Abstract
A prospective cohort study was used to assess whether Salmonella fecal shedding in commercial feedlot cattle treated with antimicrobials for respiratory disease was associated with subsequent adverse health outcomes. Feces were collected per rectum from cattle that were examined for apparent respiratory disease, had a rectal temperature ≥40 °C, and subsequently received antimicrobial treatment. Salmonella were recovered from 918 (73.7%) of 1 245 fecal samples and weekly prevalence estimates ranged from 49 to 100% over the 3-month study. Genotypic and phenotypic characteristics of Salmonella strains in the population were determined. Serogroup E Salmonella were most common (73.3%), followed by C1 (11.0%), C3 (8.6%), and B (1.1%). Predominant serotypes were Orion (46.5%), Anatum (19.8%), Kentucky (8.7%), Montevideo (7.5%), and Senftenberg (4.9%). Few isolates (36/918) were positive for antimicrobial resistance-associated integron gene intI1. Phenotypic susceptibility was associated with isolate intI1 status. Crude re-pull, re-treatment and case fatality risks were higher for cattle that were Salmonella-positive versus -negative at initial treatment, but not statistically different on multivariable analysis. However, case fatality risk was higher for cattle shedding Group B Salmonella than for cattle shedding other serogroups. Lots (groups) with a higher Salmonella prevalence at first treatment had a higher proportion of mortalities occur in a hospital pen, higher overall re-treatment risks, and were more likely to be sampled later in the study. Results indicate a high prevalence of Salmonella in this population of cattle treated for apparent respiratory disease, but that effects associated with clinical outcomes may depend on the Salmonella strain.
Keywords: Salmonella, bovine respiratory disease, feedlot, cattle
1. INTRODUCTION
Salmonella enterica in beef cattle production systems may be associated with clinical or subclinical disease in cattle, and may represent a potential source of human exposure. Although Salmonella are important pathogens, the epidemiology of Salmonella in commercial feedlot systems is not well described. Certainly, both ill and apparently healthy cattle may shed Salmonella in their feces, and prevalence estimates for Salmonella in feces, on hides, and on carcasses of clinically normal feedlot cattle have been reported extensively [2–5, 12–14, 19–22]. However, data on clinical salmonellosis in feedlot cattle, transmission dynamics of Salmonella in feedlot production systems, or the impact of clinical or subclinical Salmonella infections on feedlot cattle health and performance are extremely sparse.
Salmonella infections in cattle can cause a variety of clinical signs depending on the strain of Salmonella, the type of cattle, and many other factors [6, 26, 36, 42]. The clinical effects of Salmonella in commercial feedlots are poorly defined as descriptions of clinical salmonellosis in cattle are almost exclusively based on observations of dairy cattle or beef cattle in different production environments [18, 19, 26, 29, 30, 36, 41, 42]. Salmonellosis in cattle often results in fever and diarrhea, but dyspnea, respiratory syndromes, and sudden death, with or without diarrhea, can occur with some Salmonella strains [6, 29, 36, 42]. It has been suggested that salmonellosis could be confused with “pneumonic pasteurellosis” and that salmonellosis is one of the few diseases of cattle that are currently increasing in prevalence [18, 36]. Although Salmonella can directly or indirectly affect morbidity, mortality, and production efficiency in cattle [6, 26, 36, 42], the clinical and subclinical effects are largely undocumented for commercial feedlot cattle.
In commercial feedlots, the most common cause of morbidity, mortality, and antimicrobial therapy is bovine respiratory disease complex (BRDC) [38]. The overall disease incidence in feedlots, primarily due to BRDC, tends to peak early in the feeding period, and evidence suggests that the fecal prevalence of Salmonella may peak then as well [10, 22, 24, 26]. The diagnosis and therapy for BRDC is often based on clinical observations with limited diagnostics [26]. Potential relationships between Salmonella and BRDC have not been documented. Depending on a variety of pathogen, host, and environmental factors, Salmonella may cause subclinical infections or primary disease, predispose animals to other diseases, or result in fecal shedding or salmonellosis as sequela to other diseases [29, 36, 42]. We hypothesized that the Salmonella status of cattle in commercial feedlots may affect the clinically and economically important disease outcomes that are often associated with BRDC.
Our primary objective was to determine whether fecal shedding ofSalmonella in commercial feedlot cattle treated with antimicrobials for BRDC was associated with an increase in incidence risks for health outcomes that are frequently monitored in feedlot production systems. Our secondary objectives were to determine the prevalence of Salmonella in clinically ill feedlot cattle as they were treated for BRDC, and to describe the genotypic and phenotypic characteristics of the Salmonella isolated, which then enables evaluations of the conditional effects on cattle health.
2. MATERIALS AND METHODS
2.1. Study population and study design
This study was conducted in a large west Texas commercial feedlot with a one-time capacity of approximately 70 000 cattle. This feedlot had no documented evidence of clinical salmonellosis within the last three years. Our primary objective was addressed by performing a prospective cohort study in which cattle were categorized as Salmonella-positive or -negative based on the recovery of Salmonella in fecal samples at the time of initial treatment for BRDC, and then were monitored until slaughter for adverse health effects. Newly received cattle that were individually removed from their home pen (“pulled”), examined for apparent respiratory disease, had a rectal temperature ≥40 °C, and received treatment with an injectable antimicrobial for BRDC were eligible for inclusion in the study. Cattle in arrival lots that received metaphylactic antimicrobial treatment on arrival to the feedlot were noted, but not enrolled in the study unless later treated individually. Cattle were sampled per rectum using a new plastic sleeve for each animal. Samples were packed in coolers with frozen ice-packs and shipped by overnight courier to the Pre-harvest Food Safety Laboratory in the College of Veterinary Medicine, Kansas State University. The cooperating feedlots animal health database was used to record and track study cattle, and recover associated data.
2.2. Salmonella isolation and identification
Samples were processed and cultured according to methods previously reported to be sensitive for the isolation of Salmonella from cattle feces [3]. In brief, 10 g of feces from each sample were inoculated into 90 mL of Tryptic Soy broth (Becton Dickinson Co., Sparks, MD, USA) and incubated at room temperature (~25 °C) for 2 h, then at 42 °C for 6 h, and then 10 mL of the culture was inoculated into 90 mL of Tetrathionate broth (TT) broth (Becton Dickinson). Inoculated TT broth was incubated at 37 °C for 18 h and then immunomagnetic separation (IMS) was performed with 1 mL of culture following manufacturer’s instructions (Dynal, Inc., New Hyde Park, NY, USA). The final volume of the IMS sample was adjusted to 100 μL with phosphate buffered saline (Sigma-Aldrich, St. Louis, MO, USA), added to 10 mL of Rappaport-Vassiliadis broth (Becton Dickinson), and incubated at 42 °C for 24 h. After incubation, the culture was then streaked onto Hektoen Enteric agar (Becton Dickinson) and incubated at 37 °C for 18 h. Up to three characteristic Salmonella colonies per plate were picked, subcultured onto blood agar, and incubated at 37 °C. Poly O antisera-positive isolates were serogrouped with a slide agglutination test (Becton Dickinson) and sent to the National Veterinary Service Laboratory (Ames, Iowa, USA) for serotyping. Samples were considered positive for Salmonella when a recovered isolate was morphologically typical of Salmonella, poly O antisera-positive, and positive for the invA gene based on PCR (details below).
2.3. PCR for Salmonella virulence and integron genes
We used PCR to test for two Salmonella virulence genes, invA and pagC [34], which are associated with Salmonella invasion of epithelial cells and survival within macrophages respectively [23]. In brief, a single colony was suspended in 1 mL of distilled water and boiled for 10 min; the supernatant was obtained by centrifugation (800×g, 10 min) and 1 μL used as the test sample. Primers used were: forward 5′-GTGAATTATCGCCACGTTCGG-3′; reverse: 5′-TCATCGCACCGTCAAAGGAAC-3′ for invA, and forward: 5′-TATGAGGATCACT CTCCGGTA-3′, reverse: 5′-TTCTCCAGCGGAT TCATCTA-3′ for pagC. Reaction master mix contained 1 μM of each primer, 400 μM dNTPs, 3 mM MgCl2 and 0.5 unit Taq polymerase (all Promega, Madison, WI, USA) in a final volume of 25 μL. Amplification parameters were as follows: denaturation at 94 °C for 7 min, then 30 cycles of 94 °C for 1 min, annealing temperature 55 °C for 1.5 min and 72 °C for 1 min, and a final extension at 72 °C for 5 min (7 min for pagC).
Integron class 1 presence also was determined by PCR [43]. Briefly, primer set Int-1U (5′-GTTCGGTCAAGGTTCTG-3′) and Int-1D (5′-GCCAACTTTCAGCACATG-3′) was used to amplify the conserved regions of integron-encoded integrase genes intI1. Isolates yielding a PCR product with the primer set Int-1U and Int1-1D were further amplified using primer set qacEΔ1-F (5′-ATCGCAATAGTTGGCGAAGT-3′) and sul1-B (5′-GCAAGGCGGAAACCCGCGCC-3′) to determine whether the integron contained the 3′ conserved sequence segments. Reaction master mix contained 1 μM of each primer, 400 μM dNTPs, 3 mM MgCl2 and 0.5 unit Taq polymerase (all Promega) in a final volume of 25 μL. Cycling conditions were: initial denaturation at 94 °C for 5 min, 30 amplification cycles consisting of 0.5 min at 94 °C, 0.5 min at 50 °C, 1.5 min at 72 °C, and a final round of extension for 7 min at 72 °C in a thermal cycler (TaKaRa Bio Inc., Otsu, Shiga, Japan). For primer set qacEΔ1-F/sul1-B, conditions were modified to a primer annealing temperature of 56 °C and a final extension time of 1 min.
All primers were obtained commercially (Bio-Synthesis Inc., Lewsville, TX, USA). All PCR products were separated by 1.5% agarose gel electrophoresis in 1X TBE (Tris-Borate-EDTA; Sigma-Aldrich) buffer and visualized with ethidium bromide and ultraviolet transillumination. Presence of an appropriate sized amplicon was verified visually by comparison with a positive control (S. Typhimurium, ATCC 700408).
2.4. Phenotypic antimicrobial susceptibility determination
Susceptibility profiles were determined for all isolates positive for integron genes. In addition, profiles were determined on a subset of the integron-negative isolates by randomly choosing an equal number of isolates within each serotype. Isolates were tested for antimicrobial susceptibility according to standard protocols established by the U.S. National Antimicrobial Resistance Monitoring System (NARMS) using a Sensititre 18–24 h susceptibility system (Trek Diagnostic System Ltd., Cleveland, OH, USA). We used NARMS custom panels, CMV1AGNF, which included amikacin (0.5–32 μg/mL), amoxicillin/clavulanic acid (1.0/0.5–32/16 μg/mL), ampicillin (1–32 μg/mL), cefoxitin (0.5–32 μg/mL), ceftiofur (0.25–8 μg/mL), ceftriaxone (0.25–64 μg/mL), chloramphenicol (2–32 μg/mL), ciprofloxacin (0.0156–4 μg/mL), gentamicin (0.25–16 μg/mL), kanamycin (8–64 μg/mL), nalidixic acid (0.5–32 μg/mL), streptomycin (32–64 μg/mL), sulfizoxazole (16–512 μg/mL), tetracycline (4–32 μg/mL), and trimethoprim/sulphamethoxazole (0.12/2.38–4/76μg/mL). Breakpoints and minimum inhibitory concentrations (MICs) were based on NARMS and Clinical and Laboratory Standards Institute guidelines [33].
2.5. Data management and statistical analysis
All sample and feedlot data were recorded in spreadsheet format (Excel, Microsoft Corp., Redmond, WA, USA). Health parameters measured were those commonly captured in the feedlot’s operational database. Of particular interest were adverse health outcomes, such as mortalities, re-pulls, re-treatments and culling (of the chronically ill). Potential covariates of interest included body weight at arrival, month of placement in the feedlot, gender, and respiratory disease risk classification. The risk classification scheme used in commercial feedlots attempts to predict the level of respiratory disease for a group of cattle. Personnel in the studied feedlot classified cattle as “high” or “low” risk following their standard procedures for assessing risk based on visual appraisal, cattle source, body size, transport time and other factors. Rather than a physical pen location, a “lot” was considered the hierarchical unit for a cattle group. Lot is a standard unit that commercial feedlots maintain as they receive, manage and market a group of cattle. Feedlot data were available at the lot-level from summary files, which provide cumulative demographic, health, and performance data from arrival to slaughter. In addition, individual-level data were obtained by retrieving health records for each sampled individual from the feedlot’s operational database. All sample data and feedlot data were imported into a statistical software program for all subsequent data management and statistical analyses (SAS version 9.1, SAS Institute Inc., Cary, NC, USA). All significance testing was two-sided with p < 0.05 used for hypothesis testing.
Salmonella prevalence estimates were reported at the sample-, animal-, and lot-levels. Exact binomial 95% confidence intervals (CI) were calculated for Salmonella prevalence estimates using Proc Freq of SAS. Evaluations of unconditional associations and multivariable analyses were completed using generalized linear mixed models specified for binomial outcomes with a logit link function. In all animal-level models, lot was included as a random effect with a correlation structure to account for the lack of independence among individual cattle within lots.
Multivariable models were developed using a series of steps that are well described [15]. Briefly, unconditional associations among outcomes and each potential explanatory variable were identified initially using a screening step. Each variable associated with an outcome on screening (p < 0.2) was entered into a multivariable model, removed using a manual backward selection approach, and then re-offered into the model until all variables remaining were significant at p < 0.05. Interactions were assessed after main effects were determined. To assess potential multicollinearity, associations among factors associated with outcomes based on screening were examined [15]. Model fit was optimized by comparing fit statistics and residuals between similarly structured models [15].
Adverse health outcomes at the individual animal level were modeled as dependent variables in logistic generalized linear mixed models (Proc Glimmix), which included Salmonella results as an independent variable and lot as a random effect. Associations between the probability of an animal culturing positive for Salmonella and independent variables based on individual records retrieved from the feedlot’s operational database also were evaluated with similar models. For these analyses, only cattle from lots with at least four cattle sampled as they were initially treated for BRDC were included. For assessing associations with phenotypic or genotypic characteristics of isolates, only data on cattle shedding Salmonella as they were initially treated were used. For assessing whether cattle sampled on multiple occasions were more likely to be culture positive at different times, a similar logistic regression model was used, and a first-order autoregressive covariance structure was included to account for repeated measures on the same animal within a lot [1].
Evaluations of potential associations between estimates of Salmonella prevalence within lots at first treatment and the feedlot’s cumulative lot-level data were performed similarly. The number of culture positive and total sampled individuals within each lot were the numerator and denominator, respectively, for the dependent variable in logit models explored using generalized estimating equations (GEE) in Proc Genmod of SAS [1]. This events/trials procedure allowed us to consider that estimates of Salmonella prevalence for a given lot were more precise when more fecal samples were collected. A repeated effect was included to account for the lack of independence among observations within a lot. All lots with data on cattle sampled as they were first treated for BRDC were included. The independent variables for this analysis were all lot-level cumulative data available from post-slaughter lot summaries. Score statistics for Type 3 GEE analysis and GEE parameter estimates were used for interpretation of fixed effects.
3. RESULTS
3.1. Description of study population
All study cattle were sampled during the weeks of September 11th through December 5th of 2005. Cattle were in lots ranging from 45 to 312 animals (mean = 141.2, median = 120). Most lots were characterized by feedlot personnel as being a high (n = 85) or low (n = 47) risk for respiratory disease, but three lots were not classified. Lots classified as high risk were treated with metaphylactic antimicrobials on arrival, whereas low risk lots were not. Three study lots contained Holstein cattle, while others contained beef cattle that were either in steer (n = 94), heifer (n = 35) or mixed lots (n = 3). Average body weight at arrival within lots ranged from 171 to 430 kg (mean = 269.5, median = 250.5). The number of days that cattle were fed until shipped for slaughter ranged from 119 to 298 days (mean = 215.7, median = 226). Overall morbidity, mortality, and cull risks for study lots were 17.9% (range 0.4 to 59.2%), 3.8% (range 0 to 21.1%), and 1.3% (range 0 to 10.0%) respectively. Of the treated cattle that we studied, 23.5% were treated for BRDC more than once and 13.4% died (BRDC case fatality). The majority of mortalities (69.7%) for studied lots occurred in a hospital pen with the remainder occurring in their home pen.
3.2. Salmonella prevalence and characterization
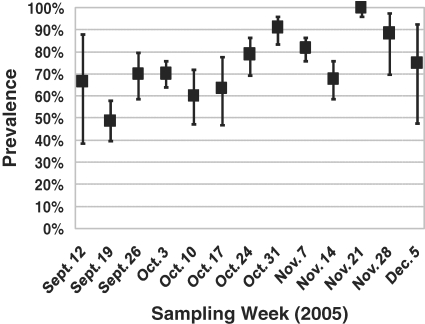
A total of 1 245 fecal samples were collected and cultured, and 918 were positive for Salmonella(73.7%; CI: 71.2–76.2%). Over the 3-month sampling period, weekly sample prevalence estimates ranged from 48.8 to 100% (Fig. 1). Of the total isolates, 673 (73.3%) serogrouped in E, 101 (11.0%) in C1, 79 (8.6%) in C2/C3, 10 (1.1%) in B, and 55 (6.0%) did not agglutinate with the slide agglutination test (groups B to E). Sixteen serotypes were detected, but 91.5% of the isolates were classified to six primary serotypes and their variants: Orion, Anatum, Kentucky, Montevideo, Senftenberg, and Mbandaka (Tab. I).
Figure 1.
The temporal distribution of sample prevalence estimates and corresponding confidence intervals for fecal shedding of Salmonella in commercial feedlot cattle that were sampled as they were treated for apparent respiratory disease.
Table I.
Salmonella serotypes recovered from feces of commercial feedlot cattle that were sampled as they were treated for apparent respiratory disease.
| Serotypes | Serogroup | Number of isolates | % of total |
|---|---|---|---|
| Orion (var.)* | E | 427 | 46.5 |
| Anatum (var.)* | E | 182 | 19.8 |
| Kentucky | C3 | 80 | 8.7 |
| Montevideo | C1 | 69 | 7.5 |
| Senftenberg | E | 45 | 4.9 |
| Mbandaka | C1 | 37 | 4.0 |
| Lille | C1 | 7 | 0.8 |
| Meleagridis | E | 7 | 0.8 |
| Typhimurium (Copenhagen) | B | 6 | 0.7 |
| Reading | B | 4 | 0.4 |
| Others and non-typeable | – | 54 | 5.9 |
| Total | 918 | 100 |
Includes all variants [Orion 15+ (18), Orion 15+ , 34+ (408), Orion (1)] [Anatum 15+ (4), Anatum 15+, 34+ (5), Anatum (173)].
All the isolates (918) were positive for both invA and pagC genes. Thirty-six isolates from 25 lots were positive for the class 1 integron gene and these belonged to serotypes Orion (16), Typhimurium (5), Anatum (5), Kentucky (2), Mbandaka (1), and non-typeable (7). An equal number of integron-negative isolates of each serotype were tested for antimicrobial susceptibilities, except for Typhimurium in which only one integron-negative isolate was available and tested. Roughly a third of integron-positive isolates were susceptible to all of the antimicrobials on the panel (11/36) whereas 16 isolates (44.4%) were categorized as intermediate/resistant to multiple antimicrobials (2 to 7). Only six Salmonella Typhimurium (Copenhagen) were isolated in this study; five were integron-positive with a penta-drug resistance phenotype ACSSuT (A = ampicillin, C = chloramphenicol, S = streptomycin, Su = sulfonamides, T = tetracyclines) and one was integron-negative and susceptible to all antimicrobials. Overall, most of the integron-negative isolates (28/32) were susceptible to all the antimicrobials tested, whereas four isolates were categorized as resistant to one antimicrobial (Tetracycline or Kanamycin). The probability of a Salmonella isolate being susceptible to all antimicrobials on the panel was significantly higher for integron-negative than integron-positive isolates (p < 0.01). The probability of detecting an integron-positive Salmonella isolate was not significantly different from lots with high and low respiratory risk classifications.
3.3. Salmonella prevalence and feedlot data
Only 1 215 of the 1 245 total samples could be linked to corresponding feedlot data. These samples were from 1 153 cattle in 135 arrival lots. Of the 1 215 samples for which feedlot data were available, 1 118 (92.0%) were from cattle being treated for the first time, 87 (7.2%) from cattle being treated for the second time, three (0.2%) from cattle being treated for the third time, and seven (0.6%) from cattle with missing treatment information. Up to 44 samples were collected from a lot (mean = 8.8, median = 6) and the number of samples collected per lot was significantly associated with the number of cattle pulled and treated as expected given the sampling strategy. Of the individual cattle sampled (n = 1 153), most were sampled once (1 092), few sampled twice (60), and one animal sampled three times. Corresponding prevalence estimates were 73.1% (CI: 70.3–75.7%), 80.8% (CI: 72.6–87.4%), and 66.7% (CI: 9.4–99.2%), respectively. Salmonella prevalence did not differ significantly between different sampling times for individual cattle sampled on multiple occasions.
Sample prevalence for Salmonella within lots ranged from 0 to 100% (mean = 74.9%, median = 80%). In ten lots (7.4%), no Salmonella isolates were recovered, but only one sample was collected from each of these lots. The lowest sample prevalence for a lot with more than one sample was 33.3% (5/15). Prevalence was not significantly different for samples from lots deemed high risk (73.7%) versus low risk (75.3%) for respiratory disease on arrival.
3.4. Salmonella status and subsequent individual animal health events
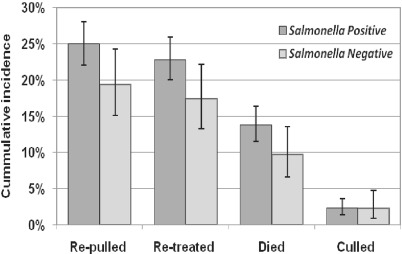
For cattle sampled as they were being treated for the first time (n = 1 118), the crude re-pull, re-treatment, and mortality risks were associated with Salmonella status. However, neither the re-pull (p = 0.27), re-treatment (p = 0.33), mortality (BRDC case fatality risk) (p = 0.34), nor chronic risk (culled before slaughter) (p = 0.75) was statistically different for Salmonella-positive versus -negative cattle in multivariable models (Fig. 2). There also was no difference between groups when assessing the probability of an animal having at least one of these adverse outcomes (p > 0.3). Prevalence of fecal shedding in these cattle was not associated with rectal temperature, the number of times they were pulled or treated, entry weight of the lot, risk category at entry, gender, lot size, or the number of days on feed at sampling (all p values > 0.2). For cattle that subsequently died, there was no association between the probability of culturing Salmonella positive and probability of dying in a hospital versus home pen.
Figure 2.
Cumulative incidence of adverse health outcomes occurring during the feeding period for feedlot cattle that were previously identified as positive or negative for Salmonella based on fecal culture at initial treatment for apparent respiratory disease. Percentages were derived from probability estimates from logistic regression models accounting for the arrival lot of each individual.
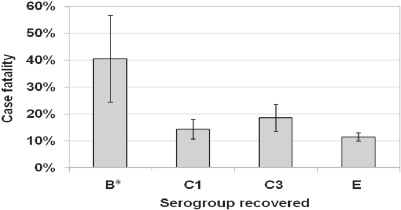
For the subset of cattle that were culture-positive for Salmonella when they were first treated for BRDC (n = 756), the re-treatment, re-pull, and cull risk did not differ statistically based on the serogroup of Salmonella recovered. However, the case fatality risk was significantly higher for cattle shedding Group B Salmonella than for cattle shedding other serogroups (Fig. 3). Due to the diversity of serotypes and corresponding sparse data, similar analyses based on Salmonella serotype could not be evaluated statistically.
Figure 3.
Percentage of respiratory disease cases that subsequently died by serogroup of Salmonella previously recovered from feces obtained when the cattle were first treated. Percentages were derived from probability estimates from a logistic regression model accounting for the arrival lot of each individual. * Significantly higher (p < 0.05) than others.
3.5. Cumulative lot information and within-lot Salmonella prevalence
For the within-lot prevalence analysis, there were 132 lots in which 1 118 cattle were sampled as they were first treated for BRDC. Several factors were unconditionally associated with within-lot Salmonella prevalence including gender (higher in heifers), days on feed (negative association), percentage pulled and treated (positive), percentage pulled and treated more than once (positive), case fatality risk (positive), the proportion of mortalities that occurred in the hospital pen (positive) versus the home pen, and the month in which most cattle were sampled (higher later in the fall) as well as the arrival month (higher later). However, other factors such as mean initial body weight, BRDC risk classification, lot size, overall morbidity, overall mortality, and cull risk were not associated with Salmonella prevalence (all p values > 0.2). The final multivariable model indicated that within-lot prevalence was higher for lots in which the re-treatment risk and the proportion of mortalities occurring in a hospital pen were higher, and for lots that were primarily sampled later in the study (Tab. II).
Table II.
Final multivariable model of lot-level factors associated with the within-lot prevalence of Salmonella in feces of commercial feedlot cattle that were sampled as they were first treated for apparent respiratory diseasea.
| Factors | P-values | Odds Ratios (OR) | 95% Confidence Intervals for OR |
|---|---|---|---|
| Percentageb of treated cattle within a lot | |||
| that were subsequently retreated | < 0.01 | 18.8 | 3.35, 105.63 |
| Percentageb of mortalities within a lot that occurred in a hospital pen | = 0.03 | 1.86 | 1.16, 2.97 |
| Month the lot was primarily sampled | < 0.01 | – | – |
| November | 2.23 | 1.51, 3.29 | |
| October | 1.67 | 1.22, 2.27 | |
| September | - - - - - - - -Reference- - - - - - - - |
Based on results of a lot-level logistic regression model of 132 lots with 1 118 total observations.
Units are ten percentage points.
4. DISCUSSION
In this study, most feedlot cattle that were clinically ill and treated for BRDC were shedding Salmonella in their feces. Although prevalence was high, we could not determine whether prevalence was higher in clinically ill cattle as we did not sample clinically normal cattle. Our primary intent was to investigate potential associations that had not been previously evaluated – whether ill cattle treated for BRDC with antimicrobials were more likely to have adverse clinical outcomes if they were Salmonella positive. Our ability to address the issue adequately may have been compromised by the fact that relatively few cattle were Salmonella negative and thus able to serve as a comparison group. There were higher percentages of adverse clinical outcomes for Salmonella-positive versus -negative cattle, yet none was statistically different after controlling for potential clustering within lots. Results from our assessments of mortalities based on serogroup of Salmonella recovered indicate that serogroup may be an important factor affecting relationships between Salmonella shedding and BRDC case outcomes. Another important finding from this study was that the distribution of mortalities and the overall re-treatment risk at the lot-level were associated with the within-lot prevalence of Salmonella in cattle as they are first treated for BRDC. Although we only studied one feedlot, some inferences may be drawn from the study population as the type of cattle, production system, and the overall disease measures were typical of large commercial feedlots.
Our prevalence estimates of Salmonella seem higher than most estimates reported for feedlot cattle, although reported estimates often vary tremendously and typically represent clinically normal animals. National Animal Health Monitoring Systems surveyed USA feedlots in 1994 and 1999/2000 and provided estimates of Salmonella fecal prevalence in clinically normal cattle of 5.5% and 6.3% for individual cattle, 26% and 22.2% for pens, and 38% and 50.7% for feedlots [14, 19, 32]. A higher prevalence was found in cattle from the southern region of the USA in 1994 [19], and a similar trend was seen in 1999/2000 [14]. Although different detection methods were used, recent studies in Texas had 45% and 50% estimates of fecal prevalence [31, 39], which are much higher than those above and more similar to the prevalence that we found herein. Salmonella prevalence can vary considerably over time within and between feedlots [37]. Fecal cultures can underestimate the true prevalence of Salmonella due to imperfect diagnostic sensitivity and the potential for positive cattle to shed Salmonella at low concentrations or intermittently [35, 36, 41]. Regional differences, time-dependent factors, and imperfect detection methods may explain why prevalence estimates vary widely. However, our study population was clearly different from the aforementioned studies given that cattle were clinically ill in this study and apparently healthy in the other studies.
Although Salmonella may be commonly isolated from feedlot cattle, there are many serotypes and their potential virulence may differ. The frequency distribution of serotypes in this study does not parallel the distribution of serotypes reported for human or cattle disease in the USA National Salmonella Surveillance System1. However, our data is consistent with surveys of clinically normal feedlot cattle in the USA, which have shown that many recovered isolates of Salmonella are dissimilar from the serotypes most commonly associated with human and cattle disease [14, 21, 31]. Serotypes Newport and Typhimurium are common causes of salmonellosis, and multidrug resistant strains are of particular concern for public health and production agriculture [7, 8, 11, 13, 40]. Only six Typhimurium were isolated in this study and five showed the penta-drug resistance phenotype ACSSuT. We recovered no Salmonella Newport. We detected a high proportion of Group E Salmonella and variants of serotypes Orion and Anatum in particular. While these serotypes seem overrepresented and may reflect strains indigenous to the studied feedlot, these results are not unexpected given that others have reported high proportions of Group E Salmonella from feedlot cattle [14, 31], and that national surveys indicate Salmonella strains may cluster within cattle operations [12, 14]. Because we did not recover some potentially important serotypes, results of our analysis of serogroup-specific case fatality risks should be interpreted with caution. When Figure 3 and Table I are interpreted concurrently, it is apparent that the difference in case fatally risk essentially reflects a difference between cattle shedding serotypes Typhimurium and Reading (the only two in serogroup B) and cattle shedding all other recovered serotypes. We cannot conclude that the difference between serogroups would hold if other strains of Salmonella were present. It does indicate however, that cattle treated with antimicrobials for BRDC may be more likely to subsequently die if shedding certain Salmonella strains.
We found that only a small percentage (3.9%) of the Salmonella isolates were positive for the integron class 1 gene. As expected, these isolates were much less susceptible to antimicrobials than the integron-negative isolates. Integron class 1 is an integrative mobilizable element capable of horizontal gene transfer, and may contain genes that confer resistance to ampicillin, chloramphenicol/ florfenicol, aminoglycosides, sulfonamides, and tetracyclines [9, 17]. It has been suggested that the distribution of antimicrobial resistant Salmonella in feedlot cattle may be associated with serotype and reflective of the clonal nature of isolates, the ability of some serotypes to acquire resistance determinants, and/or the presence of an integron [14]. Data from our study indicate that resistance to antimicrobials was relatively uncommon in the Salmonella strains isolated. In addition, we found that the Salmonella recovered from ill cattle in lots that received metaphylactic antimicrobials on arrival (lots designated as high risk for respiratory disease) were not more likely to be integron-positive than Salmonella from other lots. Thus, metaphylactic antimicrobial use did not appear to select for potentially resistant Salmonella strains in this study population.
We hypothesized that Salmonella infections may affect the clinically and economically important disease outcomes often associated with BRDC. Although there were numerically higher percentages of adverse health outcomes for Salmonella positive cattle, none of these were statistically different. Therefore, we can conclude either that a relationship did not exist or that we had insufficient statistical power to identify differences related to overall Salmonella shedding given the potential misclassification and clustering of observations within lots. Although we had data on over 1 000 cattle, we had just over 130 lots and for many lots there were few observations. In addition, a relatively small percentage of cattle cultured negative for Salmonella. Given the imperfect diagnostic sensitivity of culture, our ability to adequately evaluate the potential impact of Salmonella also may have been hindered by misclassifying truly Salmonella positive cattle as Salmonella negative. With one-time sampling of a small volume of feces, imperfect sensitivity of culture, and an apparent prevalence over 70%, it may be conceivable that the true prevalence of Salmonella shedding in this population approached 100%. We did not identify any significant risks factors at the individual animal level. This also may simply be because Salmonella shedding, in general, was extremely common across all segments of the population or because some cattle were shedding commensal or non-virulent strains. Studies of dairy cattle have indicated that it is possible for Salmonella to become established in herds in which clinical salmonellosis or impacts on performance may or may not be apparent [25, 36]. Because associations may depend on the Salmonella strain, grouping all Salmonella positive cattle together may be too crude of a measurement for adequate assessments. However, further classifying Salmonella into more specific categories can result in sparse data sets that impede adequate statistical comparisons. Unfortunately our extremely unequal ratio between some serogroups and others (e.g. B to E), and few isolates of potentially virulent serotypes such as Typhimurium, prevented us from further defining which Salmonella variants are more likely to impact health outcomes.
We found that several lot-level factors were associated with the prevalence of Salmonella in the treated cattle within a lot. Surprisingly, there was no difference in prevalence between the respiratory risk classifications, which could represent several potential Salmonella risk factors such as cattle source, weight, and transport time. In addition, all high risk lots received metaphylactic antimicrobials on arrival to the feedlot. Several independent variables unconditionally associated with prevalence also were associated with each other (data not shown), but the final model identified that re-treatment percentage, percentage of hospital mortalities, and month of sampling were associated with Salmonella prevalence within a lot. There is no published literature on similar studies. There is evidence that hospital pens and treatment or processing facilities may offer an opportunity for Salmonella to disseminate within a feedlot as indirect transmission (fomites and environmental contamination) may be more important than direct transmission in controlling salmonellosis in cattle [28, 30, 36]. Our data indicate that approximately 75% of all cattle entering a hospital setting were shedding Salmonella. In an outbreak of nosocomial Salmonella infections documented in a feedlot, the more days that cattle spent in the hospital pen the higher the Salmonella prevalence [27]. Our data indicate that lots with a higher prevalence of Salmonella in cattle initially treated for BRDC have a higher percentage of cattle that needed to be re-treated and a higher percentage of mortalities occur in a hospital pen versus a home pen. These outcomes, re-treatment and death, would have occurred after initial treatment and sampling in our study. Typically, hospital pen mortalities represent clinically ill cattle, whereas mortalities in home pens represent cattle not recognized as ill or those previously treated and not eligible for further treatment. One of the concerns when analyzing aggregate data such as these is the potential for an ecological bias/fallacy [16]. Therefore, the factors that we have found to be associated with the within-lot prevalence of Salmonella may not be risk factors at the individual level, and thus should only be interpreted at the lot-level. The differences in prevalence that we found across the months of sampling may be due to temporal effects associated with changes in cattle populations or pen environments. There is evidence that Salmonella recovery from feedlot cattle can vary over time even within the same season [37].
The health and production effects of Salmonella within feedlots are poorly documented, but risk factors that are associated with salmonellosis and Salmonella shedding in cattle and other livestock, such as commingling, transport, temporary feed restriction/change, and intensive management are common in feedlot production systems [6, 18, 29, 30, 36]. We found that Salmonella were common in cattle treated for BRDC in the commercial feedlot studied. With high levels of Salmonella exposure and potentially low levels of immunity, many different strains of Salmonella may be capable of causing disease. However, our data indicate that some strains were more likely than others to be associated with high BRDC case fatality risks. Although Salmonella may cause disease, predispose animals to disease, or result in salmonellosis as sequela to other diseases, the isolation of some strains from cattle feces may not be associated with clinical or subclinical effects. Further evaluation of associations with cattle health and production practices, may allow us to develop effective programs for reducing impacts associated with Salmonella in beef cattle productions systems.
Acknowledgements
This work supported by USDA Animal Health 1433 funds through the Kansas Agricultural Experiment Station (Publication No. 08-94-J), by the Department of Diagnostic Medicine/Pathobiology, College of Veterinary Medicine, Kansas State University, and by Cactus Research, Ltd., Cactus, Texas. We thank Neil Wallace, X. Shi, Amy Ecker, Elita Holland, Kip Butler and Brett Beier for help in laboratory and Clay Wheeler, Scott Dodson, Victor Mendoza and Soloman Tapia for sample collections.
Footnotes
Centers for Disease Control, PHLIS Surveillance Data, Salmonella, available at http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm [consulted 15 July 2008].
REFERENCES
- 1.Agresti A. Agresti A. Introduction to Categorical Data Analysis. John Wiley & Sons, Inc.; New York, NY: 1996. SAS and SPSS for categorical data analysis; pp. 267–279. [Google Scholar]
- 2.Barham A.R., Barham B.L., Johnson A.K., Allen D.M., Blanton J.R. Jr, Miller M.F.. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J. Food Prot. 2002;65:280–283. doi: 10.4315/0362-028x-65.2.280. [DOI] [PubMed] [Google Scholar]
- 3.Barkocy-Gallagher G.A., Berry E.D., Rivera-Betancourt M., Arthur T.M., Nou X., Koohmaraie M.. Development of methods for the recovery of Escherichia coli O157:H7 and Salmonella from beef carcass sponge samples and bovine fecal and hide samples. J. Food Prot. 2002;65:1527–1534. doi: 10.4315/0362-028x-65.10.1527. [DOI] [PubMed] [Google Scholar]
- 4.Beach J.C., Murano E.A., Acuff G.R.. Prevalence of Salmonella and Campylobacter in beef cattle from transport to slaughter. J. Food Prot. 2002;65:1687–1693. doi: 10.4315/0362-028x-65.11.1687. [DOI] [PubMed] [Google Scholar]
- 5.Beach J.C., Murano E.A., Acuff G.R.. Serotyping and antibiotic resistance profiling of Salmonella in feedlot and nonfeedlot beef cattle. J. Food Prot. 2002;65:1694–1699. doi: 10.4315/0362-028x-65.11.1694. [DOI] [PubMed] [Google Scholar]
- 6.Belknap E.B., Navarre C.B.. Differentiation of gastrointestinal diseases in adult cattle. Vet. Clin. North Am. Food Anim. Pract. 2000;16:59–86. doi: 10.1016/S0749-0720(15)30137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besser T.E., Gay C.C., Gay J.M., Hancock D.D., Rice D., Pritchett L.C., Erickson E.D.. Salmonellosis associated with S. Typhimurium DT104 in the USA. Vet. Rec. 1997;140:75. [PubMed] [Google Scholar]
- 8.Besser T.E., Goldoft M., Pritchett L.C., Khakhria R., Hancock D.D., Rice D.H.. et al. Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol. Infect. 2000;124:193–200. doi: 10.1017/s0950268899003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd D., Cloeckaert A., Chaslus-Dancla E., Mulvey M.R.. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 2002;46:1714–1722. doi: 10.1128/AAC.46.6.1714-1722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrier D.E., Purdy C.W., DeLoach J.R.. Effects of marketing stress on fecal excretion of Salmonella spp. in feeder calves. Am. J. Vet. Res. 1990;51:866–869. [PubMed] [Google Scholar]
- 11.Dargatz D.A., Wells S.J., Fedorka-Cray P.J., Akkina J.. The veterinarian’s role in diagnosis, treatment, and prevention of multidrug resistant Salmonella Typhimurium DT104. Bov. Pract. 1998;32:1–6. [Google Scholar]
- 12.Dargatz D.A., Fedorka-Cray P.J., Ladely S.R., Ferris K.E.. Survey of Salmonella serotypes shed in feces of beef cows and their antimicrobial susceptibility patterns. J. Food Prot. 2000;63:1648–1653. doi: 10.4315/0362-028x-63.12.1648. [DOI] [PubMed] [Google Scholar]
- 13.Dargatz D.A., Fedorka-Cray P.J., Ladely S.R., Ferris K.E., Green A.L., Headrick M.L.. Antimicrobial susceptibility patterns of Salmonella isolates from cattle in feedlots. J. Am. Vet. Med. Assoc. 2002;221:268–272. doi: 10.2460/javma.2002.221.268. [DOI] [PubMed] [Google Scholar]
- 14.Dargatz D.A., Fedorka-Cray P.J., Ladely S.R., Kopral C.A., Ferris K.E., Headrick M.L.. Prevalence and antimicrobial susceptibility of Salmonella spp. isolates from US cattle in feedlots in 1999 and 2000. J. Appl. Microbiol. 2003;95:753–761. doi: 10.1046/j.1365-2672.2003.02034.x. [DOI] [PubMed] [Google Scholar]
- 15.Dohoo I., Martin W., Stryhn H. Dohoo I., Martin W., Stryhn H. Veterinary Epidemiologic Research. AVC Inc.; Charlottetown, PEI: 2003. Model-building strategies; pp. 317–334. [Google Scholar]
- 16.Dohoo I., Martin W., Stryhn H. Dohoo I., Martin W., Stryhn H. Veterinary Epidemiologic Research. AVC Inc.; Charlottetown, PEI: 2003. Ecologic and group-level studies; pp. 561–577. [Google Scholar]
- 17.Doublet B., Butaye P., Imberechts H., Boyd D., Mulvey M.R., Chaslus-Dancla E., Cloeckaert A.. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 2004;48:2510–2517. doi: 10.1128/AAC.48.7.2510-2517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekperigin H.E., Nagaraja K.V.. Microbial food borne pathogens. Salmonella. Vet. Clin. North Am. Food Anim. Pract. 1998;14:17–29. [PubMed] [Google Scholar]
- 19.Fedorka-Cray P.J., Dargatz D.A., Thomas L.A., Gray J.T.. Survey of Salmonella serotypes in feedlot cattle. J. Food Prot. 1998;61:525–530. doi: 10.4315/0362-028x-61.5.525. [DOI] [PubMed] [Google Scholar]
- 20.Fegan N., Vanderlinde P., Higgs G., Desmarchelier P.. A study of the prevalence and enumeration of Salmonella enterica in cattle and on carcasses during processing. J. Food Prot. 2005;68:1147–1153. doi: 10.4315/0362-028x-68.6.1147. [DOI] [PubMed] [Google Scholar]
- 21.Fluckey W.M., Loneragan G.W., Warner R., Brashears M.M.. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 2007;70:551–556. doi: 10.4315/0362-028x-70.3.551. [DOI] [PubMed] [Google Scholar]
- 22.Frost A.J., O’Boyle D., Samuel J.L.. The isolation of Salmonella spp. from feed lot cattle managed under different conditions before slaughter. Aust. Vet. J. 1988;65:224–225. doi: 10.1111/j.1751-0813.1988.tb14464.x. [DOI] [PubMed] [Google Scholar]
- 23.Galan J.E., Curtiss R. III. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galland J.C., House J.K., Hyatt D.R., Hawkins L.L., Anderson N.V., Irwin C.K., Smith B.P.. Prevalence of Salmonella in beef feeder steers as determined by bacterial culture and ELISA serology. Vet. Microbiol. 2000;76:143–151. doi: 10.1016/s0378-1135(00)00230-3. [DOI] [PubMed] [Google Scholar]
- 25.Gay J.M., Hunsaker M.E.. Isolation of multiple Salmonella serovars from a dairy two years after a clinical salmonellosis outbreak. J. Am. Vet. Med. Assoc. 1993;203:1314–1320. [PubMed] [Google Scholar]
- 26.Griffin D.. Feedlot diseases. Vet. Clin. North Am. Food Anim. Pract. 1998;14:199–231. doi: 10.1016/s0749-0720(15)30251-6. [DOI] [PubMed] [Google Scholar]
- 27.Hancock D.D.. Strategic laboratory use: how to use hypothesis-based laboratory testing. Proc. Annu. Conv. Am. Assoc. Bovine Pract. 1995;28:118–125. [Google Scholar]
- 28.Hardman P.M., Wathes C.M., Wray C.. Transmission of salmonellae among calves penned individually. Vet. Rec. 1991;129:327–329. doi: 10.1136/vr.129.15.327. [DOI] [PubMed] [Google Scholar]
- 29.Hawkins L.L.. Salmonellosis in feedyards: epidemiology, clinical management, and public health risk. Proc. Annu. Conv. Am. Assoc. Bovine Pract. 2000;33:133–136. [Google Scholar]
- 30.House J.K., Smith B.P.. Current strategies for managing salmonella infections in cattle. Vet. Med. 1998;93:756–764. [Google Scholar]
- 31.Loneragan G.H., Brashears M.M.. Effects of using retention-pond water for dust abatement on performance of feedlot steers and carriage of Escherichia coli O157 and Salmonella spp. J. Am. Vet. Med. Assoc. 2005;226:1378–1383. doi: 10.2460/javma.2005.226.1378. [DOI] [PubMed] [Google Scholar]
- 32.Losinger W.C., Garber L.P., Smith M.A., Hurd H.S., Biehl L.G., Fedorka-Cray P.J.. et al. Management and nutritional factors associated with the detection of Salmonella sp. from cattle fecal specimens from feedlot operations in the United States. Prev. Vet. Med. 1997;31:231–244. doi: 10.1016/s0167-5877(96)01143-9. [DOI] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Zone diameter interpretive standards and equivalent minimal inhibitory concentration breakpoints for Enterobacteriaceae. NCCLS; Wayne, PA: 2002. pp. 42–46. [Google Scholar]
- 34.Olah P.A., Sherwood J.S., Logue C.M.. Molecular analysis of Salmonella isolates recovered from processed Turkey carcasses. J. Food Prot. 2005;68:845–849. doi: 10.4315/0362-028x-68.4.845. [DOI] [PubMed] [Google Scholar]
- 35.Palmer J.E., Whitlock R.H., Benson C.E., Becht J.L., Morris D.D., Acland H.M.. Comparison of rectal mucosal cultures and fecal cultures in detecting Salmonella infection in horses and cattle. Am. J. Vet. Res. 1985;46:697–698. [PubMed] [Google Scholar]
- 36.Smith B.P. Smith B.P. Large Animal Internal Medicine. Mosby Inc.; St. Louis, Missouri, USA: 2002. Salmonellosis in ruminants; pp. 775–779. [Google Scholar]
- 37.Smith D.R., Moxley R.A., Clowser S.L., Folmer J.D., Hinkley S., Erickson G.E., Klopfenstein T.J.. Use of rope devices to describe and explain the feedlot ecology of Salmonella by time and place. Foodborne Pathog. Dis. 2005;2:61–69. doi: 10.1089/fpd.2005.2.61. [DOI] [PubMed] [Google Scholar]
- 38.Smith R.A.. Impact of disease on feedlot performance: a review. J. Anim. Sci. 1998;76:272–274. doi: 10.2527/1998.761272x. [DOI] [PubMed] [Google Scholar]
- 39.Stephens T.P., Loneragan G.H., Thompson T.W., Sridhara A., Branham L.A., Pitchiah S., Brashears M.M.. Distribution of Escherichia coli O157 and Salmonella on hide surfaces, the oral cavity, and in feces of feedlot cattle. J. Food Prot. 2007;70:1346–1349. doi: 10.4315/0362-028x-70.6.1346. [DOI] [PubMed] [Google Scholar]
- 40.Tollefson L., Angulo F.J., Fedorka-Cray P.J.. National surveillance for antibiotic resistance in zoonotic enteric pathogens. Vet. Clin. North Am. Food Anim. Pract. 1998;14:141–150. doi: 10.1016/s0749-0720(15)30285-1. [DOI] [PubMed] [Google Scholar]
- 41.Warnick L.D., Crofton L.M., Pelzer K.D., Hawkins M.J.. Risk factors for clinical salmonellosis in Virginia, USA cattle herds. Prev. Vet. Med. 2001;49:259–275. doi: 10.1016/s0167-5877(01)00172-6. [DOI] [PubMed] [Google Scholar]
- 42.Wray C., Davies R.H. Wray C., Wray A. Salmonella in Domestic Animals. CABI Publishing; New York, NY, USA: 2000. Salmonella infections in cattle; pp. 169–190. [Google Scholar]
- 43.Zhang H., Shi L., Li L., Guo S., Zhang X., Yamasaki S., Miyoshi S., Shinoda S.. Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from healthy humans in China. Microbiol. Immunol. 2004;48:639–645. doi: 10.1111/j.1348-0421.2004.tb03473.x. [DOI] [PubMed] [Google Scholar]