Abstract
We reported the presence in human cells of a noncoding mitochondrial RNA that contains an inverted repeat (IR) of 815 nucleotides (nt) covalently linked to the 5′ end of the mitochondrial 16S RNA (16S mtrRNA). The transcript contains a stem-loop structure and is expressed in human proliferating cells but not in resting cells. Here, we demonstrate that, in addition to this transcript, normal human proliferating cells in culture express 2 antisense mitochondrial transcripts. These transcripts also contain stem-loop structures but strikingly they are down-regulated in tumor cell lines and tumor cells present in 17 different tumor types. The differential expression of these transcripts distinguishes normal from tumor cells and might contribute a unique vision on cancer biology and diagnostics.
Keywords: differential expression in cancer, RNAs with stem-loop structures
Recently, we described a novel human mitochondrial transcript of 2,374 nt that contains a long inverted repeat (IR) linked to the 5′ end of the 16S mitochondrial rRNA (16S mtrRNA) (1, 2), which we designated noncoding mitochondrial RNA or ncmtRNA (2). The IR generates a stem-loop structure with an 820-bp double-stranded region and a 40-nt loop (2). In situ hybridization (ISH) showed that the ncmtRNA is overexpressed in several tumor cell lines but not in nondividing cells, suggesting that the ncmtRNA may play a role in cell proliferation (2).
Because the results described before were obtained using tumor cell lines (2), we asked whether the ncmtRNA (from now on, sense ncmtRNA, SncmtRNA) is expressed in normal proliferating cells. Here we show that ISH of human umbilical vein endothelial cells (HUVEC), foreskin keratinocytes (HFK) (3), and human tonsil endothelial cells (HUTEC) (4) also express the SncmtRNA. However, and in striking contrast with tumor cell lines, ISH of normal proliferating cells revealed expression of antisense transcripts. These molecules were identified as 2 unique transcripts containing IRs linked to the 5′ region of the antisense 16S mtRNA transcribed from the L-strand of the mtDNA (1, 2). We named these transcripts antisense ncmtRNA-1 (ASncmtRNA-1) and antisense ncmtRNA–2 (ASncmtRNA-2). Finally, we show that the antisense transcripts are also expressed in proliferating cells present in normal human tissues but are down-regulated in cells present in human tumors of different types and patients.
Results
Expression of the Sense ncmtRNAs.
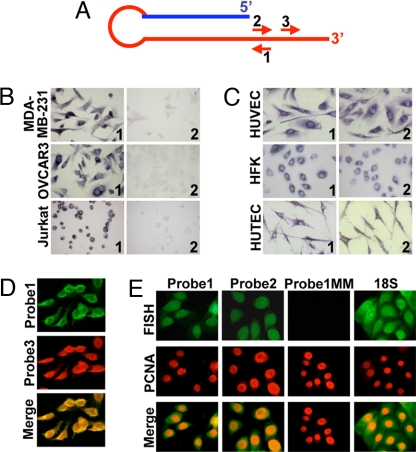
Confirming our previous report, ISH on tumor cell lines with probe 1 (Fig. 1A) revealed strong expression of the SncmtRNA in MDA-MB-231 (breast carcinoma), OVCAR3 (ovary carcinoma), and Jurkat (leukemia) cells (Fig. 1B, Left). Control hybridizations with sense probe 2 were negative (Fig. 1B, Right). A similar expression pattern was obtained with 8 additional tumor cell lines (2). The normal proliferating cells HUVEC, HFK, and HUTEC also express this transcript (Fig. 1C, Right) but, in striking contrast with tumor cells, sense probe 2 also yielded a strong hybridization signal (Fig. 1C, Left). Double-FISH using probes 1 (FITC) and 3 (rhodamine) (Fig. 1A) revealed that each HUVEC cell expressing the SncmtRNA was also expressing the target of the sense probe (Fig. 1D). To confirm the proliferative status of the cells expressing the SncmtRNA and the putative antisense transcript, FISH was carried out together with proliferating cell nuclear antigen (PCNA) detection. HUVEC cells expressing SncmtRNA or the antisense transcript (FITC) were also expressing PCNA (rhodamine) (Fig. 1E). No hybridization signal was obtained using probe 1MM, containing 7 mismatches (Fig. 1E), without probe, or with a probe specific to a salmon sequence. As expected, these cells also express the 18S rRNA (Fig. 1E).
Fig. 1.
Expression of the SncmtRNA in human cells. (A) SncmtRNA and position of probes used for ISH. Probe 1 is complementary to the transcript, and probes 2 and 3 are sense. (B) ISH of the indicated tumor cells with probe 1 (panels 1) or control sense probe 2 (panels 2) (×100). (C) ISH of HUVEC, HFK, and HUTEC with probe 1 (panels 1) or 2 (panels 2). (D) Colocalization of SncmtRNA and the antisense RNA on HUVEC cells. Dig-labeled probe 1 was used for hybridization to SncmtRNA and detected with FITC-coupled anti-dig antibody (Left). The ASncmtRNA was detected with biotin-labeled probe 3 and rhodamine-coupled streptavidin (Center). (E) HUVEC cells were subjected to FISH using dig-labeled probes 1, 2, 1MM (probe 1 with 7 mismatches), or a probe complementary to 18S rRNA. Detection was carried out with FITC-coupled anti-dig antibody. PCNA expression was also determined by fluorescent immunocytochemistry.
The Antisense Transcripts Contain IRs.
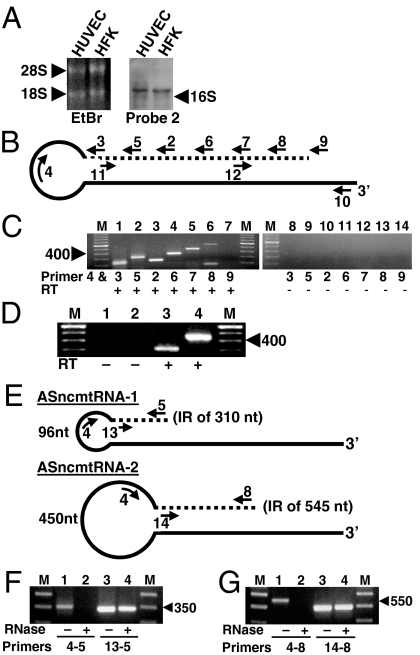
Northern blot using probe 2 on total RNA of HUVEC and HFK revealed a band corresponding to the antisense transcript that migrated together with the 18S rRNA, corresponding to a transcript of about 2,000 nt (Fig. 2A). Because the size of the putative antisense transcript is larger than the 16S rmtRNA (Fig. 2A), we asked whether this transcript would have a stem-loop structure similar to that of SncmtRNA (2). Accordingly, we designed a hypothetical antisense RNA containing an IR (dotted line) linked to the 5′ of the antisense 16S mtRNA (solid line) (Fig. 2B). To amplify this putative transcript by RT-PCR, reverse primer 4, targeted to positions 35 to 59 of the antisense 16S mtrRNA, was used in combination with forward primers 3, 5, 2, 6, 7, 8, and 9 (Fig. 2B) (Table S1). Amplification of HUVEC RNA yielded amplicons of around 240 and 330 bp using primer 4 with primers 3 and 5, respectively (Fig. 2C, lanes 1 and 2). Sequencing of the 330-bp amplicon (Fig. 2C, lane 2) revealed that a fragment of 281 nt of the sense 16S mtRNA (IR) was linked to the 5′ end of the antisense 16S mtRNA (Dataset S1 and GenBank accession no. EU_863789). The sequence of the 240-bp amplicon (Fig. 2C, lane 1) showed that this fragment was part of the same RNA. We named this transcript antisense ncmtRNA-1 or ASncmtRNA-1. Although with primers 4 and 2 one would expect an amplification fragment longer than the one obtained with primers 4 and 5, an amplicon of around 240 bp was obtained (Fig. 2C, lane 3). The sequence of this amplicon revealed that we were dealing with a second antisense transcript. When using primer 4 with primers 6, 7, and 8, amplicons of about 350, 450, and 560 bp, respectively, were obtained (Fig. 2C, lanes 4, 5, and 6). No amplification product was obtained using primers 4 and 9 (Fig. 2C, lane 7). The sequence of the 560-bp amplicon revealed that an IR of 501 nt was linked to the 5′ of the antisense 16S mtRNA (Dataset S2 and GenBank accession no. EU863790). The sequence of the other amplicons revealed that they were part of the 560-bp amplicon. We named this transcript antisense ncmtRNA-2 or ASncmtRNA-2. A second amplicon of about 160 bp, obtained with primers 4 and 8 (Fig. 2C, lane 6), was not further investigated. No amplification was obtained without reverse transcriptase (Fig. 2C, lanes 8–14). The same sequences of AS ncmtRNA-1 and ASncmtRNA-2 were obtained with RNA from HFK and PHA-stimulated lymphocytes. To define the 5′ end of the ASncmtRNAs, total RNA from HFK was digested with RNase A and the isolated stems were used as template for 5′RACE using primer 11 or 12 (Fig. 2B) (see SI Text). The sequence of the amplicon obtained with primer 11 and the anchor primer revealed that the IR of the ASncmtRNA-1 of 281 nt was extended by 29 additional nucleotides (Dataset S1). A similar experiment revealed that the IR of 501 nt of the ASncmtRNA-2 was extended in 44 nt (Dataset S2). The position of the IRs of ASncmtRNA-1 and ASncmtRNA-2 with respect to the complete sequence of the 16S mtrRNA is shown in Fig. S1.
Fig. 2.
The antisense transcripts contain stem-loop structure. (A) Northern blot of total HUVEC and HFK RNA with probe 2. Position of 16S mtrRNA is indicated. (B) Theoretical structure of the antisense transcript. The 5′ end of the antisense 16S mtrRNA (solid line) is linked to a fragment of the sense 16S mtrRNA or IR (dotted line), forming a stem and a loop. Position of reverse (Under the lines) and forward primers (Above the lines) are indicated (see Materials and Methods). (C) PCR amplification of HUVEC cDNA using primer 4 with primers 3 (lanes 1 and 8), 5 (lanes 2 and 9), 2 (lanes 3 and 10), 6 (lanes 4 and 11), 7 (lanes 5 and 12), 8 (lanes 6 and 13), and 9 (lanes 7 and 14). Lanes 1–7 show results with reverse transcriptase and lanes 8–14 are without the enzyme. Lane M, 100-bp ladder. (D) HUVEC cDNA was synthesized with gene-specific primer 10 and PCR amplified using primer pairs 4 and 5 (lanes 1 and 3) or 4 and 7 (lanes 2 and 4), respectively. Lane M, 100-bp ladder. No amplification was observed without reverse transcriptase (lanes 1 and 2). (E) Schematic structure of the ASncmtRNA-1 and ASncmtRNA-2, showing the size of the loops, the length of the double-stranded regions, and the primers used for amplification. (F and G) HUVEC RNA was incubated in 2× SSC without (odd lanes) or with RNase A (even lanes) and RNA was recovered and amplified. (F) RT-PCR with primers 4 and 5 (lanes 1 and 2) or 13 and 5 (lanes 3 and 4). Lane M, 50-bp ladder. (G) RT-PCR with primers 4 and 8 (lanes 1 and 2) or 14 and 8 (lanes 3 and 4). Lane M, 100-bp ladder.
To determine whether the IRs were linked to the complete antisense 16S mtRNA, cDNA was synthesized with primer 10 (Fig. 2B), complementary to the 3′ end of the antisense 16S mtrRNA. Amplification of this cDNA using primer 4 with primer 5 or 7 (Fig. 2D) yielded the expected amplicons of 330 bp (ASncmtRNA-1) (lane 3) or 450 bp (ASncmtRNA-2) (lane 4). The presence of ASncmtRNAs -1 and -2 in HUVEC was further confirmed by S1 protection assays (Fig. S2).
The Antisense ncmtRNAs Contain Stem-Loop Structures.
The IR of the ASncmtRNA-1 forms in theory a 310-bp stem and a 96-nt loop (Fig. 2E and Dataset S1). Similarly, the IR of the ASncmtRNA-2 forms a 545-bp stem and a 450-nt loop (Fig. 2E and Dataset S2). Because the stems should be resistant to RNase A digestion, HUVEC RNA was mock treated or treated with RNase A and RNA was recovered and amplified by RT-PCR, using the primers described in Fig. 2E for each transcript. As expected, primers 4 and 5 (ASncmtRNA-1) or primers 4 and 8 (ASncmtRNA-2) gave rise to amplicons of 330 and 560 bp, respectively (Fig. 2 F and G, lanes 1). Amplification of these fragments was abolished by RNase A digestion (Fig. 2 F and G, lanes 2), indicating that the loops were digested by the enzyme. However, amplification between primers 13 and 5 (ASncmtRNA-1) (Fig. 2F) or 14 and 8 (ASncmtRNA-2) (Fig. 2G) targeted to 3′ and 5′ of each stem was not affected by RNase A treatment. (Fig. 2 F and G, compare lanes 3 and 4). As expected, neither stem was amplified when the RNA was digested with the double-stranded specific RNase III from Escherichia coli (Fig. S3) (5).
Expression of the SncmtRNA and ASncmtRNAs.
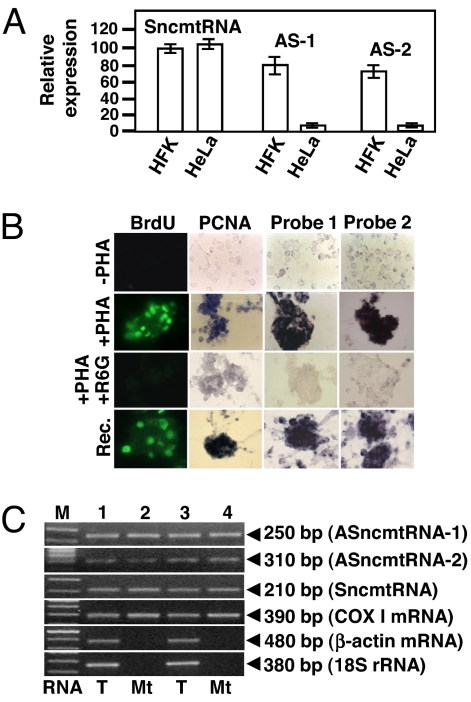
Down-regulation of the ASncmtRNAs in tumor cells shown by ISH was further confirmed by qRT-PCR. The amplified region for each transcript encompassed part of the 5′ of the 16S mtrRNA (SncmtRNA) or part of the 5′ of the antisense 16S RNA (ASncmtRNAs) and part of the 3′ end of each IR (see Materials and Methods). As shown in Fig. 3A, the relative expression levels of the SncmtRNA, normalized to GAPDH mRNA, were similar in HFK and in HeLa cells. The Ct values for GAPDH mRNA in HFK cells were 21, 21, and 22 cycles, compared to 22, 22, and 23 cycles for HeLa cells. On the other hand, the relative expression levels of both ASncmtRNAs in HFK cells were superior to the relative expression of the same transcripts in HeLa cells. (Fig. 3A). Similar results were obtained with HUVEC cells and other tumor cell lines.
Fig. 3.
Expression of the SncmtRNA and the ASncmtRNAs. (A) qRT-PCR, relative expression of S- and ASncmtRNAs -1 and -2 in HFK and HeLa cells was normalized to GAPDH mRNA. Results are the average of 3 determinations. (B) Expression of S- and ASncmtRNAs is inhibited by rhodamine 6G. Isolated lymphocytes were incubated alone (−PHA) or with PHA (+PHA) in the presence of BrdU. Another group of cells was incubated with PHA and rhodamine 6G. At 48 h, cells were fixed and subjected to immunodetection of BrdU or PCNA and to ISH to detect S- and ASncmtRNAs. After incubation with rhodamine 6G, the drug was eliminated and cells were incubated with BrdU and PHA for another 24 h (recovery, rec.). Incorporation of BrdU, PCNA expression, and expression of the ncmtRNAs was determined as described before. (C) Total RNA (T) from HFK (lanes 1 and 2) and PHA-stimulated lymphocyte (lanes 3 and 4) or RNA from RNase A-treated isolated mitochondria (Mt) (2) was used for RT-PCR. Amplicons of 250 bp (ASncmtRNAS-1), 310 bp (ASncmtRNA-2), 210 bp (SncmtRNA), 390 bp (COX I mRNA), 480 bp (β-actin mRNA), and 360 bp (18S rRNA) were obtained.
Similar to SncmtRNA (2), expression of the ASncmtRNAs is induced in PHA-stimulated lymphocytes (Fig. 3B). Together with expression of these transcripts, PHA-stimulated cells also incorporate BrdU and express PCNA (Fig. 3B). We confirmed by qRT-PCR that in resting lymphocytes (lymph), the relative expression of SncmtRNA and the ASncmtRNAs -1 and -2 was quite low compared to their relative expression in PHA-stimulated lymphocytes (Fig. S4).
Then we asked whether rhodamine 6G, which disables mitochondrial function and inhibits cell proliferation (6), would inhibit expression of these RNAs. Treatment of stimulated cells with rhodamine 6G suppresses BrdU incorporation, PCNA expression, and expression of SncmtRNA and the ASncmtRNAs (Fig. 3B). The effect was reversible because 24 h after eliminating the drug, expression of the transcripts, and BrdU incorporation and PCNA expression were reestablished (Fig. 3B).
The sequences of the ASncmtRNAs exhibit 99.9% identity with the 16S gene of the mtDNA. Therefore, we determined whether the ASncmtRNAs were present in isolated mitochondria from HFKs and PHA-stimulated lymphocytes. The S- and ASncmtRNAs were amplified from RNA extracted from mitochondria treated externally with RNase A (2) (Fig. 3C). As expected, COX 1 mRNA was also present in treated mitochondria, but not the cytoplasmic transcripts 18S rRNA and β-actin mRNA (Fig. 3C).
The ASncmtRNAs Are Down-Regulated in Solid Tumors.
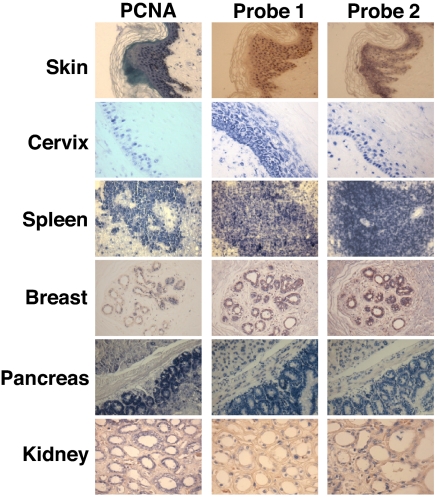
Because the previous studies were carried out with cell lines, we wondered whether the differential expression of the ncmtRNAs would be observed as well in human tissues. Expression of ncmtRNAs was determined by ISH in serial sections of biopsies containing regions of normal tissues (Fig. 4 and Table 1). A parallel section of each tissue was used to determine PCNA expression (7). Expression of SncmtRNA and the ASncmtRNAs was confined to PCNA-expressing proliferating cells (Fig. 4). Note that no expression of these transcripts or PCNA was found in stromal cells surrounding the proliferating tissues (Fig. 4). Similar expression patterns were observed in 74 different samples obtained from different patients (Table 1).
Fig. 4.
Expression of SncmtRNA and ASncmtRNAs in normal human tissues. Parallel biopsy sections containing normal tissue were subjected to PCNA immunodetection and ISH for S- and ASncmtRNAs. Expression of the ncmtRNAs was confined to PCNA-expressing proliferating cells. Stromal tissue was negative for PCNA expression and ISH. (magnification 40×).
Table 1.
Expression of SncmtRNA and the ASncmtRNAs in normal and tumor tissues
| ncmtRNAs |
|||
|---|---|---|---|
| No. of cases | S | AS | |
| Normal tissues | |||
| Skin | 14 (2) | + | + |
| Cervical epithelium | 22 (3) | + | + |
| Sebaceous gland | 9 | + | + |
| Esophagus epithelium | 4 | + | + |
| Kidney | 6 | + | + |
| Breast | 8 (1) | + | + |
| Pancreas | 5 | + | + |
| Spleen | 6 | + | + |
| Total | 74 | ||
| Tumor tissues | |||
| Breast carcinoma | 64 (7) | + | − |
| Prostate carcinoma | 27 (3) | + | − |
| Lung carcinoma | 14 (1) | + | − |
| Colon carcinoma | 17 (1) | + | − |
| Gallbladder carcinoma | 11 | + | − |
| Gastric carcinoma | 22 (4) | + | − |
| Uterine cervix* carcinoma | 27 (2) | + | − |
| Brain tumors† | 12 (1) | + | − |
| Kidney carcinoma | 9 (1) | + | − |
| Hepatocarcinoma | 5 | + | − |
| Thyroid carcinoma | 12 | + | − |
| Ovary carcinoma | 8 | + | − |
| Bladder carcinoma | 12 (2) | + | − |
| Melanomas | 12 (1) | + | − |
| Lymphoma | 6 | + | − |
| Small intestine | 3 | + | − |
| Carcinoma | − | ||
| Leukemia‡ | 12 | + | − |
| Total | 273 | ||
Numbers in parentheses indicate biopsies that yielded weak or negative hybridization signals.
*CIN II, CIN III, and squamous carcinoma.
†Glyoblastomas, astrocytomas, and meningiomas.
‡Seven blood smears of CLL and 5 smears of AML.
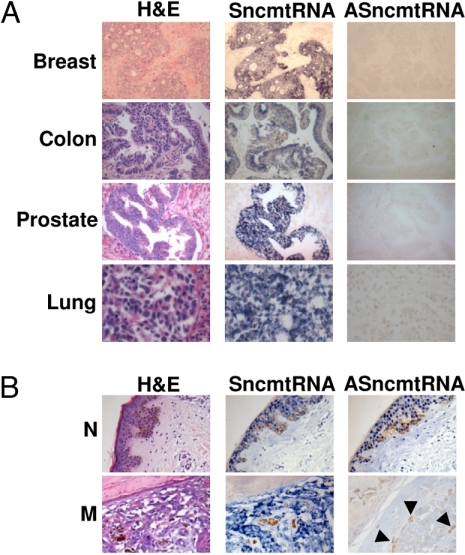
In striking contrast to normal proliferating tissues, cells present in human tumors down-regulate the expression of the ASncmtRNAs. The SncmtRNA is expressed in breast, colon, prostate, and lung tumor (Fig. 5A), but ISH performed on parallel sections rendered negative results for ASncmtRNAs (Fig. 5A). The tumor tissue was identified after staining with hematoxylin-eosin (H&E) (Fig. 5A). The same expression pattern was observed in 12 additional tumors and leukemia (Table 1 and Fig. S5). ISH can discriminate between normal and tumor tissue in the same biopsy as illustrated in Fig. 5B. Normal skin surrounding a melanoma lesion expresses the SncmtRNA and the ASncmtRNAs (Fig. 5B, normal, N), but the melanoma lesion only expresses the SncmtRNA (Fig. 5B, melanoma, M). A summary of the expression of the SncmtRNA and the ASncmtRNAs in 273 biopsies of 16 different types of tumor and patients is shown in Table 1 and Fig. S5. Numbers in parentheses represent samples from each biopsy type where the ISH signal was weak or negative. On average, about 11% of the samples revealed weak or no signal.
Fig. 5.
The ASncmtRNAs are down-regulated in tumor tissues. (A) Serial tumor sections were subjected to ISH to detect S- and ASncmtRNAs. A parallel section was stained with hematoxylin and eosin (H&E). (B) Comparison of normal and tumor tissue in the same biopsy. Normal skin (N) surrounding the melanoma expresses the S- and the ASncmtRNAs. The melanoma in the same biopsy expresses the SncmtRNA and down-regulates the ASncmtRNA. Arrowheads show melanin (magnification 40×).
Discussion
The ASncmtRNAs.
Here we describe the presence in human cells of 2 mitochondrial transcripts (ASncmtRNAs) containing stem-loop structures similar to that of the previously described SncmtRNA (2). Interestingly, the sequences of the ASncmtRNA-1 and ASncmtRNA-2 are not present in the human EST database (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). As discussed in relation to the SncmtRNA (3), the absence of these sequences in the EST database might be the result of replication slippage (2, 8).
Differential Expression of the ncmtRNAs.
Regarding expression of the SncmtRNA and the ASncmtRNAs, 3 different phenotypes of human cells can be defined. Normal proliferating cells express both families of transcripts. In striking contrast, tumor cells express the SncmtRNA and down-regulate the ASncmtRNAs. Finally, neither of these transcripts is expressed in nondividing cells. Down-regulation of the ASncmtRNAs was observed in 15 different tumor cell lines (ref. 2 and this work) and in tumor cells present in 273 cancer biopsies corresponding to 17 different cancer types (Table 1). On average, we observed that about 11% of the biopsies yielded weak or negative hybridization signals (see Table 1). This result was independent of the type of tumor or patient. Poor hybridization signals have been reported before and are the result of problems in tissue handling, fixation times, and formalin concentration (9–11). Although the biopsies analyzed here constitute a somewhat modest sampling, one is tempted to hypothesize that down-regulation of the ASncmtRNAs is a universal characteristic of tumor cells. In fact, similar expression patterns are observed in normal and tumor mouse cells. The SncmtRNA and the ASncmtRNAs are expressed in normal proliferating mouse cells but not in nondividing cells. In contrast, mouse tumor cells express the SncmtRNA and down-regulate the ASncmtRNA.
Function of SncmtRNA and the ASncmtRNAs.
Expression of the SncmtRNA correlates with the replicative state of the cell and it may play a role in the regulation of the cell cycle. SncmtRNA is expressed in any proliferating cell independently whether we are dealing with a regulated or a dysfunctional cell cycle. Recently, we showed that aphidicolin reversibly blocks proliferation in G1, along with down-regulation of SncmtRNA expression (2). Similarly, rhodamine 6G, an inhibitor of mitochondrial function and cell proliferation (6), reversibly blocks DNA synthesis, PCNA expression, and expression of the SncmtRNA and the ASncmtRNAs. However, how this molecule participates in the regulation of cell cycle remains unknown and warrants future research.
As to the function of the ASncmtRNAs, our understanding is less clear. One hypothetical role might be the regulation of the SncmtRNA function, for example, by partial base pair interaction. However, the fact that the ASncmtRNAs are always down-regulated in tumor cells, suggests that hypothetically the ASncmtRNAs might function as a unique mitochondria-encoded tumor suppressor. Classical tumorigenesis models postulate alteration of tumor suppressor genes by chromosomal rearrangements, deletions, and mutations (12). Recently, several reports have shown that other ncRNAs (13), especially microRNAs (miR), can function as tumor suppressors (14). In about 65% of patients with B-cell chronic lymphocytic leukemia (CLL), miR-15a and miR-16–1 are down-regulated (15). Interestingly, miR-15a and miR-16–1 negatively regulate the expression of the antiapoptotic protein BCL2 (16). Similarly, a close correlation between down-regulation of another miRNA, let-7, and lung cancer was reported (17). The authors proposed that let-7 is a unique tumor suppressor and show regulation of the oncoprotein Ras by overexpression of let-7 (17). The designation of tumor suppressor is based on experimental evidence showing the role of this class of molecules in cell cycle control. At present we are studying how the interference of the SncmtRNA or the ASncmtRNAs with antisense oligonucleotides or siRNAs affects the cell cycle of normal proliferating cells.
Materials and Methods
Cell Culture.
HeLa, MDA-MB-231, OVCAR3, Jurkat, and HUVEC cells were cultured according to American Type Culture Collection recommendations. HFK and HUTEC were cultured as described before (3, 4). Isolated human lymphocytes were cultured for 48 h without or with 10 μg/mL phytohaemagglutinin (PHA) as described before (2). Incorporation of 5-bromo-2-deoxyuridine (BrdU) poststimulation with PHA was determined with a FITC-coupled anti-BrdU antibody (KPL). PHA-stimulated lymphocytes were also incubated with 1 μg/mL of rhodamine 6G in the same culture medium plus 50 μg/mL uridine and 1 mM sodium pyruvate (2).
RNA Isolation and Amplification.
Total RNA from cells and isolated mitochondria was extracted as described before (4, 18, 19). RNA was treated with TurboDNA-free (Ambion), according to manufacturer's directions. Reverse transcription was carried out on 50 to 100 ng of freshly prepared RNA as described before (2, 18, 19). The cDNA was PCR amplified and amplicons were cloned and sequenced (2). Sequence and position of primers are listed in Table S1. Northern blot with dig-labeled probe 2 was done as described before (2). For quantitative RT-PCR (qRT-PCR) we used a mastermix containing 1× brilliant II SYBR Green QPCR MasterMix, 1 μL of cDNA, 0.4 μL of ROX (diluted 1/200), and 0.2 μL of each primer set (0.1 μM). The cDNA was synthesized with 50 to 100 ng of total RNA in a final volume of 20 μL. Amplification was carried out with primers 3 and 4 for ASncmtRNA-1, 2 and 4 for ASncmtRNA-2, and 15 and 16 for SncmtRNA (Table S1). The amplified regions encompassed part of the 5′ region of the 16S RNA (sense or antisense) and part of the 3′ of the corresponding IR. The amplification reaction consisted of an initial incubation at 95 °C for 10 min, followed by 40 cycles of 30 s at 95 °C, 40 s at 58 °C, and 40 s at 72 °C. Results were normalized to GAPDH mRNA amplification with the corresponding primers (Table S1) (20). For amplification of digested RNA, total RNA (20 μg/mL) from HUVEC cells was dissolved in 2× SSC and 50 μL of this dilution were incubated in 25 μg/mL RNase A for 10 min at 25 °C (2, 21). Under these conditions, the enzyme does not digest the double-stranded genomic RNA of the virus IPN (2). Digested RNA was extracted and used for RT-PCR amplification (2).
In Situ Hybridization. Cells cultured in 8-well chamber slides (Lab-Tek, NUNC) for 24 to 48 h or resting or PHA-stimulated human lymphocytes, were fixed in 4% p-formaldehyde for 10 min (2). ISH was done as described, using oligonucleotide probes labeled at the 3′ end with digoxigenin-11-dUTP (2, 22). For double FISH, cells were hybridized with dig-labeled probe 1 and biotin-labeled probe 3. After hybridization and washing, cells were incubated in ISH blocking solution for 30 min at room temperature (RT) and then with a mixture of FITC-conjugated anti-digoxigenin antibody and rhodamine-conjugated steptavidin (Molecular Probes), diluted 1:250 in the same solution. After 3 washes, cells were mounted in fluorescent mounting media (DAKO). Results were analyzed on a fluorescence Olympus BX51 microscope. Confocal microscopy was performed on an LSM 5 Zeiss microscope equipped with a 63× objective and analysis was carried out using the Zeiss LSM 5 Image Browser software. For tissue samples, ≈5-μm-thick serial paraffin sections were collected on silanized slides (DAKO) and deparaffinized in 2 consecutive 5-min xylene washes. One section was stained with hematoxylin and eosin. The others were rehydrated in two 3-min washes of 100 and 95% ethanol each, and once in DEPC-treated distilled water for 5 min (2, 11). Sections were then incubated in 2.5 μg/mL of Proteinase K (Invitrogen) at RT for 20 min and then washed twice for 3 min in DEPC-treated water, immersed in 96% ethanol for 10 s, and air dried (2, 11). Hybridization was carried out with dig-labeled probes 1 and 13 to detect the SncmtRNA and probes 2 and 3 for the ASncmtRNAs (Table S1). Hybridization mixtures contained 35 pmoles/mL of each probe in hybridization solution as described before (2, 11). Hybridization, washing, and color development were carried out as described before (2) with modifications (see SI Text).
PCNA Expression.
Tissue sections were incubated for 15 min at 95 °C in Target Retrieval Solution (DAKO) diluted 1:10 in distilled water. Sections were then incubated with anti-PCNA antibody, followed by alkaline phosphatase-coupled anti-mouse IgG (KPL), diluted 1:250 in blocking solution. Color reaction was developed as described above. For localization studies, cells were subjected to FISH as described above, with the exception that monoclonal antibody against PCNA (DAKO) was added with the FITC-coupled anti-dig antibody. After incubation, cells were washed 3 times for 5 min in PBS and incubated at RT for 30 min with rhodamine-coupled anti-mouse IgG antibody, diluted 1:250 in blocking solution. Cells were finally washed 3 times for 5 min in PBS, mounted in fluorescent mounting media, and analized as described above.
Supplementary Material
Acknowledgments.
We apologize to our many colleagues whose work was either shortly summarized or not cited because of space limitations. This work was supported by Millennium Scientific Initiative no. P-77–09F, Chile, and Grants DID-32–03, DID-26–04, and DID-57–04; Universidad Andrés Bello, Santiago, Chile; Grant D04I1338, Fondef, Conicyt; and the CCTE-PFB16 Program, Conicyt, Chile.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU863789 and EU863790).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903086106/DCSupplemental.
References
- 1.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 2.Villegas J, et al. Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res. 2007;35:7336–7347. doi: 10.1093/nar/gkm863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boccardo E, Nova F, Broker TR, Chow LT, Villa LL. HPV-18 confers resistance to TNF-alpha in organotypic cultures of human keratinocytes. Virology. 2004;328:233–243. doi: 10.1016/j.virol.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 4.Reyes LJ, Escobar P, Bono MR, Rosemblatt M. Adhesion of B cell lines to endothelial cells from human lymphoid tissue modulates tyrosine phosphorylation and endothelial cell activation. J Immunol. 2002;169:5881–5888. doi: 10.4049/jimmunol.169.10.5881. [DOI] [PubMed] [Google Scholar]
- 5.Robertson HD. Escherichia coli ribonuclease III. Methods Enzymol. 1990;181:189–202. doi: 10.1016/0076-6879(90)81121-a. [DOI] [PubMed] [Google Scholar]
- 6.Felty Q, Singh KP, Roy D. Estrogen-induced G1/S transition of G0-arrested estrogen-dependent breast cancer cells is regulated by mitochondrial oxidant signaling. Oncogene. 2005;24:4883–4893. doi: 10.1038/sj.onc.1208667. [DOI] [PubMed] [Google Scholar]
- 7.Fuss J, Linn S. Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J Biol Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 8.Viguera E, Canceill D, Ehrlich SD. In vitro replication slippage by DNA polymerases from thermophilic organisms. J Mol Biol. 2001;312:323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 9.Tbakhi A, et al. Fixation conditions for DNA and RNA in situ hybridization: A reassessment of molecular morphology dogma. Amer J Pathol. 1998;152:35–41. [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie JW, et al. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160:449–457. doi: 10.1016/S0002-9440(10)64864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henke RT, Kim SE, Maitra A, Paik S, Wellstein A. Expression analysis of mRNA in formalin-fixed, paraffin-embedded archival tissues by mRNA in situ hybridization. Methods. 2006;38:253–262. doi: 10.1016/j.ymeth.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031–2045. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 13.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Villegas J, et al. A novel chimeric mitochondrial RNA localized in the nucleus of mouse sperm. DNA Cell Biol. 2000;19:579–588. doi: 10.1089/104454900439809. [DOI] [PubMed] [Google Scholar]
- 19.Villegas J, Müller I, Arredondo J, Pinto R, Burzio LO. A putative RNA editing from U to C in a mouse mitochondrial transcript. Nucleic Acids Res. 2002a;30:1895–1901. doi: 10.1093/nar/30.9.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acid Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young PG, Attardi G. Characterization of double-stranded RNA from HeLa cell mitochondria. Biochem Biophys Res Commun. 1975;65:1201–1207. doi: 10.1016/s0006-291x(75)80357-3. [DOI] [PubMed] [Google Scholar]
- 22.Villegas J, Araya P, Bustos-Obregon E, Burzio LO. Localization of the 16S mitochondrial rRNA in the nucleus of mammalian spermatogenic cells. Mol Hum Reprod. 2002;8:977–983. doi: 10.1093/molehr/8.11.977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.