochondrial DNA (mtDNA) variation performed at the highest level of molecular resolution (52 entire mitochondrial genomes), that the most likely homeland for U5b3—a haplogroup present at a very low frequency across Europe—was the Italian Peninsula. In contrast to mtDNA haplogroups that expanded from other refugia, the Holocene expansion of haplogroup U5b3 toward the North was restricted by the Alps and occurred only along the Mediterranean coasts, mainly toward nearby Provence (southern France). From there, ∼7,000–9,000 years ago, a subclade of this haplogroup moved to Sardinia, possibly as a result of the obsidian trade that linked the two regions, leaving a distinctive signature in the modern people of the island. This scenario strikingly matches the age, distribution, and postulated geographic source of a Sardinian Y chromosome haplogroup (I2a2-M26), a paradigmatic case in the European context of a founder event marking both female and male lineages.
Main Text
According to the archaeological evidence, modern humans first entered Southwest Asia ∼45–50 thousand years ago (kya), and Europe soon afterwards. The first modern Europeans came from the Levant,1 but an almost concomitant arrival of related groups in European Russia from interior western Asia via the Caucasus or along the eastern coast of the Caspian Sea might have also occurred.2,3 These findings are consistent with the proposal that modern Europeans might have developed from related groups living in several regional enclaves in the same broad geographic area of Southwest Asia4 and the observation that mitochondrial DNA (mtDNA) variation in all modern European populations is completely embedded in the western Eurasian portion of the mtDNA phylogeny.5
Approximately 20 ky after the arrival of their ancestors from Southwest Asia, Europeans faced dramatic and rapid climatic changes, which peaked with the Last Glacial Maximum (LGM), centered at ∼21 kya. Major gaps in the archaeological record reveal an abandonment of North and Central Europe6 and a contraction of the human range to southern European regions that served as refugia.7,8 The deglaciation sequence began with the Bølling warming about 15 kya but stabilized only at the end of the Younger Dryas cold snap 11.6 kya.9–12 In the refugia, human genetic variation was affected by drift and founder events, but the effects were probably strongest for mtDNA and Y chromosome because of their uniparental transmission and reduced effective population size. Thus, pre-LGM mtDNA and Y chromosome haplotypes were differently preserved (or lost) in the various refugia, but at the same time new haplotypes arose as a result of the occurrence of novel mutations. When the climate improved and Paleolithic populations from European refugia repopulated the continent, some of these novel (or differently preserved) haplotypes also spread. They subsequently gave rise to new star-like haplogroups in the phylogeny, marking the expansion range from each refugium.
In the last 10 years, numerous studies have evaluated the distribution and extent of variation of haplogroups in European populations, and evidence of the overwhelming importance of the Franco-Cantabrian refugium for the repeopling of much of Western and Northern Europe at the beginning of the Holocene has been obtained by the age estimates and geographic distributions of mtDNA haplogroups H1, H3, V, and U5b1b.5,13–21 Y chromosome haplogroups R1b1b2-M269, I1-M253, and I2b1-M223 support the important role of the Franco-Cantabrian refuge zone,22–24 whereas other Y haplogroups (I2a1-M423 and R1a1-M17) reveal that the Balkan and Ukrainian refuge zones were also major genetic sources25–30 for the human recolonization of Europe.
In addition to the refugia mentioned above, another glacial refugium in Europe was the Italian Peninsula.8 However, neither mtDNA nor Y chromosome studies have yet been able to identify haplogroups marking expansions from this area, thus suggesting a marginal role, if any, of this southern European area in the postglacial repeopling of Europe.
Haplogroup U5 is one of the most ancient mtDNA haplogroups found in Europe. It evolved mainly within Europe where it spread after being involved in the first settlement of the continent by modern humans.4,31 Its phylogeny is characterized by two branches—U5a and U5b—which are common in most European populations,19,32,33 with U5b further split into U5b1 and U5b2.19 In 2006, a third uncommon branch, named U5b3, harboring the control-region motif 16169A-16192-16235-16270-16519-150 was detected only in Sardinia,34 an island that remained unconnected with the mainland even when the sea level was lowest during the LGM35 and that was probably the last of the large Mediterranean islands to be colonized by modern humans.36
To shed some light on the origin of haplogroup U5b3, we surveyed a wide range of European (and neighboring) populations for the presence of U5 mtDNAs lacking the diagnostic markers of haplogroups U5b1 and U5b2. For all subjects involved, an appropriate informed consent was obtained and institutional review boards at the Universities of Pavia, Tartu, Santiago de Compostela, at the Rambam Health Care Campus, and at the Sorenson Molecular Genealogy Foundation approved all procedures. Several mtDNAs with this feature were identified in Sardinia, in agreement with the presence of U5b3 in the island, but others were detected, at a very low frequency, also in other regions. With the exception of most mtDNAs from Sardinia, which harbored the previously described U5b3 control-region motif, almost all other U5 mtDNAs were characterized by a different but related control-region motif (16192-16270-16304-150).
To define the phylogenetic relationships between the U5b3 mtDNAs from Sardinia and the U5 mtDNAs with the related control-region motif, we completely sequenced a total of 43 mtDNAs and, together with nine previously published sequences (Table S1 available online), incorporated them in a phylogeny of haplogroup U5 (Figure 1). All sequences clustered in a U5 clade that is defined by a transition at np 7226 in the coding region—a mutation whose presence can be easily tested at the population level by a survey with the restriction enzyme DdeI. This clade splits into different minor subsets with a clear star-like pattern, including one branch that corresponds to the previously defined U5b3. This finding prompted us to revise the nomenclature and name the entire clade as U5b3, six of its main subsets as U5b3a-f, and the branch encompassing the Sardinian mtDNAs as U5b3a1a (Figure 1).
Figure 1.
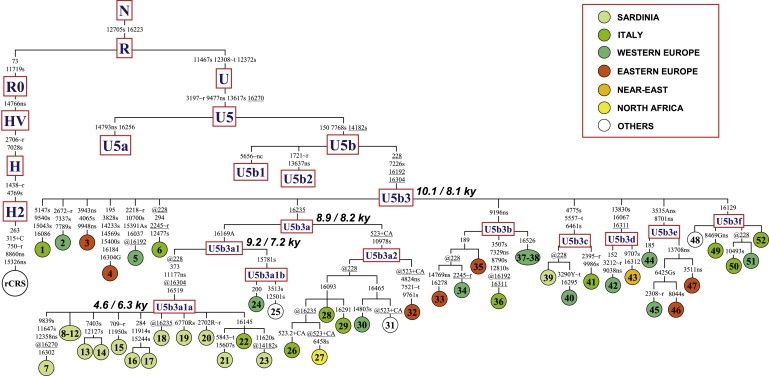
Detailed Tree of U5b3 in the Context of Haplogroup U5
The tree includes 52 complete mtDNA sequences and illustrates sub-haplogroup affiliations. The position of the revised Cambridge reference sequence (rCRS)51 is indicated for reading off sequence motifs. MtDNAs were selected through a preliminary sequence analysis of the control region and an RFLP survey in order to include the widest possible range of internal variation of haplogroup U5b3. The sequencing procedure and phylogeny construction were performed as described elsewhere.4,14,15 Sequences 1–9, 13–14, 18–19, 21, 24–52 are new while the others have been previously reported (Table S1). Mutations are shown on the branches; they are transitions unless a base is explicitly indicated. The prefix “@” designates reversions, whereas suffixes indicate: transversions (to A, G, C, or T), indels (+, d), gene locus (∼t, tRNA; ∼r, rRNA; ∼nc, noncoding region outside of the control region), synonymous or nonsynonymous changes (s or ns), and heteroplasmies (R, Y). Recurrent mutations are underlined. The variation in number of Cs at np 309 was not included in the phylogeny: sequences 2, 4–5, 24, 30, 34–38, 47–49, 51–52 harbored 309+C, whereas sequence 50 harbored 309+CC. Additional information regarding each mtDNA is available on Table S1. Time estimates shown for clades are averaged distance (ρ) of each haplotype with respect to the respective root. The first value has been obtained by considering one coding-region substitution every 4610 years, whereas the second one assumes 7650 years per synonymous transition.
When all coding-region base substitutions are considered,37 the average sequence divergence (±SE computed as in Saillard et al.38) of the 52 coding region sequences from the root of U5b3 is 2.19 ± 0.44 substitutions (Table 1)—a value virtually identical to those reported for haplogroups H1 (2.11 ± 0.23) and H3 (2.14 ± 0.28).15 This finding indicates that U5b3 expanded at about the same time as H1 and H3. Table 1 reports also the average sequence divergences calculated by using only synonymous transitions.39 Because the mutation rate of Mishmar et al.37 is probably an overestimate, mainly caused by partial saturation of some synonymous mutations,40 and that of Kivisild et al.39 represents an underestimate,41 we used the intermediate global coalescence time of modern human mtDNA recently proposed by Perego et al.42 as a reference point for the internal calibration of both approaches. Accordingly, we converted the haplogroup sequence divergences into time estimates by using averaged time calibrations corresponding to 4610 years per coding-region substitution and 7650 years per synonymous transition (Table 1). With this approach, the coalescence time estimates for the entire U5b3 are between 10.1 ky and 8.1 ky.
Table 1.
Averaged Divergence of Relevant Nodes in the U5b3 Phylogeny of Figure 1
| Clade | No. of MtDNAs | All Coding-Region Base Substitutions |
Only Synonymous Transitions |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ρa | σb | Tc(ya) | ΔT(ya) | ρa | σb | Tc(ya) | ΔT(ya) | ||
| U5b3 | 52 | 2.192 | 0.439 | 10,107 | 2,026 | 1.058 | 0.217 | 8,091 | 1,658 |
| >U5b3a | 26 | 1.923 | 0.744 | 8,865 | 3,429 | 1.077 | 0.357 | 8,238 | 2,729 |
| > > U5b3a1 | 19 | 2.000 | 0.942 | 9,220 | 4,340 | 0.947 | 0.279 | 7,247 | 2,131 |
| > > > U5b3a1a | 17 | 1.000 | 0.294 | 4,610 | 1,356 | 0.824 | 0.276 | 6,300 | 2,111 |
The average number of base substitutions in the mtDNA coding region (between positions 577 and 16023) from the root sequence type.
Standard error calculated from an estimate of the genealogy.38
Taking into account the limits of previous estimates reported by Mishmar et al.37 for all coding-region base substitutions and by Kivisild et al.39 for only synonymous transitions, we here employed a rate recently proposed by Perego et al.42 With three decimal digits throughout, their rounded values were 5140 years per coding-region substitution and 6760 years per synonymous transition, respectively. The rho-estimated (average distance of the haplotypes of a clade from the respective root) human coalescence times are then 202 ky according to Mishmar et al.37 and 160 ky according to Kivisild et al.39 The postulated time obtained as the arithmetic mean of both estimates is ∼181 ± 21 ky. Thus, ages estimated considering all the coding-region substitution have to be decreased by a factor of 181/202 ≈0.90, whereas the estimates based only on synonymous transitions have to be increased by a factor of 181/160 ≈1.13. Given that 5140 × 181/202 ≈4610 and 6760 × 181/160 ≈7650, we obtained the averaged calibrations.
To evaluate the distribution of haplogroup U5b3 in modern European (and neighboring) populations, we performed a survey of all U5 control-region motifs reported in almost 35,000 subjects from 81 population samples. For published and unpublished data sets for which only hypervariable segment I (HVS-I) data were available, U5 mtDNAs were affiliated within U5b3 when lacking 16189 or 16256 and harboring 16304. The presence or absence of the mutations 16169A, 16192, and 16235 was also considered. The results of this survey are reported in Table S2 and illustrated in the spatial distribution of Figure 2. Haplogroup U5b3 is virtually absent in the Near East (the single U5b3 mtDNA found in Iraq was completely sequenced) and North Africa and is rare in Europe where, with the exception of the frequency peak in Sardinians (3.8%), its frequency barely reaches 1% only in some Mediterranean populations.
Figure 2.
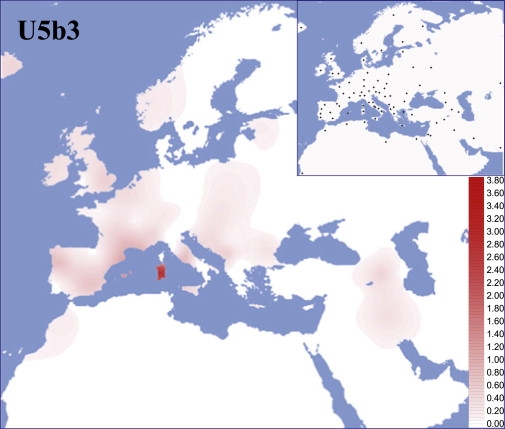
Spatial Frequency Distribution of Haplogroup U5b3 and Geographical Locations of Populations Surveyed
Populations and corresponding frequency values are listed in Table S2.
Out of the 55 U5b3 mtDNAs detected in Sardinians, all but one (sequence n. 39 in Figure 1) are characterized by the diagnostic control-region motif of sub-haplogroup U5b3a1a, whose coalescence time estimate is between 4.6 ky and 6.3 ky (Figure 1 and Table 1). The phylogeny of Figure 1 includes 17 complete sequences belonging to this sub-haplogroup and, with the possible exception of sequence n. 22 that is classified as a generic “Italian” without regional details,43 all are from Sardinia. A search for the U5b3a1a control-region motif in published data sets revealed only two matches (both 16169A-16192-16235-16270) outside Sardinia, one in Sicily44 and one in Rome.45 Details concerning the ancestry of the two subjects are not available, but the geographic proximity of Sardinia to the areas where they were detected makes it likely that they represent recent events of gene flow from the island. This would mean that U5b3a1a has arisen in situ in Sardinia after the arrival of an U5b3a1 founder mtDNA from somewhere else in Europe and that U5b3a1a affiliation is a marker of maternal Sardinian ancestry. The phylogeny of Figure 1 provides additional information concerning the entry time of the founder mtDNA; the upper limit is 9.2–7.2 ky (the age of U5b3a1 node), whereas the lower limit is 4.6–6.3 ky (the age of the U5b3a1a node), when the sub-haplogroup began to expand in Sardinia.
The phylogeny of Figure 1 also indicates a possible ancestral source for the founder(s) of the Sardinian U5b3a1a. The Sardinian-specific branch harbors a sister clade (U5b3a1b) formed by two sequences (n. 24 and 25): one from Languedoc, a region of southern France, and the other from a U.S. subject of undefined European ancestry. A search for the U5b3a1b control-region motif (16169A-16192-16235-16270-16304) was able to detect only one additional mtDNA from the southwestern (French-speaking) part of Switzerland,46 matching such a motif. This preliminary observation suggests a stronger link between Sardinia and southern France than with other European regions, including continental Italy. Archaeological data from the period 5–10 kya show that the Monte Arci region of western Sardinia (Oristano province) was one of the four Mediterranean sources (together with the small islands of Palmarola, Lipari, and Pantelleria) of obsidian, the “black gold” of the Neolithic. In particular, a blooming trade of obsidian has been documented from Sardinia to other Mediterranean regions, including southern France. Moreover, it has been calculated that the obsidian employed in the Neolithic sites of the southern France was almost exclusively from a “single” Monte Arci subsource, suggesting not only a preferential link between French sites and Sardinia but also preferential transport mechanisms, different from those connecting Sardinia with other Mediterranean regions (Corsica and northern Italy) where this selection of specific subsources has not been detected.47
What about the ancestral homeland of the entire haplogroup U5b3? Its divergence is virtually identical to that reported for H1 and H3, thus indicating a population expansion at about the same time. Haplogroups H1 and H3 diffused from the Franco-Cantabrian refuge zone when climatic conditions improved;15,18 therefore, it is possible that also the founder U5b3 sequence expanded from the same area and the three haplogroups were involved in the same demographic processes. However, there is also an alternative scenario: the expansion of U5b3 could have still occurred at the same time as H1 and H3 when climatic conditions in Europe changed, but from a distinct geographical source. With consideration to the modern range distribution of U5b3 (Figure 2), the only other potential candidate for the latter scenario is the glacial refuge in the Italian Peninsula.
To discriminate between the two possibilities, we measured the extent of U5b3 variation in different geographical areas by employing all available HVS-I (nps 16024–16365) data. A total of 152 U5b3 mtDNAs were detected, encompassing 40 HVS-I haplotypes, and their relationships are illustrated in the network of Figure 3. As expected, despite the frequency peak, Sardinians showed a very low haplotype diversity (H = 0.570), whereas much higher H values were observed in Italy (0.877) and Iberia (0.904) (Table 2), thus confirming that the relatively high frequency of U5b3 in Sardinia is the result of a founder event after the arrival on the island. Other indices such as nucleotide diversity and average number of nucleotide differences (Table 2), which are more informative than haplotype diversity because they take into account also the extent of diversity between haplotypes, not only confirm that Italy (0.717 and 2.45, respectively) and Iberia (0.645 and 2.21, respectively) are the European regions with the highest levels of U5b3 diversity but also reveal a peak in Italy, thus indicating continental Italy as the most likely focus of the U5b3 expansion.
Figure 3.
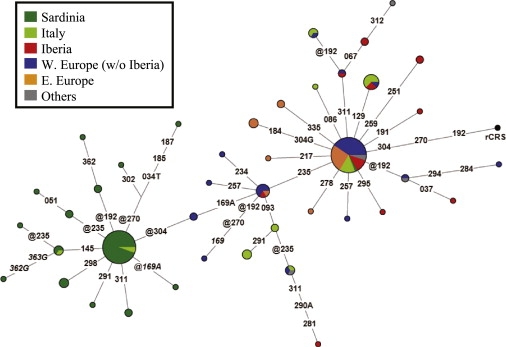
Median-Joining Network of HVS-I Haplotypes Observed in 152 U5b3 mtDNAs
Eighty-three mtDNAs are from the literature and a subset of these (N = 32) were not included in the population frequency table (Table S2) because population sample sizes were undefined. We constructed the tree by using the Network 4.510 software program (http://www.fluxus-engineering.com). The numbers (plus 16000) on the connecting branches refer to the revised reference sequence51 and indicate mutations. These are transitions unless the base change is explicitly indicated; the prefix “@” designates reversions. Mutations in italics are most likely erroneous and were disregarded in the calculation of diversity measures. The size of each circle is proportional to the haplotype frequency and geographical origins are indicated by different colors. Fifty-five mtDNAs are from Sardinia; 23 are from Italy [continental Italy (N = 20) and Sicily (N = 3)]; 17 are from Iberia [Spain (N = 11), Portugal (N = 2) and Balearic Islands (N = 4)]; 33 are from Western Europe (excluding Iberia) [Belgium (N = 1), Denmark (N = 1), England (N = 6), France (N = 4), Germany (N = 3), Iceland (N = 3), Ireland (N = 3), Netherlands (N = 2), Norway (N = 1), Scotland (N = 6), Switzerland (N = 2), and Wales (N = 1)]; 19 are from Eastern Europe [Croatia (N = 4), Bosnia (N = 2), Bulgaria (N = 1), Crete (N = 1), Czech Republic (N = 4), Estonia (N = 1), Hungary (N = 2), Montenegro (N = 1), Poland (N = 1), and Slovakia (N = 2)]; and five are “Others” [Armenia (N = 1), Iraq (N = 1), Algeria (N = 1), and Morocco (N = 2)].
Table 2.
Diversity of Haplogroup U5b3 MtDNAs in Different European Geographic Areas
| Geographic Areas | No. of MtDNAs | No. of Haplotypesa | Hb | πc | Md |
|---|---|---|---|---|---|
| Sardinia | 55 | 13 | 0.570 ± 0.080 | 0.288 ± 0.062 | 0.986 |
| Italy (continental Italy and Sicily) | 23 | 9 | 0.877 ± 0.040 | 0.717 ± 0.093 | 2.451 |
| Iberia (Spain, Portugal, and Balearic Islands) | 17 | 10 | 0.904 ± 0.055 | 0.645 ± 0.117 | 2.206 |
| Western Europe (w/o Iberia) | 33 | 13 | 0.729 ± 0.081 | 0.409 ± 0.079 | 1.398 |
| Eastern Europe | 19 | 6 | 0.655 ± 0.111 | 0.315 ± 0.074 | 1.076 |
HVS-I haplotypes (from np 16024 to np 16365).
Haplotype diversity.
Nucleotide diversity %.
Average number of nucleotide differences.
Overall, the coalescence time of U5b3 (and those of the more common haplogroups H1 and H3) appears to indicate that the major post-LGM re-expansion phase in Europe was at the beginning of the Holocene (∼11 kya) and not earlier. Whereas populations expanded geographically earlier during the warm phases of the Bølling-Allerød oscillations, the intermediate shorter-term cold phases and the Younger Dryas, in particular, led to retractions into the refugia again; it thus seems that in the Bølling-Allerød only some minor additional secondary refugia were created, which were too short-lived to leave discernible mutational marks in the mtDNA pools.
In contrast to the more common mtDNA haplogroups H1 and H3, however, the U5b3 diversity in modern Europe suggests that the glacial refuge located in the Italian Peninsula8,48 rather than the Franco-Cantabrian refuge was the ancestral expansion source for haplogroup U5b3. Postglacial expansions of refugial populations from this area toward the North were restricted not only by cold phases but also by a geographical barrier—the Alps.49 Thus, the ancestral U5b3 haplotype could have expanded (at a low frequency) outside the Italian Peninsula only along the coasts of the Tyrrhenian and Adriatic Seas, mainly toward the nearby Provence (southern France), and from there further west. The root of U5b3a1 originated probably in the Mediterranean coast of southern France and the same haplotype then went into Sardinia some 7–9 kya, possibly as a result of the obsidian trade that linked the two regions. There it expanded at the middle of the Neolithic, giving rise to an mtDNA clade (U5b3a1a) that distinctively marks the people of the island. Remarkably, the events leading to the arrival and expansion of this maternal lineage in Sardinia are not only supported but also magnified by data from male-specific lineages. Indeed ∼37% of Sardinian Y chromosomes belong to haplogroup I2a2-M26,50 a lineage rare outside Sardinia, whose age, distribution, and postulated geographic source (southern France)28 strikingly match those of mtDNA haplogroup U5b3a1—a paradigmatic case of parallel founder events for both maternal and paternal lineages in the European context.
Acknowledgments
This research received support from Fondazione Cassa di Risparmio di Foligno (to A.A. and F.P.), Fondazione Cassa di Risparmio di Perugia (to A.A. and F.P.), the European Union (European Regional Development Fund through the Centre of Excellence in Genomics), Estonian Biocentre, Estonian Science Foundation grants 7858 (to E.M.) and 6040 (to K.T.), the Swedish Collegium of Advanced Studies (to R.V.), Österreichische Forschungsförderungsgesellschaft, KIRAS Sicherheitsforschung DNATOX (to W.P.), Ministerio de Ciencia e Innovación (SAF2008-02971; to A.S.), Landau Network-Centro Volta (to N.A.-Z.), Progetti Ricerca Interesse Nazionale 2007 (Italian Ministry of the University) (to O.S. and A.T.), Ministero degli Affari Esteri (to O.S.), Compagnia di San Paolo (to O.S. and A.T.), and Fondazione Cariplo (to A.T.). We are grateful to all the donors for providing blood samples and to Cristian Capelli and the other people who helped collecting the samples.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
Network 4.510 software, http://www.fluxus-engineering.com
Accession Numbers
Previously unreported mtDNA sequences reported in this paper have been deposited in GenBank under accession numbers GQ129143–GQ129183.
References
- 1.Mellars P. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature. 2006;439:931–935. doi: 10.1038/nature04521. [DOI] [PubMed] [Google Scholar]
- 2.Goebel T. The missing years for modern humans. Science. 2007;315:194–196. doi: 10.1126/science.1137564. [DOI] [PubMed] [Google Scholar]
- 3.Anikovich M.V., Sinitsyn A.A., Hoffecker J.F., Holliday V.T., Popov V.V., Lisitsyn S.N., Forman S.L., Levkovskaya G.M., Pospelova G.A., Kuz'mina I.E. Early Upper Paleolithic in Eastern Europe and implications for the dispersal of modern humans. Science. 2007;315:194–196. doi: 10.1126/science.1133376. [DOI] [PubMed] [Google Scholar]
- 4.Olivieri A., Achilli A., Pala M., Battaglia V., Fornarino S., Al-Zahery N., Scozzari R., Cruciani F., Behar D.M., Dugoujon J.-M. The mtDNA legacy of the Levantine Early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- 5.Torroni A., Achilli A., Macaulay V., Richards M., Bandelt H.-J. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Street M., Terberger T. The last Pleniglacial and the human settlement of Central Europe: New information from the Rhineland site of Wiesbaden-Igstadt. Antiquity. 1999;73:259–272. [Google Scholar]
- 7.Straus L.G. The Upper Paleolithic of Cantabrian Spain. Evol. Anthropol. 2005;14:145–158. [Google Scholar]
- 8.Banks W.E., d'Errico F., Peterson A.T., Vanhaeren M., Kageyama M., Sepulchre P., Ramstein G., Jost A., Lunt D. Human ecological niches and ranges during the LGM in Europe derived from an application of eco-cultural niche modelling. J. Archaeol. Sci. 2008;35:481–491. [Google Scholar]
- 9.Alley R.B., Meese D.A., Shuman C.A., Gow A.J., Taylor K.C., Grootes P.M., White J.W.C., Ram M., Waddington E.D., Mayewski P.A. Abrupt increase in Greenland snow accumulation at the end of the Younger Dryas event. Nature. 1993;362:527–529. [Google Scholar]
- 10.Björck S., Kromer B., Johnsen S., Bennike O., Hammarlund D., Lemdahl G., Possnert G., Rasmussen T.L., Wohlfarth B., Hammer C.U., Spurk M. Synchronized terrestrial-atmospheric deglacial records around the North Atlantic. Science. 1996;274:1155–1160. doi: 10.1126/science.274.5290.1155. [DOI] [PubMed] [Google Scholar]
- 11.Peteet D. Sensitivity and rapidity of vegetational response to abrupt climate change. Proc. Natl. Acad. Sci. USA. 2000;97:1359–1361. doi: 10.1073/pnas.97.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alley R.B., Marotzke J., Nordhaus W.D., Overpeck J.T., Peteet D.M., Pielke R.A., Jr., Pierrehumbert R.T., Rhines P.B., Stocker T.F., Talley L.D. Abrupt climate change. Science. 2003;299:2005–2010. doi: 10.1126/science.1081056. [DOI] [PubMed] [Google Scholar]
- 13.Torroni A., Bandelt H.-J., D'Urbano L., Lahermo P., Moral P., Sellitto D., Rengo C., Forster P., Savontaus M.L., Bonné-Tamir B. MtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am. J. Hum. Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torroni A., Bandelt H.-J., Macaulay V., Richards M., Cruciani F., Rengo C., Martinez-Cabrera V., Villems R., Kivisild T., Metspalu E. A signal, from human mtDNA, of postglacial recolonization in Europe. Am. J. Hum. Genet. 2001;69:844–852. doi: 10.1086/323485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achilli A., Rengo C., Magri C., Battaglia V., Olivieri A., Scozzari R., Cruciani F., Zeviani M., Briem E., Carelli V. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster P. Ice ages and the mitochondrial DNA chronology of human dispersals: A review. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:255–264. doi: 10.1098/rstb.2003.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble C., Davies W., Pettitt P., Richards M. Climate change and evolving human diversity in Europe during the last glacial. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:243–253. doi: 10.1098/rstb.2003.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loogväli E.-L., Roostalu U., Malyarchuk B.A., Derenko M.V., Kivisild T., Metspalu E., Tambets K., Reidla M., Tolk H.-V., Parik J. Disuniting uniformity: A pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol. Biol. Evol. 2004;21:2012–2021. doi: 10.1093/molbev/msh209. [DOI] [PubMed] [Google Scholar]
- 19.Achilli A., Rengo C., Battaglia V., Pala M., Olivieri A., Fornarino S., Magri C., Scozzari R., Babudri N., Santachiara-Benerecetti A.S. Saami and Berbers - an unexpected mitochondrial DNA link. Am. J. Hum. Genet. 2005;76:883–886. doi: 10.1086/430073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira L., Richards M., Goios A., Alonso A., Albarrán C., Garcia O., Behar D.M., Gölge M., Hatina J., Al-Gazali L. High resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005;15:19–24. doi: 10.1101/gr.3182305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Álvarez-Iglesias V., Mosquera-Miguel A., Cerezo M., Quintáns B., Zarrabeitia M.T., Cuscó I., Lareu M.V., García O., Pérez-Jurado L., Carracedo A. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS ONE. 2009;4:e5112. doi: 10.1371/journal.pone.0005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semino O., Passarino G., Oefner P.J., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S. The genetic legacy of Paleolithic Homo sapiens in extant Europeans: A Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 23.Wells R.S., Yuldasheva N., Ruzibakiev R., Underhill P.A., Evseeva I., Blue-Smith J., Jin L., Su B., Pitchappan R., Shanmugalakshmi S. The Eurasian heartland: A continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zei G., Lisa A., Fiorani O., Magri C., Quintana-Murci L., Semino O., Santachiara-Benerecetti A.S. From surnames to the history of Y chromosomes: The Sardinian population as a paradigm. Eur. J. Hum. Genet. 2003;11:802–807. doi: 10.1038/sj.ejhg.5201040. [DOI] [PubMed] [Google Scholar]
- 25.Passarino G., Semino O., Magri C., Al-Zahery N., Benuzzi G., Quintana-Murci L., Andellnovic S., Bullc-Jakus F., Liu A., Arslan A. The 49a,f haplotype 11 is a new marker of the EU19 lineage that traces migrations from northern regions of the Black Sea. Hum. Immunol. 2001;62:922–932. doi: 10.1016/s0198-8859(01)00291-9. [DOI] [PubMed] [Google Scholar]
- 26.Barać L., Peričić M., Martinović Klarić I., Rootsi S., Janićijevć B., Kivisild T., Parik J., Rudan I., Villems R., Rudan P. Y chromosomal heritage of Croatian population and its island isolates. Eur. J. Hum. Genet. 2003;11:535–542. doi: 10.1038/sj.ejhg.5200992. [DOI] [PubMed] [Google Scholar]
- 27.Cinnioğlu C., King R., Kivisild T., Kalfoglu E., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K. Excavating Y-chromosome haplotype strata in Anatolia. Hum. Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 28.Rootsi S., Magri C., Kivisild T., Benuzzi G., Help H., Bermisheva M., Kutuev I., Barać L., Peričić M., Balanovsky O. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europa. Am. J. Hum. Genet. 2004;75:128–137. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peričić M., Barać Lauc L., Martinović Klarić I., Rootsi S., Janićijevć B., Rudan I., Terzić R., Čolak I., Kvesić A., Popović D. High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations. Mol. Biol. Evol. 2005;22:1964–1975. doi: 10.1093/molbev/msi185. [DOI] [PubMed] [Google Scholar]
- 30.Battaglia V., Fornarino S., Al-Zahery N., Olivieri A., Pala M., Myres N.M., King R.J., Rootsi S., Marjanovic D., Primorac D. Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. Eur. J. Hum. Genet. 2008 doi: 10.1038/ejhg.2008.249. in press. Published online December 24, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards M., Macaulay V., Hickey E., Vega E., Sykes B., Guida V., Rengo C., Sellitto D., Cruciani F., Kivisild T. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 32.Finnilä S., Lehtonen M.S., Majamaa K. Phylogenetic network for European mtDNA. Am. J. Hum. Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tambets K., Rootsi S., Kivisild T., Help H., Serk P., Loogväli E.L., Tolk H.V., Reidla M., Metspalu E., Pliss L. The western and eastern roots of the Saami - the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am. J. Hum. Genet. 2004;74:661–682. doi: 10.1086/383203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraumene C., Belle E.M.S., Castrì L., Sanna S., Mancosu G., Cosso M., Marras F., Barbujani G., Pirastu M., Angius A. High resolution analysis and phylogenetic network construction using complete mtDNA sequences in Sardinian genetic isolates. Mol. Biol. Evol. 2006;23:2101–2111. doi: 10.1093/molbev/msl084. [DOI] [PubMed] [Google Scholar]
- 35.Shackleton J.C., van Andel T.H., Runnels C.N. Coastal paleogeography of the central and western Mediterranean during the last 125,000 years and its archaeological implications. J. Field Archaeol. 1984;11:307–314. [Google Scholar]
- 36.Sondaar P.Y. Palaeolithic Sardinians: Paleontological evidence and methods. In: Balmuth M.S., Tykot R.H., editors. Sardinian and Aegean chronology. Oxbow Books; Oxford: 1998. pp. 45–51. [Google Scholar]
- 37.Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saillard J., Forster P., Lynnerup N., Bandelt H.-J., Nørby S. MtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am. J. Hum. Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kivisild T., Shen P., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamm E., Kivisild T., Reidla M., Metspalu M., Smith D.G., Mulligan C.J., Bravi C.M., Rickards O., Martinez-Labarga C., Khusnutdinova E.K. Beringian standstill and spread of Native American founders. PLoS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achilli A., Perego U.A., Bravi C.M., Coble M.D., Kong Q.-P., Woodward S.R., Salas A., Torroni A., Bandelt H.-J. The phylogeny of the four pan-American mtDNA haplogroups: Implications for evolutionary and disease studies. PLoS ONE. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perego U.A., Achilli A., Angerhofer N., Accetturo M., Pala M., Olivieri A., Hooshiar Kashani B., Ritchie K.H., Scozzari R., Kong Q.-P. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 43.Ingman M., Kaessmann H., Pääbo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 44.Vona G., Ghiani M.-E., Calò C.-M., Vacca L., Memmì M., Varesi L. Mitochondrial DNA sequence analysis in Sicily. Am. J. Hum. Biol. 2001;13:576–589. doi: 10.1002/ajhb.1096. [DOI] [PubMed] [Google Scholar]
- 45.Turchi C., Buscemi L., Previderè C., Grignani P., Brandstätter A., Achilli A., Parson W., Tagliabracci A., Ge.F.I. Group Italian mitochondrial DNA database: Results of a collaborative exercise and proficiency testing. Int. J. Legal Med. 2008;122:199–204. doi: 10.1007/s00414-007-0207-1. [DOI] [PubMed] [Google Scholar]
- 46.Dimo-Simonin N., Grange F., Taroni F., Brandt-Casadevall C., Mangin P. Forensic evaluation of mtDNA in a population from south west Switzerland. Int. J. Legal Med. 2000;113:89–97. doi: 10.1007/pl00007715. [DOI] [PubMed] [Google Scholar]
- 47.Tykot R.H. Chemical fingerprinting and source tracing of obsidian: The central Mediterranean trade in black gold. Acc. Chem. Res. 2002;35:618–627. doi: 10.1021/ar000208p. [DOI] [PubMed] [Google Scholar]
- 48.Mussi M. Plenum Publishing Company; New York: 2001. Earliest Italy. An overview of the Italian Palaeolithic and Mesolithic. [Google Scholar]
- 49.Taberlet P., Fumagalli L., Wust-Saucy A.G., Cosson J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- 50.Contu D., Morelli L., Santoni F., Foster J.W., Francalacci P., Cucca F. Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse Sardinian population: Inference for association scans. PLoS ONE. 2008;3:e1430. doi: 10.1371/journal.pone.0001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


