Abstract
Viral internal ribosomal entry sites (IRESs) mediate end-independent translation initiation. There are 4 major structurally-distinct IRES groups: type 1 (e.g., poliovirus) and type 2 (e.g., encephalomyocarditis virus), which are dissimilar except for a Yn-Xm-AUG motif at their 3′ borders, type 3 (e.g., hepatitis C virus), and type 4 (dicistroviruses). Type 2–4 IRESs mediate initiation by distinct mechanisms that are nevertheless all based on specific noncanonical interactions with canonical components of the translation apparatus, such as eukaryotic initiation factor (eIF) 4G (type 2), 40S ribosomal subunits (types 3 and 4), and eIF3 (type 3). The mechanism of initiation on type 1 IRESs is unknown. We now report that domain V of type 1 IRESs, which is adjacent to the Yn-Xm-AUG motif, specifically interacts with the central domain of eIF4G. The position and orientation of eIF4G relative to the Yn-Xm-AUG motif is analogous in type 1 and 2 IRESs. eIF4G promotes recruitment of eIF4A to type 1 IRESs, and together, eIF4G and eIF4A induce conformational changes at their 3′ borders. The ability of mutant type 1 IRESs to bind eIF4G/eIF4A correlated with their translational activity. These characteristics parallel the mechanism of initiation on type 2 IRESs, in which the key event is binding of eIF4G to the J–K domain adjacent to the Yn-Xm-AUG motif, which is enhanced by eIF4A. These data suggest that fundamental aspects of the mechanisms of initiation on these unrelated classes of IRESs are similar.
Keywords: coxsackievirus, enterovirus 71, poliovirus, translation initiation, RNA–protein interaction
Translation initiation on most eukaryotic mRNAs occurs by the scanning mechanism. First, a 43S preinitiation complex comprising a 40S ribosomal subunit, eukaryotic initiation factor 2 (eIF2)·GTP/Met-tRNAMetI, eIF3, eIF1, and eIF1A is loaded onto mRNA by eIF4A, eIF4B, and eIF4F (1). eIF4F consists of eIF4A (DEAD-box RNA helicase), eIF4E (cap-binding protein), and eIF4G (which binds eIF4E, eIF4A, and eIF3). eIFs 4A/4B/4F unwind the cap-proximal region of mRNA, allowing 43S complexes to attach. After that, 43S complexes scan the 5′ UTR to the start codon, where they form 48S initiation complexes with established codon–anticodon base-pairing.
The genomes of several families of RNA viruses contain internal ribosomal entry sites (IRESs), which can mediate end-independent initiation. Viral IRESs are classified into 4 major groups, based on their sequence and structure: type 1 [e.g., poliovirus (PV)], type 2 [e.g., encephalomyocarditis virus (EMCV)], type 3 (e.g., hepatitis C virus), and type 4 (e.g., cricket paralysis virus). The mechanisms of initiation on type 2, 3, and 4 IRESs are distinct, but are all based on specific interactions of IRESs with components of the translation apparatus. Type 3 IRESs interact specifically with 40S subunits and eIF3, which allows direct attachment of 43S complexes to the initiation codon (2). Initiation on type 4 IRESs also involves direct binding to 40S subunits, but does not require eIFs or Met-tRNAMetI (3). The ≈450-nt-long type 2 IRESs have 4 major domains (H, I, J-K, and L) and a Yn-Xm-AUG motif at their 3′ border, in which a pyrimidine-rich tract is separated by a ≈20-nt spacer (Xm) from the AUG codon (see Fig. 2I), which can serve as an initiation codon. Initiation on type 2 IRESs depends on binding of eIF4G to the J-K domain adjacent to the Yn-Xm-AUG motif, which is enhanced by eIF4A (4–6). eIF4G/eIF4A restructure the region of ribosomal attachment (7) and promote binding of 43S complexes to the IRES.
Fig. 2.
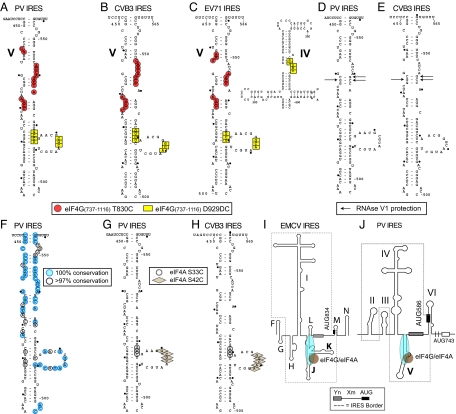
Sites of interaction of elF4G and elF4A with CVB3, EV71, and PV Type 1 IRESs. (A–C) Sites of directed hydroxyl radical cleavage from positions 830 and 929 of eIF4G737–1116 mapped onto PV Domain V (A), CVB3 domain V (B), and EV71 domains IV and V (C). (D and E) Nucleotides protected by eIF4G from RNase V1 cleavage mapped onto PV (D) and CVB3 domain V (E). (F) Sequence conservation in domain V of enteroviruses. (G and H) Sites of directed hydroxyl radical cleavage from Cys-33 and Cys-42 of eIF4A mapped onto PV (G) and CVB3 domain V (H). (I and J) Models of the type 2 EMCV IRES (I) and the type 1 PV IRES (J), showing the proximity and orientation of the bound eIF4G/eIF4A complex relative to the Yn-Xm-AUG motif.
Type 1 IRESs occur in PV, coxsackievirus type B3 (CVB3), enterovirus 71 (EV71), and other members of the Enterovirus genus of Picornaviridae (e.g., refs. 8–10). They are ≈450 nt long and have 4 major domains (II, IV, V, and VI) (Fig. 1A), but share little homology with type 2 IRESs except for a Yn-Xm-AUG motif at their 3′ border, in which the AUG triplet is naturally silent (10, 11). Instead, depending on the virus, initiation occurs 30–150 nt downstream of this motif.
Fig. 1.
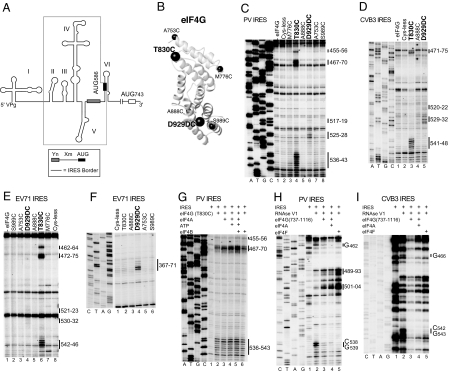
Specific binding of eIF4G to type 1 IRESs identified by directed hydroxyl radical cleavage and enzymatic footprinting. (A) Model of the PV1M 5′ UTR, showing its type 1 IRES (boxed), the Yn-Xm-AUG motif, the initiation codon AUG743, and domains I–VI. (B) Model of eIF4G's central domain (Protein Data Bank ID code 1HU3), with spheres labeled to show the positions of cysteines used to tether Fe(II)-BABE. (C–G) Primer extension analysis of directed hydroxyl radical cleavage of PV (C and G), CVB3 (D), and EV71 (E and F) IRESs from Fe(II)-tethered eIF4G in IRES/eIF4G complexes in the absence (C–E) and presence of eIF4A/eIF4B (G), as indicated. (H and I) Primer extension analysis of residues in PV (H) and CVB3 (I) IRESs protected from RNase V1 cleavage by eIF4G, eIF4G/eIF4A, and eIF4F, as indicated. Sites of hydroxyl radical cleavage and residues with altered sensitivity to RNase V1 cleavage are indicated to the right. Lanes A, T, G, and C depict sequences generated from the same primers.
The mechanism of initiation on type 1 IRESs has not been solved, and little is known about its requirements for canonical factors or their roles in this process. Inhibition of PV translation by dominant-negative mutant eIF4A, which sequesters eIF4G into inactive complexes, and the fact that eIF4F rescues translation more effectively than WT eIF4A (12) suggest a direct but as-yet undefined role for eIF4A and eIF4G in PV translation. eIF4E is not required (13), and consistently, PV translation is not impaired by the cleavage of eIF4G into an N-terminal domain (NTD) that binds eIF4E and a C-terminal domain (CTD) that binds eIF3 and eIF4A that occurs in infected cells (14). Cross-linking of domain V of the PV IRES to eIF4G in rabbit reticulocyte lysate (RRL) has been reported (15). In the same study, domain V additionally cross-linked to eIF3 and eIF4B.
Because initiation on type 2–4 IRESs occurs by distinct mechanisms that all are based on primary interactions of IRESs with eIFs or 40S subunits, we looked for similar interactions that could promote recruitment of 43S complexes to type 1 IRESs. We report that, like type 2 IRESs, type 1 IRESs bind specifically to eIF4G, and its orientation and position on type 1 and 2 IRESs relative to the Yn-Xm-AUG motif are analogous. eIF4G recruits eIF4A to type 1 IRESs, and together they induce changes at the IRES 3′ borders analogous to those reported for type 2 IRESs (7). This finding suggests that fundamental aspects of the mechanisms of initiation on these unrelated classes of IRESs are similar.
Results
Mapping the eIF4G/Type 1 IRES Interaction by Directed Hydroxyl Radical Cleavage.
Directed hydroxyl radical cleavage allows localization of RNA-bound proteins. Fe(II) is conjugated to unique cysteines on their surface via the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE) (16). Fenton chemistry generates hydroxyl radicals near the tethered iron that cleave adjacent RNA. Cleavage sites are identified by primer extension.
We used this technique to determine the orientation of eIF4G's central domain on a type 2 IRES (7). The central domain of eIF4G consists of 5 pairs of α-helices (ref. 17 and Fig. 1B). Six eIF4G737–1116 mutants with unique surface-exposed cysteines (Fig. 1B) were fully active in 48S complex formation on the EMCV IRES in an in vitro-reconstituted system and induced specific cleavage in its J–K domain in eIF4G/IRES complexes (7).
We used these mutants to study potential eIF4G/type 1 IRES interactions. Fe(II)-conjugated eIF4G737–1116 Cys mutants were incubated with CVB3, EV71, and PV IRESs (prepared as described in SI Text). Hydroxyl radicals generated from 2 positions in eIF4G737–1116 induced cleavage in domain V in all 3 IRESs (Figs. 1 C–E and 2 A–C and (Figs. S1 and S2). Cys-830, located between helices 2b and 3a, cleaved strongly at the base of domain V (PV, nucleotides 455–456, 467–470, and 536–543; CVB3, nucleotides 471–475 and 541–548; and EV71, nucleotides 462–464, 472–475, and 542–546), whereas Cys-929, located after helix 4b, induced weak cleavage in domain V's central loop (PV, nucleotides 517–519 and 525–528; CVB3, nucleotides 520–522 and 529–532; and EV71, nucleotides 521–523 and 530–532) (Fig. 1 C–E). These data indicate that eIF4G737–1116 interacts specifically with domain V of type 1 IRESs with its C terminus directed toward the apex and the N terminus toward the base of this domain. Cleavage from Cys-929 also occurred at nucleotides 367–371 in the apex of EV71 IRES domain IV (Fig. 1F and Fig. 2C). There was no evidence from footprinting experiments that eIF4G binds to this region of type 1 IRESs (see below), and these cleavages may instead reflect the proximity of the apices of domain IV and eIF4G-bound domain V of the EV71 IRES, indicating that it adopts a more compact conformation than PV and CVB3 IRESs.
Cleavage in PV IRES domain V from eIF4G-Cys-830 was slightly enhanced by eIF4A with/without ATP or eIF4B (Fig. 1G), which may have been caused by enhancement of eIF4G's interaction with type 1 IRESs [as has been shown for type 2 IRESs (5)], but could also reflect eIF4A-induced conformational changes in the IRES.
Enzymatic Footprinting Analysis of the Interaction of eIF4G with Type 1 IRESs.
Enzymatic footprinting was used to confirm eIF4G's interaction with type 1 IRESs. eIF4G737–1116 protected nucleotides 462 and 538–539 in PV domain V from RNase V1 cleavage (Fig. 1H, lane 3). Protection by eIF4G737–1116/eIF4A or eIF4F was slightly stronger than by eIF4G alone (Fig. 1H, lanes 4 and 5), which again suggests that eIF4A might enhance the eIF4G/IRES interaction. In CVB3 domain V, eIF4G737–1116, eIF4G737–1116/eIF4A, and eIF4F all protected nucleotides 466 and 542–543 (Fig. 1I). No protection was apparent elsewhere in the PV and CVB3 IRESs (e.g., Fig. S3). These sites of protection mapped to opposite strands of a short helix in domain V (Fig. 2 D and E) and correlated with a site of directed hydroxyl radical cleavage (Fig. 2 A–C). eIF4G/eIF4A, and to a greater extent eIF4F, enhanced RNase V1 cleavage at the apex of PV domain V (nucleotides 489–493 and 501–504) (Fig. 1H), suggesting that their binding might also induce conformational changes in this region. The eIF4G binding-site maps to the highly-conserved basal region of domain V of type 1 IRESs (ref. 10 and Fig. 2F), so that all type 1 IRESs likely interact specifically with the central domain of eIF4G.
eIF4G-Mediated Recruitment of eIF4A to Type 1 IRESs.
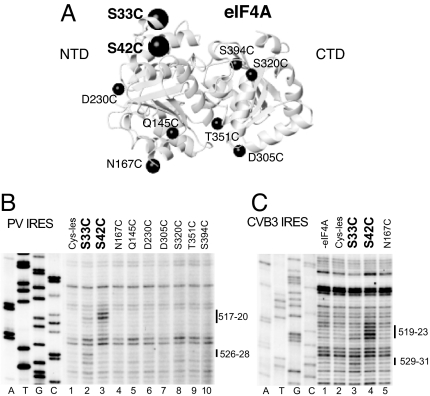
Directed hydroxyl radical cleavage and enzymatic footprinting data suggested that eIF4G recruits eIF4A to type 1 IRESs. We therefore investigated eIF4A's position in IRES/eIF4G/eIF4A complexes by directed hydroxyl radical cleavage, using a panel of 9 eIF4A cysteine mutants (Fig. 3A). eIF4A consists of 2 domains joined by a flexible linker (Fig. 3A). The eIF4A-NTD binds to the C-terminal helix of the central domain of eIF4G, whereas the eIF4A-CTD binds to the N-terminal 2 HEAT-repeats and a flexible N-terminal region (18).
Fig. 3.
Recruitment of eIF4A to type 1 IRESs by eIF4G. (A) Model of eIF4A (Protein Data Bank ID code 3EX7), with spheres labeled to show the positions of cysteines used to tether Fe(II)-BABE. (B and C) Primer extension analysis of directed hydroxyl radical cleavage of PV (B) and CVB3 (C) IRESs from Fe(II)-tethered eIF4A in IRES/eIF4G/eIF4A complexes. Sites of hydroxyl radical cleavage are indicated to the right. Lanes A, T, G, and C depict sequences generated from the same primers.
Hydroxyl radicals generated from 2 positions in the eIF4A-NTD induced cleavage of PV and CVB3 IRESs in domain V's central loop and an adjacent helix (Figs. 2 G and H and 3 B and C). Cys-42 induced cleavage at PV nucleotides 517–520 and CVB3 nucleotides 519–523, whereas Cys-33 induced weaker cleavage at PV nucleotides 518–519 and 526–528 and at CVB3 nucleotides 529–531 (Fig. 3 B, lanes 2 and 3 and C, lanes 3 and 4). No cleavage occurred in the absence of eIF4G (Fig. S4). The overlap between the sites of cleavage induced by eIF4A-Cys-33 and eIF4A-Cys-42 (Fig. 2 G and H) and those induced by eIF4G-Cys-929 (Fig. 2 A–C) is consistent with the proximity of these residues in eIF4G/eIF4A complexes (18).
eIF4G/eIF4A Induce Conformational Changes at the 3′ Border of Type 1 IRESs.
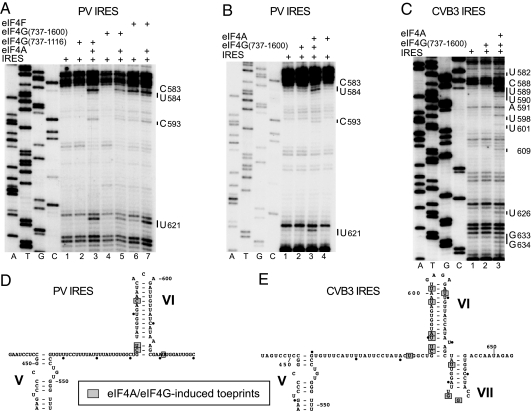
Although no cleavage was observed around the Yn-Xm-AUG motif at the 3′ border of type 1 IRESs from eIF4G or eIF4A (Figs. S1, S2, and S5), in toe-printing experiments, incubation of the PV IRES with eIF4F or with eIF4A and either eIF4G737–1116 or eIF4G737–1600 (which contain 1 or both binding sites for eIF4A, respectively) together led to the appearance of stops in domain VI and immediately downstream of it (nucleotides 583–584, 593, and 621) (Fig. 4A, B, and D). In analogous experiments, incubation of the CVB3 IRES with eIF4G737–1600/eIF4A also led to the appearance of toe prints in domain VI and its vicinity (nucleotides 582, 588–591, 598, 601, 609, 626, and 633–634) (Fig. 4 C and E). Taking into account the lack of directed hydroxyl radical cleavage in domain VI from eIF4G (Figs. S1, S2, and S5) or eIF4A, and the lack of protection from RNase V1 cleavage in regions downstream of domain V by eIF4G/eIF4A (Fig. S3), such toe prints in domain VI and its vicinity are more consistent with induction by eIF4G/eIF4A of conformational changes at the 3′ border of type 1 IRESs than with direct interaction of eIF4G/eIF4A with these regions.
Fig. 4.
Conformational changes induced by eIF4G/4A at the 3′ border of PV and CVB3 IRESs. (A–C) Toe-printing analysis was done on PV (A and B) and CVB3 (C) IRESs in the presence of ATP and eIFs as indicated. Lanes A, T, G, and C depict sequences generated from the same primers. The positions of stops caused by the presence of eIFs are indicated to the right. (D and E) The positions of stops are mapped onto PV domain VI (D) and CVB3 domains VI and VII (E) (25, 26).
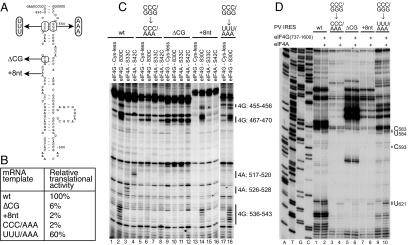
Binding of eIF4G/eIF4A to Domain V Is Required for PV IRES Function.
The functional importance of the interactions of eIF4G and eIF4A with the PV IRES was addressed by analyzing the effects on these interactions of mutations in domain V (Fig. 5A) that impaired IRES-mediated translation (Fig. 5B). The defect in translation of the ΔCG463–464 mutant was consistent with previous reports (e.g., ref. 19). The GGG548–550 → AAA mutation strongly reduced cleavage of the IRES from eIF4G-Cys-830 and from eIF4A-Cys-33 and eIF4A-Cys-42 (Fig. 5C, lanes 2–4 and 6–8), indicating that it significantly impaired binding of eIF4G, and consequently, eIF4A. This defect was restored in the CCC453–456/GGG548–550 → UUU/AAA double mutant (Fig. 5C, lanes 17 and 18). A deletion (ΔCG463–464) in a helix that is directly protected by eIF4G from RNase V1 cleavage also abrogated cleavage from eIF4G-Cys-830, and eIF4A-Cys-33 and eIF4A-Cys-42, and thus by implication, prevented their binding to the IRES (Fig. 5C, lanes 9–12). These observations are consistent with the results of directed hydroxyl radical probing and enzymatic footprinting, which mapped eIF4G's site of interaction to the basal region of domain V. Importantly, an 8-nt insertion (A468 + 8 nt) abrogated only cleavage from eIF4A-Cys-33 and eIF4A-Cys-42 without significantly affecting cleavage from eIF4G-Cys-830 (Fig. 5C, lanes 13–16), indicating that this insertion impairs recruitment of eIF4A to the eIF4G/IRES complex. Consistently, all mutations that affected binding of eIF4G or recruitment of eIF4A to the IRES also strongly reduced the appearance in toe-printing experiments of stops at nucleotides 583–584, 593, and 621 (Fig. 5D). Thus, the ability of IRES variants to interact with the eIF4G/eIF4A complex correlated with their translational activity.
Fig. 5.
Determinants of binding of eIF4G/eIF4A to domain V of the PV IRES. (A and B) The effects of mutations in PV domain V (A) were assayed by translation in vitro (B). Values represent the translational activity of IRES mutants as a percentage of the value for the WT IRES, and are the mean of 3 assays. (C) Primer extension analysis of directed hydroxyl radical cleavage of PV IRES mutants in eIF4G/IRES complexes by Fe(II)-tethered eIF4G737–1116(Cys-830), and in eIF4G/eIF4A/IRES complexes by Fe(II)-tethered eIF4A-Cys-33 and eIF4A-Cys-42, as indicated. (D) Toe-printing analysis of conformational changes induced by eIF4G/4A at the 3′ border of PV IRES mutants. Lanes A, T, G, and C depict sequences generated from the same primer.
Discussion
We report that eIF4G specifically recognizes domain V of CVB3, EV71, and PV IRESs, binds to them with the same orientation, and, importantly, does so independently of other factors. These data extend the observation that eIF4G cross-links to PV IRES domain V in RRL (15). eIF4G's binding site is limited to the near-universally-conserved base of domain V (Fig. 2F and ref. 10). Mutations in this region severely impair translation and consequently impair or even abrogate virus growth (Fig. 5; refs. 9 and 19–21). Importantly, the introduction of the attenuating Sabin 3 mutation into domain V of the WT PV IRES at nucleotide 469 reduced cross-linking of eIF4G and eIF4B to this domain in RRL (15). These data and the fact that compensatory mutations that restored base-pairing in the base of PV domain V restored IRES-mediated translation and the IRES's ability to interact with eIF4G (Fig. 5) strongly support the importance of this interaction for type 1 IRES function.
Type 1 and 2 IRESs are unrelated, and the only similarity between them is the Yn-Xm-AUG motif at their 3′ borders. It is therefore significant that the position and orientation with which the central domain of eIF4G binds type 1 and 2 IRESs relative to the Yn-Xm-AUG motif are analogous. Thus, in both cases, eIF4G interacts with domains (V in type 1 IRESs and J–K in type 2 IRESs) that are adjacent to this motif (Fig. 2 I and J), and in both instances its C terminus is oriented toward the apex and the N terminus is oriented toward the base of these domains (this paper and ref. 7). As with type 2 IRESs (5), eIF4G recruits eIF4A to type 1 IRESs, and eIF4A may in turn enhance the eIF4G-type 1 IRES interaction (e.g., Fig. 1 G and H). eIF4A is critical for IRES function, and the failure to recruit it by the PV (A468 + 8 nt) IRES mutant correlated with its severe loss of activity, even though it retained the ability to bind eIF4G. The correlation between the ability of the PV IRES to recruit eIF4A and its ability to mediate internal initiation is consistent with the dependence of type 1 IRESs on eIF4A (12).
eIF4G and eIF4A together induced strong toe prints at the 3′ border of type 1 IRESs (in domain VI and its immediate vicinity) that likely indicate conformational changes in the IRES, because no direct interaction of eIF4G or eIF4A with this region of the IRES was detected in hydroxyl radical cleavage or footprinting experiments. The finding that recruitment of eIF4G/eIF4A to domain V led to these changes in domain VI is consistent with observations suggesting that domains V and VI are functionally linked and might interact structurally, such as their synergy in promoting UV cross-linking to the IRES of a 36-kDa protein in cellular extracts (22) and determining PV neurovirulence (23). Importantly, binding of eIF4G/eIF4A to the type 2 EMCV IRES induced analogous toe prints at its 3′ border (7). These conformational changes induced at the 3′ borders of type 1 and 2 IRESs could be essential for subsequent attachment of 43S complexes to these regions.
The observations reported here are therefore consistent with a model for initiation in which eIF4G binds to the IRES adjacent to the site of ribosomal entry and recruits eIF4A, which can then remodel the IRES so that it adopts a conformation to which a 43S complex can attach productively. This model is directly analogous to the proposed mechanism of initiation on type 2 IRESs, which is similarly based on binding of eIF4G to a domain of the IRES adjacent to the site of ribosomal attachment, recruitment of eIF4A, enhancement of eIF4G's binding to the IRES by eIF4A, and eIF4A/eIF4G-induced conformational changes at the 3′ border of the IRES (4–7). Thus, despite the lack of sequence and structural similarity between type 1 and 2 IRESs, several fundamental aspects of the mechanisms of initiation on them are nevertheless similar.
Although our data strongly suggest that binding of eIF4G/eIF4A is a key step in initiation on type 1 and 2 IRESs, it is important to note that it is not sufficient for recruitment of 43S complexes on type 2 IRESs (and likely also not for type 1 IRESs). Thus, even though the individual J–K domain of the type 2 EMCV IRES can bind eIF4G/eIF4A, it is unable to recruit the 40S subunit in the absence of the other IRES domains (see ref. 7 for a discussion). Moreover, although the eIF3–eIF4G interaction may contribute to recruitment of 43S complexes to both classes of IRESs, our determination that eIF4G deletion mutants that are unable to bind eIF3 can nevertheless support initiation on the EMCV IRES (5) suggests that additional (and as-yet unidentified) interactions of type 2 IRESs with the translational apparatus are required for productive recruitment of 43S complexes. The actual mechanism of recruitment of the 40S subunit to eIF4G/eIF4A-bound type 1 and 2 IRESs therefore remains unresolved.
Analyses of type 2, 3, and 4 IRESs have established 3 fundamentally different modes of IRES-mediated initiation, each involving distinct primary interactions with components of the translation apparatus. By contrast, we report here that there are fundamental underlying similarities between the mechanisms of initiation on the unrelated type 1 and 2 IRESs. These observations suggest that there are likely only a limited number of possible mechanisms of IRES-mediated initiation and IRES/factor interactions that drive them, and thus that other IRESs with structural properties that are distinct from those of the 4 major IRES groups may nevertheless use similar mechanisms to mediate initiation.
Methods
In Vitro Translation Assays.
PV mRNAs were translated for 1 h at 30 °C in cell-free HeLa extracts (11). Translation products were analyzed by SDS/PAGE and quantified with a PhosphoImager.
Purification of Initiation Factors.
eIF4F was purified from RRL (Green Hectares), and recombinant eIF4A, eIF4B, eIF4G737–1116, and eIF4G737–1600 were expressed and purified from Escherichia coli DE3 (4, 7, 24).
Footprinting Analysis of eIF4G on PV or CVB IRES Complexes.
PV/CVB3 IRES (1 pmol) was incubated alone or with 2 pmol of eIF4G (with/without 3 pmol of eIF4A) or 2 pmol of eIF4F in 40 μL of buffer [20 mM Tris (pH 7.5), 100 mM KAc, 1 mM DTT, 2.5 mM MgCl2, and 0.25 mM spermidine] for 10 min at 37 °C, and then digested with RNase V1 (0.28 × 10−3 units/mL) for 10 min at 37 °C. Cleavage sites were mapped by using avian myeloblastosis virus reverse transcriptase (AMV-RT) and appropriate primers (Table S1).
Fe(II)-BABE Modification of eIF4GI737–1116 and eIF4A Mutants.
Derivatization of single-cysteine eIF4G737–1116 and eIF4A mutants and mock-derivatization of cysteineless eIF4G737–1116 and eIF4A with Fe(II)-BABE was done as described (7).
Directed Hydroxyl Radical Probing.
Complexes were formed by incubating 4 pmol of [Fe(II)-BABE]-eIF4G737–1116 or 8 pmol of [Fe(II)-BABE]-eIF4A and 4 pmol of eIF4G737–1600 with 2 pmol of CVB3, EV71 or PV IRESs in 40 μL of buffer [20 mM Tris·HCl (pH 7.5), 80 mM KCl, 2.5 mM MgAc] at 37 °C for 10 min. When indicated, 8 pmol of eIF4A, 8 pmol of eIF4B, and 1 mM ATP were added to reactions containing [Fe(II)-BABE]-eIF4G737–1116. To generate hydroxyl radicals, the reaction mixture was supplemented with 0.05% H2O2 and 5 mM ascorbic acid and incubated on ice for 10 min. Reactions were quenched by adding 20 mM thiourea and 110 μL of stop solution (300 mM β-mercaptoethanol, 0.2% SDS, 10 mM EDTA). Cleavage sites were analyzed by primer extension using AMV-RT.
Toe-Printing Assay.
Complexes were assembled by incubating 2 pmol of PV or CVB3 RNA for 10 min at 37 °C in 40 μL of buffer [1 mM DTT, 80 mM KCl, 20 mM Tris (pH 7.5), 2.5 mM MgOAc, 40 units RNase OUT, 0.25 mM spermidine, 1 mM ATP] with combinations of eIF4F (4 pmol), eIF4G737–1116 (8 pmol), or eIF4G737–1600 (8 pmol), and eIF4A (20 pmol) as indicated. CVB3 (nucleotides 701–682) or PV1 (nucleotides 706–687) 32P-labeled primer was added to the mixture for 5 min and chilled on ice. Primer extension analysis was done by adding 1 μL of MgOAc (360 mM), 4 μL of dNTPs (5 mM), and 5 units AMV-RT to reaction mixtures, followed by incubation for 2 h at 20 °C. cDNAs were analyzed by electrophoresis through 6% polyacrylamide sequencing gels.
Sequence Alignment.
A total of 241 enterovirus IRES domain V sequences were identified, aligned in BLAST searches (www.ncbi.nlm.nih.gov/BLAST) of viral sequences in the GenBank nonredundant database, and used to determine nucleotide identities.
Supplementary Material
Acknowledgments.
We thank M. Arita (National Institute of Infectious Diseases, Tokyo), E. Dobrikova (Duke University Medical Center, Durham, NC), and M. Gromeier (Duke University Medical Center, Durham, NC) for plasmids. This work was supported by National Institutes of Health Award AI-51340 and American Heart Association (Heritage Affiliate) Award 0755900T.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900153106/DCSupplemental.
References
- 1.Pestova TV, Lorsch J, Hellen CUT. In: Translational Control in Biology and Medicine. Mathews M, Sonenberg N, Hershey J, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–120. [Google Scholar]
- 2.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 4.Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilipenko EV, et al. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 7.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 9.Bonderoff JM, Lloyd RE. CVB translation: Lessons from the polioviruses. Curr Top Microbiol Immunol. 2008;323:123–147. doi: 10.1007/978-3-540-75546-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova TV, Hellen CUT, Wimmer E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology. 1994;204:729–737. doi: 10.1006/viro.1994.1588. [DOI] [PubMed] [Google Scholar]
- 12.Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svitkin YV, et al. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 15.Ochs K, et al. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J Virol. 2003;77:115–122. doi: 10.1128/JVI.77.1.115-122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 17.Marcotrigiano J, et al. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 18.Marintchev A, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dildine SL, Stark KR, Haller AA, Semler BL. Poliovirus translation initiation: Differential effects of directed and selected mutations in the 5′ noncoding region of viral RNAs. Virology. 1991;182:742–752. doi: 10.1016/0042-6822(91)90615-i. [DOI] [PubMed] [Google Scholar]
- 20.Svitkin YV, Cammack N, Minor PD, Almond JW. Translation deficiency of the Sabin type 3 poliovirus genome: Association with an attenuating mutation C472 → U. Virology. 1990;175:103–109. doi: 10.1016/0042-6822(90)90190-3. [DOI] [PubMed] [Google Scholar]
- 21.Macadam AJ, Stone DM, Almond JW, Minor PD. The 5′ noncoding region and virulence of poliovirus vaccine strains. Trends Microbiol. 1994;2:449–454. doi: 10.1016/0966-842x(94)90803-6. [DOI] [PubMed] [Google Scholar]
- 22.Haller AA, Semler BL. Stem-loop structure synergy in binding cellular proteins to the 5′ noncoding region of poliovirus RNA. Virology. 1995;206:923–934. doi: 10.1006/viro.1995.1015. [DOI] [PubMed] [Google Scholar]
- 23.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 25.Bailey JM, Tapprich WE. Structure of the 5′ nontranslated region of the coxsackievirus B3 genome: Chemical modification and comparative sequence analysis. J Virol. 2007;81:650–668. doi: 10.1128/JVI.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilipenko EV, et al. Conserved structural domains in the 5′ untranslated region of picornaviral genomes: An analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.