Abstract
Double-strand breaks (DSBs) in DNA are lethal unless repaired. Faithful repair requires processing of the DSB ends and interaction with intact homologous DNA, which can produce genetic recombinants. To determine the role of nucleases in DSB end-processing and joint molecule resolution, we studied recombination at the site of a single DSB, generated by induction of the I-SceI endonuclease, during meiosis of fission yeast lacking Rec12 (Spo11 homolog) and, hence, other DSBs. We find that in the presence of the MRN (Rad32-Rad50-Nbs1) complex efficient recombination requires Ctp1, the ortholog of the nuclease Sae2, but not the nuclease activity of MRN. In the absence of MRN, exonuclease 1 (Exo1) becomes the major nuclease required for efficient recombination. Our data indicate that MRN enables access of Ctp1 to the DSB but blocks access of Exo1. In our assay, the Rad16-Swi10 nuclease, required for nucleotide excision-repair, is required for efficient recombination, presumably to remove heterologous DNA at the end of the I-SceI cut site. Another nuclease, the Mus81-Eme1 Holliday junction resolvase, is required to generate crossovers accompanying gene conversion at the I-SceI cut site. Additional, previously published evidence indicates that these 5 nucleases play similar roles in wild-type fission yeast meiotic recombination and in the repair of spontaneous and damage-induced mitotic DSBs. We propose that in wild-type meiosis MRN, in conjunction with Ctp1, removes the covalently attached Rec12 protein from the DNA end, which is then resected by Ctp1 and other activities to produce the single-stranded DNA necessary for further steps of DSB repair.
Keywords: DNA resection, DSB repair, S. pombe, Sae2
Genome integrity is essential for cell viability. Genomic DNA double-strand breaks (DSBs), introduced by cellular processes or by exogenous DNA damaging agents, must be repaired before chromosome segregation to ensure that each daughter cell receives a complete genomic complement. Faithful repair of a DSB (i.e., without loss or gain of DNA) requires interaction of the broken DNA with an intact homologous DNA duplex, which can provide a template for DNA synthesis and replacement of nucleotides lost at the site of the DSB. If the 2 DNA molecules differ at or near the DSB, this interaction can produce genetic recombinants between the sites of difference (genetic markers).
An essential step in DSB repair by homologous recombination is thought to be the generation of long single-stranded (ss) DNA with a 3′ end at the DSB and the coating of this ss DNA with Rad51 protein or its prokaryotic homolog RecA (1) (see Discussion). The resulting protein-DNA filament can engage an intact duplex and form base-pairs with its complement by displacing 1 strand of the target duplex to form a ss D-loop, one of several types of joint DNA molecules arising during homologous recombination. The D-loop can be enlarged by DNA synthesis primed by the invading 3′ end. Although numerous fates of the D-loop have been postulated (1), one fate is pairing with its complement in the initially broken DNA to form a crossed-strand structure, a Holliday junction (HJ). Resolution of the HJ can produce intact DNA potentially recombinant for markers at or near the DSB site (by gene conversion within or near the region of the D-loop) or for more distant markers flanking the D-loop (by crossing over).
Cells have multiple mechanisms for DSB repair, depending on the nature and context of the DNA breakage. In most postulated mechanisms of repair by homologous recombination, nucleases are essential for at least 2 steps: generation of ss DNA ends at the DSB and resolution of the joint molecule to form a crossover (see Discussion). Cells contain a wide variety of nucleases, and sorting out their roles in recombination requires a combination of genetics and biochemistry. We address here the roles of 5 nucleases in recombinational repair of a DSB during meiosis, the formation of haploid gametes from diploid precursor cells.
During meiosis, DSBs are introduced by the Spo11 protein as part of the program to ensure proper segregation of homologous chromosomes (homologs) at the first meiotic division (2). A topoisomerase II-like protein thought to act as a dimer, Spo11 makes a DSB by 2 transesterification reactions, in which a tyrosine residue of each Spo11 subunit attacks a phosphodiester bond in DNA and becomes covalently linked to the 5′ end of the DNA (2). In the budding yeast Saccharomyces cerevisiae and in mice Spo11 is subsequently released from the DSB attached to a short oligonucleotide (3). In S. cerevisiae the responsible endonuclease requires the MRX (Mre11-Rad50-Xrs2) nuclease complex (3), but the role of MRX in this reaction and the identity of nucleases in subsequent steps are unclear.
To identify the nucleases required for DSB resection and repair, we have studied recombination at the site of a single DSB induced during meiosis of the fission yeast Schizosaccharomyces pombe. The S. pombe Spo11 and MRX homologs are designated Rec12 and MRN (Rad32-Rad50-Nbs1), respectively (4, 5). In addition to MRN, we studied other nucleases. Ctp1 is a homolog of S. cerevisiae Sae2, a nuclease that interacts with MRX (6, 7). Exonuclease 1 (Exo1) is a double-stranded-specific exonuclease that generates long 3′ ss tails (8). The Rad16-Swi10 complex, homolog of the human XPF-ERCC1 complex, is involved in excision of damaged nucleotides in DNA (9), and Mus81-Eme1 is a HJ resolvase (10–13). To study the steps of meiotic DSB repair after Rec12 removal, we created a meiotic DSB without an attached protein by inducing the site-specific endonuclease I-SceI (14) in the absence of Rec12. Because S. pombe has only 3 chromosomes, abundant viable gametes (spores) are formed in the absence of meiotic recombination (i.e., by nearly random segregation of chromosomes) (15, 16). High viable spore yields allowed us to measure recombination between markers at and near the I-SceI site as a measure of repair of this specific DSB. Our results, which reveal interactions among these nucleases and a critical regulatory role for the MRN complex, along with previously published information, allow conclusions about DSB repair in wild-type meiosis and repair of mitotic DSBs as well.
Results
An I-SceI Cut Site Is a Meiotic Recombination Hotspot.
To generate a single DSB during meiosis, we inserted into the S. pombe ade6 gene 80 bp of DNA containing the 18-bp recognition site for the I-SceI homing endonuclease (14) to generate ade6-3061. A similar insertion, ade6-3062, differs from ade6-3061 only by a single bp change that blocks I-SceI cutting (14). By selecting Ade+ (recombinant) spores, we measured recombination between these alleles and ade6-52, a single bp change located 772 bp from the insertion site (Fig. 1). To supply I-SceI and to eliminate all other (Rec12-dependent) DSBs, we replaced the rec12 coding sequence with that of I-SceI (17); rec12 is detectably expressed only during meiosis (18, 19), as expected for a protein with a meiosis-specific role. In meiotic crosses between strains containing ade6-3061 and ade6-52, we observed 1,120 ± 80 Ade+ spores per million total viable spores (Table 1 and Fig. 2). Crosses between ade6-3062 and ade6-52 produced only 6.1 ± 0.8 Ade+ per million spores [supporting information (SI) Table S1]. Thus, the I-SceI cut site, ade6-3061, is a strong hotspot of meiotic recombination relative to the noncuttable allele ade6-3062. [In each of the mutants studied below, crosses of ade6-3062 x ade6-52 also produced very few Ade+ recombinants (see Table S1).]
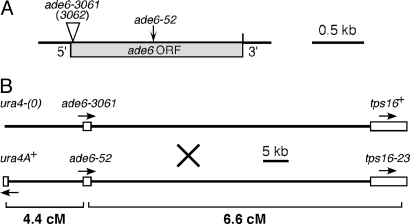
Fig. 1.
Genetic markers used in this study. (A) ade6 alleles. Both ade6-3061 (cleavable by I-SceI) and ade6-3062 (noncleavable by I-SceI) are 80 bp insertions (see Table S2); not drawn to scale. ade6-52 (G1670A) is a missense mutation. (B) Markers flanking ade6 for crosses in Fig. 4. tps16 is synonymous with ags1. See ref. 27 for details about these markers. Open boxes indicate ORFs, whose transcriptional directions are indicated by arrows. The diagram is drawn to physical scale.
Table 1.
Ctp1 or Exo1, each modulated by MRN, is required for efficient DSB-promoted recombination
| exo1 | rad32 | ctp1 | I-SceI induced recombinants* |
|
|---|---|---|---|---|
| rad50+ | rad50Δ | |||
| + | + | + | 1120 ± 80 (36) | 1800 ± 200 (20) |
| Δ | + | + | 1100 ± 92 (15) | 440 ± 64 (11) |
| + | D65N | + | 1390 ± 240 (6) | — |
| Δ | D65N | + | 1130 ± 220 (3) | — |
| + | + | Δ | 230 ± 24 (8) | 1380 ± 150 (12) |
| Δ | + | Δ | 150 ± 38 (8) | 390 ± 74 (6) |
*Ade+/million viable spores in crosses of ade6-3061 (I-SceI cut site) × ade6-52, as mean ± SEM of (n) crosses. See Results for statistical significance of the differences between pairs of data. In most cases, crosses with ade6-3062 (noncuttable allele) gave < 10 Ade+/million viable spores (Table S1). —, not determined, because in cells Rad32 (Mre11) has no known activity in the absence of Rad50 (24).
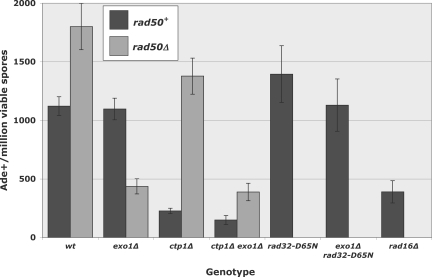
Fig. 2.
Recombination at a single I-SceI cut site depends on multiple nucleases. Data are the frequency (mean ± SEM) of Ade+ recombinants from crosses with the I-SceI-cuttable allele ade6-3061 and ade6-52 (Fig. 1A) in the indicated mutant background. Data are from Table 1 and Results (for rad16Δ).
DSBs at the I-SceI Cut Site Arise During Meiotic Replication, Earlier than Rec12-dependent DSBs.
To test directly for DSBs at the ade6-3061 site, as predicted by its hotspot activity, we analyzed DNA from meiotically induced cells by Southern blot hybridization (Fig. 3 and Fig. S1). At about the time of DNA replication (≈2 h after meiotic induction) (Fig. S2), DSBs at the I-SceI site were first detectable (see Fig. 3 and Fig. S1). The fraction of DNA broken at the I-SceI site increased to a maximum of ≈0.6% at ≈3 h and then steadily decreased to a nearly undetectable level at ≈5 h. As expected, no DSBs were detectable at the noncuttable allele ade6-3062 (Fig. S3). I-SceI-dependent DSBs arose earlier than wild-type (Rec12-dependent) DSBs at the natural meiotic break site mbs1, which were first detectable at 3 to 3.5 h after meiotic induction (Fig. S4) (20). Both types of DSBs, however, largely disappeared by 5 h. I-SceI-dependent DSBs may arise earlier than wild-type Rec12-dependent DSBs because Rec12 requires nearly a dozen other proteins to effect DSB formation (21), whereas I-SceI acts alone (14).
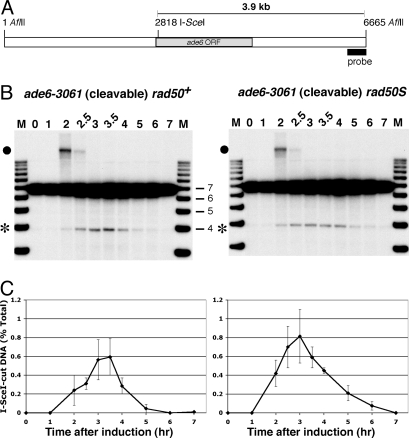
Fig. 3.
An I-SceI-dependent DSB arises early in meiosis and is repaired in the rad50S mutant background. (A) Diagram of the 6.7 kb AflII fragment on chromosome 3 containing ade6 with the location of the probe used in B. (B) Southern blot of DNA prepared at the indicated times after meiotic induction and digested with AflII. Digested DNA was separated on agarose gels, blotted, and hybridized with the probe indicated in A. (Left) Strain GP6308 (ade6-3061 rad50+ rec12-172∷I-SceI). (Right) Strain GP6306 (ade6-3061 rad50S rec12-172∷I-SceI). Size markers (kb) are in lane M. The asterisk (*) indicates the position of the 3.9 kb meiosis-specific I-SceI broken fragment. The bullet (●) indicates the position of replication intermediates at 2 hr, deduced from flow cytometry of cellular DNA content (Fig. S2) and the apparent mass of the species. A replicate experiment is shown in Fig. S1. (C) Quantification of I-SceI-dependent meiotic DSBs. The radioactivity on the blots in B was quantified with a phosphorimager. For each strain, data are the percent of broken DNA (mean and range of 2 inductions) at each time relative to the sum of all radioactivity in the corresponding lane and after subtraction of the uninduced (0 hr) value at the position of the I-SceI-specific band.
The rad50S mutation alters the Rad50 subunit of the MRN complex such that wild-type (Rec12-dependent) DSBs accumulate, with Rec12 protein covalently linked to the ends (22, 23). I-SceI-dependent DSBs, however, did not accumulate in the rad50S mutant (see Fig. 3 and Fig. S1). These data show that the activity blocked by the rad50S mutation is not required for processing of a DSB free of an attached protein. These results suggest that the rad50S mutant is specifically blocked in removal of Rec12 from meiotic DSBs (see Discussion).
MRN Nuclease Complex Is Not Required for Recombination, But in Its Absence Exo1 Strongly Promotes Recombination.
Rad50 is essential for repair of wild-type (Rec12-dependent) DSBs (24). We therefore tested a requirement for the MRN complex in repair of I-SceI-dependent DSBs, by measuring recombination in a rad50Δ mutant. Rad50 is a large polypeptide to which the other components of the complex bind. In the absence of Rad50, the Mre11 (Rad32) subunit, which in isolation has nuclease activity (25), apparently has no activity in cells because the phenotypes of rad50Δ and mre11Δ (or rad32Δ) mutants, alone or in combination, are the same (24). In contrast to our initial expectation of a requirement for MRN, there was a slight (1.6-fold) increase in the recombinant frequency in the rad50Δ mutant compared to wild-type (see Table 1 and Fig. 2) (P < 0.005). This result shows that the MRN complex is not required for I-SceI-dependent recombination.
Exo1 is meiotically induced and has the appropriate substrate specificity and activity to generate a ss DNA tail with a 3′ end at a DSB (8). We found, however, that the exo1Δ mutation had no discernible effect on recombination at the I-SceI DSB in rad50+ cells (see Table 1 and Fig. 2) (P > 0.8). In contrast, in rad50Δ cells Exo1 was clearly required for efficient recombination: the Ade+ frequency was reduced by a factor of 4.1 in exo1Δ rad50Δ relative to rad50Δ, or by a factor of 2.5 relative to wild-type (exo1+ rad50+) (see Table 1 and Fig. 2) (P < 0.001 for each comparison). These results show that Exo1 is not required for I-SceI-dependent recombination when the MRN complex is present, as shown previously for Rec12-dependent recombination (26). For wild-type levels of recombination Exo1 is required, however, when the MRN complex is absent. We propose an explanation for these results in the Discussion.
MRN Nuclease Activity Is Not Required for Recombination, Even in the Absence of Exo1.
The MRN nuclease domain resides in the Rad32 subunit (Mre11 homolog) (4, 25) and, by analogy with the corresponding S. cerevisiae mutation, the Rad32 D65N amino acid change inactivates this nuclease (4). The rad32-D65N mutation had no significant effect on I-SceI-dependent recombination (see Table 1 and Fig. 2) (P > 0.3). This result contrasts sharply with that in wild-type (Rec12-dependent) meiosis: the rad32-D65N mutation reduces the viable spore yield by a factor of 103 to 104 and is suppressed by rec12Δ (27). This result suggests that the Rad32 nuclease is required specifically for the removal of Rec12 from DSBs (see Discussion).
The recombinant frequency was also not significantly changed when both Exo1 and Rad32 nucleases were inactivated (see Table 1 and Fig. 2) (P > 0.97). Thus, although Exo1 is required for efficient recombination when the MRN complex is absent, it is not required when just the nuclease activity of MRN is absent. This result suggested that some nuclease other than Exo1 and Mre11 is required for recombination in the presence of MRN.
Ctp1 Nuclease Is Required for Efficient Recombination But only in the Presence of the MRN Complex.
To identify the nuclease suggested by the results above, we tested the ctp1Δ mutant (6). Ctp1 is a distantly related homolog of Sae2, an S. cerevisiae protein that is required for repair of wild-type (Spo11-dependent) meiotic DSBs (2, 3) and that has endonuclease activity modulated by the MRX complex (7). The S. pombe ctp1Δ mutant had significantly reduced I-SceI-dependent recombination, about 5 times less than that of wild-type (ctp1+) (see Table 1 and Fig. 2) (P < 0.001). In the absence of Rad50, however, the recombinant frequency was only slightly, and not significantly (P > 0.1), reduced by the ctp1Δ mutation. In the exo1Δ background, ctp1Δ also had a much greater effect in the presence of Rad50 (7.3-fold reduction; P < 0.001) than in its absence (11% reduction; P > 0.63). Thus, Ctp1 appears to depend on Rad50 for its action.
Paradoxically, in the ctp1Δ exo1Δ background, removing Rad50 increased the recombinant frequency by a factor of 2.6 (P < 0.03). Thus, the activities of Ctp1, Exo1, and perhaps other nucleases are clearly modulated by Rad50. We discuss these interactions later in the text.
Rad16-Swi10 Nuclease Complex Is Required for Efficient Recombination with Heterology at the DSB.
Each end of the I-SceI-dependent DSB studied here has ≈40 bp of sequence heterology relative to the ade6+ DNA at that site (see Fig. 1 and Table S2). This heterology is likely to impede strand invasion promoted by Rad51 or subsequent steps and might need to be removed by a nuclease. We previously found that the Rad16-Swi10 nuclease is required for efficient recombination between ade6-52 and a palindrome at the same site in the ade6 gene as the I-SceI site used here (27). DSBs arise at the palindrome during replication just as they do at the I-SceI site (27) and also leave heterologies at the ends. Similar to the palindrome results, rad16Δ reduced recombination by a factor of about 3 in crosses with the I-SceI DSB: rad16Δ mutant crosses gave 390 ± 38 Ade+ per million viable spores, compared to 1,120 ± 80 for rad16+ crosses (see Table 1 and Fig. 2) (P < 0.002). Because the I-SceI insertion inactivates the ade6 gene, it was not possible to test a requirement for Rad16 in the absence of heterology, but meiotic and mitotic recombination between single bp (point) mutations in ade6 and ade7 is not dependent on Rad16 (28) (Table S3). Thus, the available evidence indicates that Rad16 is required for efficient recombination only when there is DNA heterology at the DSB end.
Mus81-Eme1 HJ Resolvase Is Required for Crossing over and Stimulates Gene Conversion at the I-SceI DSB.
The assays above, for Ade+ recombinants, measured intragenic recombination. Where tested, ade6 intragenic recombination is entirely gene conversion (nonreciprocal recombination) (29). Gene conversion is often accompanied by reciprocal recombination (crossing over), about 65 to 80% of the time in S. pombe (17, 30). To determine if the I-SceI-dependent Ade+ recombinants also were accompanied by a crossover, we measured recombination of the ura4A+ and tps16-23 markers flanking ade6 (see Fig. 1). Among the selected Ade+ spores, 38% had an exchange between ura4A+ and tps16-23 (Fig. 4); among the total spore population, <1% are expected because the cells are rec12Δ (15) and the I-SceI DSB arises at a low level (see Fig. 3). Thus, as in wild-type (Rec12-dependent) recombination, gene conversion is frequently accompanied by a crossover, although only about half as frequently as in wild type.
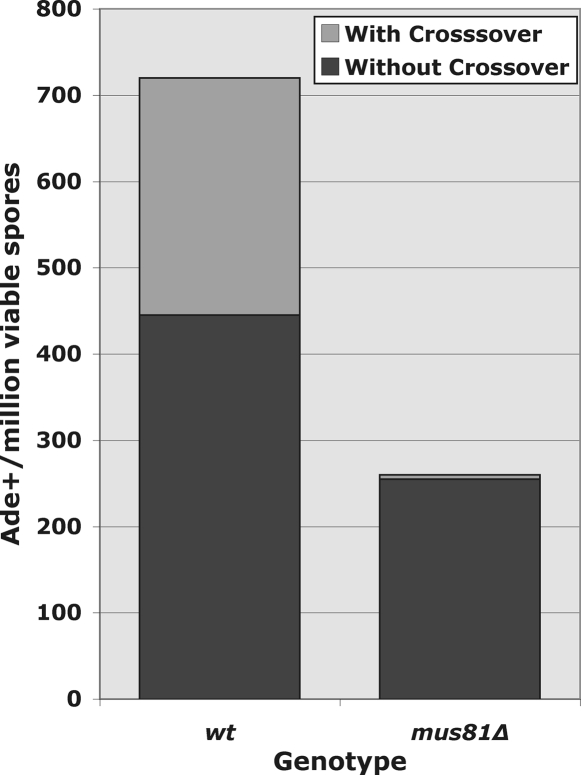
Fig. 4.
Crossing-over at an I-SceI-dependent DSB depends on Mus81. The ade6 alleles crossed and the flanking markers are shown in Fig. 1B. In mus81+ cells, Ade+ recombinants are resolved into cross-overs (38%) and noncross-overs (62%). The Ade+ frequencies were 720 and 260 Ade+ per million viable spores in the mus81+ and mus81Δ crosses, respectively; 338 and 289 Ade+ spore colonies were tested for flanking marker inheritance in the mus81+ and mus81Δ crosses, respectively.
Crossovers in wild-type (rec12+) cells are dependent on the Mus81-Eme1 nuclease (11, 12). We found that I-SceI-dependent crossovers are also nearly entirely dependent on Mus81 (see Fig. 4): only 2% of the Ade+ spores from the mus81Δ mutant cross had an exchange between ura4A+ and tps16-23, as opposed to 38% in mus81+. The frequency of gene convertants (Ade+ spores) was reduced by a factor of about 3 in the mus81Δ mutant (see Fig. 4 legend).
Discussion
Our results can be summarized as follows. (i) Ctp1 is the major nuclease required for efficient meiotic recombination at a DSB made by the I-SceI endonuclease. (ii) The MRN complex is required for the action of Ctp1 but prevents the action of Exo1. (iii) Exo1 is the major nuclease in the absence of MRN. (iv) In the absence of both Ctp1 and Exo1, other nucleases can weakly promote recombination, most effectively in the absence of MRN. (v) Rad16-Swi10 is required for efficient recombination, perhaps because of the heterology at the DSB. (vi) Mus81-Eme1 is required for essentially all crossing over at the DSB.
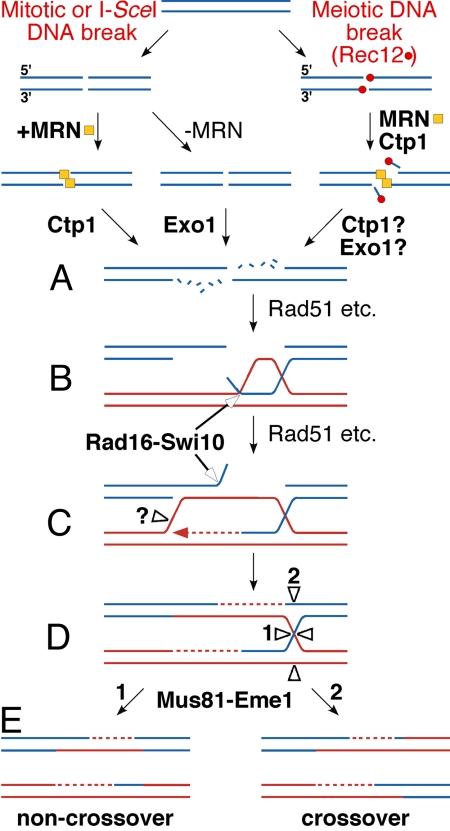
These and previously published results from S. pombe and additional species lead us to propose the following model for DSB end-processing and repair in S. pombe (Fig. 5). We interpret our results in light of established properties of the enzymes studied here and the reactions they promote (21). In our model, the MRN complex and Ctp1 bind to a DSB end. If a protein such as Rec12 is covalently linked to the DSB end, the MRN-Ctp1 combination cleaves off the protein with an oligonucleotide attached. This reaction requires the nuclease activity of MRN and does not occur in rad50S mutants. The MRN-Ctp1 combination resects the 5′-ended strand to form a long 3′ ss tail; this reaction does not require the MRN nuclease activity. If MRN is absent, Ctp1 does not act and Exo1 resects the end. In the absence of both Ctp1 and Exo1, another nuclease (or nucleases) can inefficiently resect the end; this nuclease may be Dna2 combined with the Rqh1 helicase, as shown for their homologs in S. cerevisiae mitotic cells (31). Resection by this additional nuclease is most active in the absence of MRN, which may block access to DSB ends by this nuclease as well as by Exo1. Aided by Rad51 and other strand-exchange proteins, the 3′ tail forms a D-loop, which is converted into a HJ. Mus81-Eme1 resolves the HJ to form recombinant DNA with either the parental (noncrossover) or the nonparental (crossover) configuration of markers flanking the region of strand exchange. Below, we discuss the implications of our findings for mitotic recombination and DSB repair and for meiotic recombination promoted by Rec12, which unlike I-SceI endonuclease remains covalently linked to the DSB ends.
Fig. 5.
Model for nucleolytic processing of DSB ends in S. pombe. Nucleases are indicated by bold type. Each line is one strand of DNA, with blue designating one parental DNA and red the other. A DSB is formed in intact DNA in mitotic cells by various processes, such as replication and transcription, or in meiotic cells by the experimentally induced I-SceI endonuclease or by Rec12 (red ball), which becomes covalently attached to the DNA. MRN (yellow square) and Ctp1 are required to cleave Rec12, attached to an oligonucleotide, from the DNA. The DNA strands with 5′ ends are resected by Ctp1 or Exo1, or other unidentified nucleases acting at low level, to produce long 3′ single-stranded ends (A). Rad51 strand-exchange and accessory proteins produce a D-loop (B). Heterologous DNA is cleaved off the hybrid DNA by Rad16-Swi10, DNA synthesis (dashed line) replaces the nucleotides lost in the preceding steps, an unknown nuclease (?) cleaves the D-loop, and annealing of single-stranded DNA produces a Holliday junction (HJ) (C and D). The HJ is cleaved by Mus81-Eme1 in direction 1 or 2 to produce recombinant DNA (E) with parental (Left) or nonparental (Right) configuration of markers flanking the region of strand exchange and DNA synthesis.
Although the recombinant frequencies measured here (see Table 1 and Fig. 2) might be affected by mismatch correction, we used the same pair of ade6 markers in all of the experiments; a mutation, such as exo1Δ, that alters mismatch correction would be expected to have the same effect in all genetic backgrounds. We therefore interpret the increases and decreases in recombinant frequency among the mutants studied as a reflection of DSB end resection, a role compatible with the known enzymatic activities of MRN, Ctp1, and Exo1.
Ctp1: The Major Nuclease Required for Recombination and DSB Repair.
Among the nuclease mutants we tested, ctp1Δ had the greatest effect: it reduced the recombinant frequency by a factor of 5: that is, to 20% of the wild-type level (see Table 1 and Fig. 2). In the absence of MRN, the ctp1Δ reduction was modest (to 75% of the wild-type level) and of questionable significance (P > 0.1). Thus, the action of Ctp1 in meiotic cells, as measured by I-SceI-promoted recombination, is almost entirely dependent on the MRN complex. Purified Sae2, the S. cerevisiae homolog of Ctp1, and CtIP, the human homolog, are also activated by the MRN complex (7, 32).
The role of the MRN complex is not entirely clear, but we propose that it effectively binds Ctp1 to DSB ends and allows Ctp1 to resect those ends. Although MRN also has a nuclease domain, in the Rad32 (Mre11 homolog) subunit, this nuclease appears not to be required for I-SceI-dependent recombination (see Table 1 and Fig. 2). The rad32-D65N mutation that we used to inactivate the nuclease active site (4) leaves the MRN complex intact (33). Thus, these data indicate that Ctp1, not Rad32, is the nuclease responsible for DSB end resection at the I-SceI DSB. The rad50S mutation may indirectly block the Rad32 (Mre11) nuclease activity, because rad50S and rad32-D65N mutants have similar meiotic phenotypes (see next paragraph). In accord with this view, the rad50S mutation did not significantly affect I-SceI DSB repair (see Figs. 3 and Fig. S1), and the rad32-D65N mutation did not affect I-SceI-dependent recombination (see Table 1 and Fig. 2).
The situation is slightly different in wild-type cells, where Rec12 remains covalently linked to the DSB end (see Fig. 5, Top right) (22, 23). We suppose that Rec12, like Spo11 (3), must be cleaved off the DSB end by an endonuclease. This nuclease may be either Ctp1 or Rad32, as ctp1Δ and rad32-D65N mutants yield very few viable spores, a phenotype suppressed by rec12Δ (6, 27). The low viable spore yield of ctp1Δ and rad32-D65N mutants is similar to that of rad50Δ, rad50S, and rad32Δ mutants (4, 6, 24, 27); in contrast, exo1Δ mutants yield abundant viable spores (26). These data suggest that Ctp1 and MRN, but not Exo1, are required to cleave Rec12 from the DSB end. S. cerevisiae Sae2, the Ctp1 functional homolog, is required to cleave Spo11 off the DSB end; MRX and Exo1 were not tested (3). Direct assays for the cleavage of Rec12 off a DSB end are needed, however, for a firm conclusion. Resection after Rec12 removal may occur as we propose for I-SceI DSB ends, or may be altered by the proteins of the putative Rec12 complex.
Exo1: The Major Nuclease in the Absence of the MRN Complex.
When MRN was absent because of the rad50Δ mutation, exo1Δ reduced the recombinant frequency by a factor of 4, in contrast to ctp1Δ, which did not significantly reduce the frequency (see Table 1 and Fig. 2). When MRN was present, however, the opposite effects were seen: recombination was significantly reduced by ctp1Δ but not by exo1Δ. Thus, Exo1 is the major nuclease in the absence of the MRN complex but plays no significant role in its presence. Exo1 is the most potent double-stranded DNA exonuclease in extracts of meiotically induced cells and has the appropriate activity to produce the 3′-ended tails for strand invasion (D-loop formation) (see Fig. 5) (8, 26). Although Exo1 has a role in mismatch repair during meiotic gene conversion between some markers in wild-type S. pombe (26, 34), it has no significant role in meiotic crossing over in wild-type (26) or in gene conversion at I-SceI-induced DSBs in rad50+ cells. The data in Table 1 and Fig. 2 and the activity of Exo1 on linear DNA (8) indicate that S. pombe Exo1 is blocked by the MRN complex from access to the DSB end and plays little or no role in repair of wild-type meiotic DSBs. In contrast, S. cerevisiae exo1Δ mutants undergo meiotic crossing over at about one-half the rate of crossing over in wild-type cells (35, 36), and Exo1 aids resection of DSBs in S. cerevisiae mitotic and meiotic cells (31, 36, 37). Additional marked differences between meiotic recombination in S. cerevisiae and S. pombe have been noted (21).
Positive and Negative Roles for the MRN Complex.
The rad50Δ mutation decreases the recombinant frequency in exo1Δ (ctp1+) mutants but increases it in ctp1Δ mutants (see Table 1 and Fig. 2), indicating that Rad50, or the MRN complex, has both positive and negative roles in recombination. These and previously published data indicate that MRN aids the action of Ctp1 at a DSB end but blocks that of Exo1. Other investigators, noted below, have also reported positive and negative interactions among 4 DNA repair proteins, MRN, Ctp1, Exo1, and the Ku complex, which is involved in nonhomologous DNA end-joining. Based on these reports and our observations (see Table 1 and Fig. 2), we propose that MRN but not Ku blocks Exo1 in meiotic cells, and Ku but perhaps not MRN blocks Exo1 in mitotic cells; in both cell types, MRN aids Ctp1.
A positive interaction between MRN and Ctp1 is clear: MRN aids localization of Ctp1 to mitotic DSBs, and each protein is required for homologous recombination of linear transforming DNA in mitotic cells (6). In human mitotic cells, the Ctp1 homolog CtIP immunoprecipitates with Rad50, and the purified proteins together have Mg2+-dependent ss endonuclease activity lacking in either single protein preparation (32); a similar cooperative activity is seen with S. cerevisiae MRX and Sae2 (7). As in S. pombe mitotic cells (6), depletion in human cells of either CtIP or Mre11 reduces DSB-induced homologous recombination (32). We suggest that these results with mitotic cells differ from our results with meiotic cells (see Table 1 and Fig. 2) because Exo1, being inhibited by the Ku complex (see below), cannot resect DSB ends in mitotic cells.
A negative interaction between MRN and Exo1 is shown by the lack of an exo1Δ recombination phenotype except in rad50Δ (MRN-deficient) meiotic cells (see Table 1 and Fig. 2). In both ctp1+ and ctp1Δ strains, the recombinant frequency is higher in rad50Δ derivatives, and this recombination is largely Exo1-dependent. In mitotic cells, exo1Δ has a noticeable phenotype only in pku70Δ rad32Δ cells (6, 38), indicating that Ku also has a negative role. Note that pku70, the gene encoding one of the Ku subunits, is expressed in meiotic cells (18). The basis of this differential inhibition in mitotic and meiotic cells remains to be determined.
In S. cerevisiae the situation is different or more complex. exo1Δ single mutants have reduced meiotic and mitotic recombination (35–37), even when both MRN and Ku are present. DSB end resection is reduced by exo1Δ, partially in sae2Δ and strongly in sgs1Δ background (31, 37). Furthermore, sae2Δ mutants are only mildly sensitive to ionizing irradiation, whereas S. pombe ctp1Δ mutants are severely sensitive (6). In S. cerevisiae, MRX and Ku may only partially block the action of Exo1 and other nucleases required for DSB repair and recombination.
Biasing the Mus81-Eme1 HJ-Resolvase Toward Crossing over.
In most models of recombination, HJs are essential intermediates to crossovers (1). HJs can be resolved, however, into either crossovers or noncrossovers (see Fig. 5). If all recombination events involve the resolution of HJs, then the frequency of resolution to crossovers can be equated with the fraction of gene convertant recombinants that have crossover (nonparental) inheritance of markers flanking the region of strand exchange, the region in which gene conversion can occur (see Fig. 5). However, gene conversions without crossovers may also arise by other mechanisms, such as synthesis-dependent strand annealing (SDSA), in which HJs are not involved (1). Therefore, the fraction of observed gene convertants that have an associated crossover may underestimate the frequency with which HJs are resolved to give crossovers. The observed fraction varies markedly, depending on the organism and locus tested; fractions from 15 to 85% have been reported (39). In S. pombe, the associated fraction varies from 65 to 80% for the 4 loci tested (12, 17, 30). Equally frequent cleavage of HJs in the 2 complementary directions (see Fig. 5) would result in a fraction of 50%; this fraction would be less if SDSA contributes significantly to gene conversion events. Thus, in the context of these models, S. pombe HJ resolution is at least modestly biased toward crossing over and may be highly biased.
Genetic and physical evidence strongly indicates that Mus81-Eme1 is the major, or sole, meiotic HJ resolvase in S. pombe (10–13). The genetic evidence cited above indicates that HJ resolution by Mus81-Eme1 is biased toward crossovers in wild-type (Rec12-dependent) meiotic recombination. We found, however, that this bias is significantly reduced in I-SceI-induced recombination: only 38% of the gene convertants have an associated crossover. These crossovers are Mus81-dependent (see Fig. 4), as they are in wild type (Rec12-dependent) recombination (11, 12). In addition to greatly reducing the frequency with which crossovers are associated with gene conversions, deletion of mus81 results in a 64% reduction in gene conversion frequency (see Fig. 4). If this reduction reflects the proportion of recombination events that involve HJ resolution (e.g., as opposed to SDSA), then in the presence of Mus81 64% of all gene conversions stem from HJ resolution and the observed fraction of associated crossovers (38%) arises from these events. Thus, approximately half (38%/64%) of the I-SceI-induced HJ resolution events lead to a crossover and half lead to a noncrossover. In contrast, the proportion of recombination events involving HJs inferred here (64%) is close to the frequency of crossovers among wild-type (Rec12-dependent) convertants. This result, in turn, suggests that nearly all Rec12-induced HJs are resolved to give crossovers, while I-SceI-induced HJs are resolved at random. In accord with this view, when the bacterial RusA HJ resolvase replaces Mus81 in S. pombe meiosis (with Rec12 making DSBs), the frequency of crossovers in the ade6–arg1 genetic interval is reduced to approximately half of that when Mus81 is the resolvase (11).
We propose that the wild-type (Rec12-dependent) recombination machinery interacts with Mus81-Eme1 to bias it, perhaps very strongly, toward crossover resolution. We infer that this interaction is missing in I-SceI- and RusA-dependent meiotic recombination, reducing the bias perhaps to random. The components responsible for this bias are unknown but may be part of the putative Rec12 complex that makes DSBs (21). Alternatively, the nature of the DSB or of its processing by multiple nucleases and strand exchange proteins into HJs (see Fig. 5) may impart the bias. Further genetic and physical analyses may shed light on this problem.
Conclusion
We have shown that multiple nucleases interact, positively and negatively, to efficiently promote meiotic recombination. Genetic studies such as these provide the foundation for biochemical studies to elucidate these interactions and the molecular mechanism of genetic recombination.
Materials and Methods
Table S4 lists the S. pombe strains used and their genotypes. Meiotic crosses were conducted as described (27) (see the SI Materials and Methods). Statistical significance of the difference between pairs of values was calculated using 2-tailed, unpaired t tests assuming unequal variances. Meiotic inductions and analysis of DNA were performed as described (20).
Supplementary Material
Acknowledgments.
We thank Sue Amundsen and Neta Milman for helpful comments on the manuscript. This work was supported by research Grant GM032194 from the National Institutes of Health (to G.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902793106/DCSupplemental.
References
- 1.Haber JE. In: Recombination and Meiosis: Models, Means, and Evolution. Egel R, Lankenau D-H, editors. Berlin: Springer; 2008. pp. 1–64. [Google Scholar]
- 2.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 3.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson S, Tavassoli M, Watts FZ. Schizosaccharomyces pombe rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res. 1998;26:5261–5269. doi: 10.1093/nar/26.23.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chahwan C, Nakamura TM, Sivakumar S, Russell P, Rhind N. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol Cell Biol. 2003;23:6564–6573. doi: 10.1128/MCB.23.18.6564-6573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 9.Carr AM, et al. The rad16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:2029–2040. doi: 10.1128/mcb.14.3.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cromie GA, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81●Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 13.Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 14.Colleaux L, d'Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis L, Smith GR. Non-random homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mata J, Lyne R, Burns G, Bahler J. The transcription program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Smith GR. Transient meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 21.Cromie GA, Smith GR. In: Recombination and Meiosis: Models, Means, and Evolution. Egel R, Lankenau D-H, editors. Berlin: Springer; 2008. pp. 195–230. [Google Scholar]
- 22.Cromie GA, et al. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyppa RW, Cromie GA, Smith GR. Indistinguishable landscapes of meiotic DNA double-strand breaks in rad50+ and rad50S strains of fission yeast. PLoS Genet. 2008;4:e1000267. doi: 10.1371/journal.pgen.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 26.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 27.Farah JA, Cromie G, Steiner WW, Smith GR. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics. 2005;169:1261–1274. doi: 10.1534/genetics.104.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossenbacher-Grunder AM, Thuriaux P. Spontaneous and UV-induced recombination in radiation-sensitive mutants of Schizosaccharomyces pombe. Mut Res. 1981;81:37–48. doi: 10.1016/0027-5107(81)90085-3. [DOI] [PubMed] [Google Scholar]
- 29.Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;135:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willis N, Rhind N. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol Biol Cell. 2009;20:819–833. doi: 10.1091/mbc.E08-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudolph C, Fleck O, Kohli J. Schizosaccharomyces pombe exo1 is involved in the same mismatch repair pathway as msh2 and pms1. Curr Genet. 1998;34:343–350. doi: 10.1007/s002940050405. [DOI] [PubMed] [Google Scholar]
- 35.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita K, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehouse HLK. Genetic Recombination. New York: John Wiley & Sons; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.