Abstract
The lengths of von Willebrand factor (VWF) concatamers correlate with hemostatic potency. After secretion in plasma, length is regulated by hydrodynamic shear force-dependent unfolding of the A2 domain, which is then cleaved by a specific protease. The 1.9-Å crystal structure of the A2 domain demonstrates evolutionary adaptations to this shear sensor function. Unique among VWF A (VWA) domains, A2 contains a loop in place of the α4 helix, and a cis-proline. The central β4-strand is poorly packed, with multiple side-chain rotamers. The Tyr-Met cleavage site is buried in the β4-strand in the central hydrophobic core, and the Tyr structurally links to the C-terminal α6-helix. The α6-helix ends in 2 Cys residues that are linked by an unusual vicinal disulfide bond that is buried in a hydrophobic pocket. These features may narrow the force range over which unfolding occurs and may also slow refolding. Von Willebrand disease mutations, which presumably lower the force at which A2 unfolds, are illuminated by the structure.
Although there is great interest in the use of mechanical force to unfold protein domains and gain insights into the structural factors that govern mechanical stability, there are, as yet, few structural studies on domains that have evolved to unfold in the course of normal physiology and serve as biological force sensors (1). Von Willebrand factor (VWF) senses the high vascular flow rates (shear) found at sites of arterial bleeding and cross-links platelets to plug vessels in hemostasis. High shear activates binding of the A1 domain in VWF to platelet GPIb, and facilitates binding of VWF through its A3 domain to subendothelial collagen exposed at sites of vessel injury. The force-sensing A2 domain is located in between the A1 and A3 domains (2, 3).
Hemostatic potential greatly increases with VWF length, which is tightly regulated in vivo (2, 3). The 240,000-Mr VWF monomer is dimerized through disulfide bonds at its C terminus (C C) and then concatenated through specific disulfide bonds at its N terminus (N
C) and then concatenated through specific disulfide bonds at its N terminus (N N) into multimers up to ≈50 × 106 Mr (Fig. 1A). VWF is stored in granules in endothelial cells in an ultralarge form (ULVWF) that is secreted in response to thrombogenic stimuli. Within 2 hours after release into the circulation, ULVWF is converted by ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) to smaller multimers with a wide range of lengths that are characteristic of the circulating pool of VWF (4). Cleavage depends on hydrodynamic shear force, and occurs within the A2 domain at its Tyr1605–Met1606 bond. Genetic or acquired deficiency of ADAMTS13 causes thrombotic thrombocytopenic purpura, a life-threatening disease in which microvascular thrombi form in arterioles and capillaries. Conversely, mutations in the A2 domain cause a bleeding disorder, type 2A von Willebrand disease (VWD), in which VWF multimers are smaller in size than in healthy individuals and have less hemostatic potential (2, 5, 6).
N) into multimers up to ≈50 × 106 Mr (Fig. 1A). VWF is stored in granules in endothelial cells in an ultralarge form (ULVWF) that is secreted in response to thrombogenic stimuli. Within 2 hours after release into the circulation, ULVWF is converted by ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) to smaller multimers with a wide range of lengths that are characteristic of the circulating pool of VWF (4). Cleavage depends on hydrodynamic shear force, and occurs within the A2 domain at its Tyr1605–Met1606 bond. Genetic or acquired deficiency of ADAMTS13 causes thrombotic thrombocytopenic purpura, a life-threatening disease in which microvascular thrombi form in arterioles and capillaries. Conversely, mutations in the A2 domain cause a bleeding disorder, type 2A von Willebrand disease (VWD), in which VWF multimers are smaller in size than in healthy individuals and have less hemostatic potential (2, 5, 6).
Fig. 1.
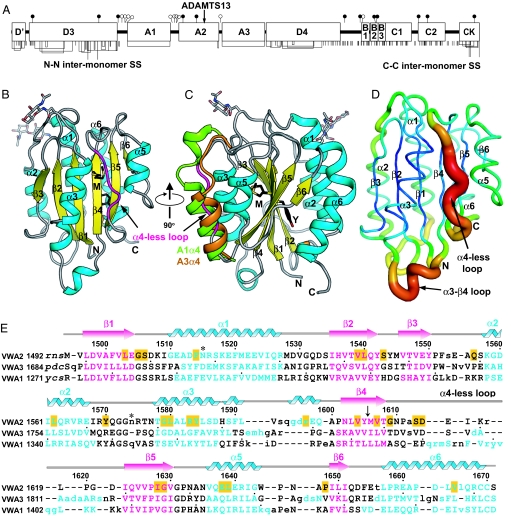
The VWF A2 domain. (A) Overall structure of mature VWF. Cysteines are shown as vertical lines and are connected for chemically defined disulfides (10). N- and O- linked glycans are closed and open lollipops, respectively. (B and C) Ribbon diagram of VWF A2 domain. N-linked glycosylation sites are shown in stick. The C-terminal disulfide bond and ADAMTS13 cleavage site residues (Y, Y1605; M, M1606) are shown as sticks. In C, the α4 helices in VWF A1 and A3 are shown after superposition of the domains. (D) Cα B-factors. Higher B-factors are represented by a thicker chain-trace and a spectral shift from blue to red. (E) Structure-based sequence alignment. β-Strands and α-helices are colored. Dots show decadal residues. The ADAMTS13 cleavage site is indicated by an arrow. Glycosylation sites are indicated by asterisks. Residues involved in VWD mutations are highlighted in yellow. Unaligned residues are lower case and residues absent in the A2 structure are italicized.
Models of the A2 domain suggest that the ADAMTS13 cleavage site is either in a loop (7), or buried in its central β-sheet (8) and, hence, inaccessible to ADAMTS13. Indeed, single-molecule laser-tweezers experiments show that the unfolded, but not folded, A2 domain is cleaved by ADAMTS13 (9). Mutations in type 2A VWD are thought to destabilize the A2 domain, enabling unfolding and, hence, cleavage at lower forces.
The crystal structure of A2 reveals interesting evolutionary adaptations to its function as a shear sensor domain that undergoes force-regulated cleavage by ADAMTS13 and shows how mutations destabilize A2 in VWD.
Results
Overall Structure of VWF A2.
A 1.9-Å crystal structure of A2 (Table 1) reveals 2 independent molecules, with a Cα atom RMSD of 0.2 Å, in the asymmetric unit. One GlcNAc residue is visible in density at each predicted N-linked site (Fig. 1 B and C). A cis-peptide bond is present between Trp1644 and Pro1645; the implications of this cis-peptide for refolding of the A2 domain are discussed below.
Table 1.
Statistics of data collection and refinement
| Data collection | |
| Space group | P21 |
| Wavelength, Å | 0.9792 |
| Cell parameters (a, b, c), Å β, ° | 55.15, 60.81, 56.3199.14 |
| Molecules/asymmetric unit | 2 |
| Total unique reflections | 29,040 |
| Rmerge* | 10.4 (80.1)† |
| I/σI | 11.4 (2.0) |
| Completeness, % | 98.8 (98.3) |
| Redundancy | 4.1 (4.1) |
| Refinement | |
| Resolution, Å | 1.90 |
| No. reflections | 27,322 |
| Rwork/Rfree, %‡ | 18.9/22.4 |
| Nonhydrogen atoms (protein/NAcGln/water) | 3,222/84/410 |
| B factors (average) (protein/NAcGln/water) | 15.6/38.2/29.6 |
| rmsd bond length, Å | 0.007 |
| rmsd bond angle, ° | 0.984 |
| Ramachandran plot (% favored/allowed/outliers)§ | 97.3/2.7/0.0 |
| PDB ID code | 3GXB |
*Rmerge=ΣhklΣi|Ii−〈I〉|/ΣhklΣi|Ii|, where Ii and 〈I〉 are the ith and mean measurement of the intensity of reflection hkl.
†Values in parentheses are for highest-resolution shell (2.00–1.90).
‡Rwork=Σhkl∥Fobs|−|Fcalc∥/Σhkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. No I/σ cutoff was applied. Rfree is the cross-validation R factor computed for the 5% test set of unique reflections.
§Residues in favored, allowed, and outlier regions of the Ramachandran plot as reported by MOLPROBITY (31).
The A2 fold resembles the VWA fold with α-helices and β-strands that largely alternate in sequence (Fig. 1). The hydrophobic β-sheet core contains β-strands in order β3-β2-β1-β4-β5-β6, with β3 antiparallel to the others. Other proteins with VWA folds contain 6 amphipathic α-helices that sequentially encircle the β-sheet (Fig. 1C and supporting information (SI) Fig. S1).
The A2 domain is immediately distinguished from other VWA domains by its lack of an α4-helix (Fig. 1 B and C). In place of the α4-helix, a long loop runs from the C terminus of the β4-strand to the N terminus of the β5-strand. To facilitate comparisons to other VWA domains, we give the α-helices in the A2 domain the same indices as the corresponding helices in the A1 and A3 domains. Furthermore, to emphasize that the long β4–β5 loop in A2 occupies the same topological position as the α4-helix in A1 and A3, we call this idiosyncratic loop the “α4-less loop.”
The ADAMTS13 cleavage site at residues Tyr1605 and Met1606 is present at the center of the β-sheet, near the middle of the central β4-strand (Fig. 1 B, C, and E). Provocatively, the cleavage site is buried by the α4-less loop.
α4-Less Loop.
The α4-less loop in A2 occupies the same position in the fold as the α4-helix in other VWA domains, between the α3 and α5 helices (Fig. 1 B and C). The side chain of Asp1614 N-caps the α3-helix by forming multiple H-bonds to the backbone nitrogens of the first few α3-helical residues (Fig. 2 A and B). Asp1806 of the A3 domain has a capping function similar to Asp1614 of A2 (Fig. 1E).
Fig. 2.
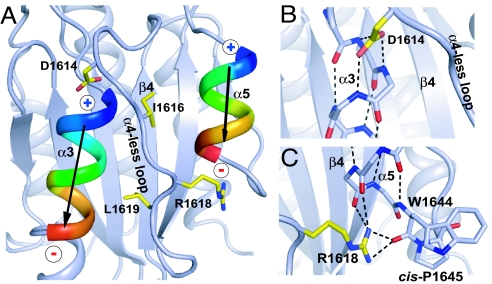
The α4-less loop environment. (A) The α3 and α5-helix dipole moments are symbolized. (B) The Asp-1614 N-cap. (C) The Arg1618 C-cap and H-bond to the cis-peptide. Key side chains and helix main chain (in B and C) are shown in stick, and H-bonds are dashed.
Because the α4-less loop has a more extended conformation than an α-helix, fewer residues are present than in the corresponding segments in A1 and A3 (Fig. 1E). A2 residues 1615–1619 have a similar topological function to the missing α4-helix, burying the hydrophobic central β-sheet and running from the C terminus to the N terminus of the β-sheet. The side chain of α4-less loop residue Ile1616 is buried beneath the loop in a contact with residue Met1606 of the ADAMTS13 cleavage site. Similarly, Leu1619 is buried in a hydrophobic contact with the central β-sheet (Fig. 2A). Furthermore, uniquely in A2, the side chain of α4-less loop residue Arg1618 C-caps the α5-helix by H-bonding to backbone carbonyl oxygens at the α5 C terminus (Fig. 2 A and C). Thus, the dipoles of the α3- and α5-helices interact with charged, capping residues Asp1614 and Arg1618, respectively (Fig. 2), which are invariant in A2 (Fig. S2). Together, the buried hydrophobic residues and helix-capping residues help stabilize the conformation of the α4-less loop.
As a surrogate for protein backbone flexibility, we examined Cα atom B factors (Fig. 1D). The α4-less loop shows the highest B factors in the A2 domain. Positive density near the backbone at residues 1617–1619 in the α4-less loop also suggests a second, minor backbone conformation here. In addition, in the central β-sheet, the short β6-strand has lower B factors than the other β-strands, consistent with unfolding from the C terminus as discussed below.
ADAMTS13 Cleavage Site.
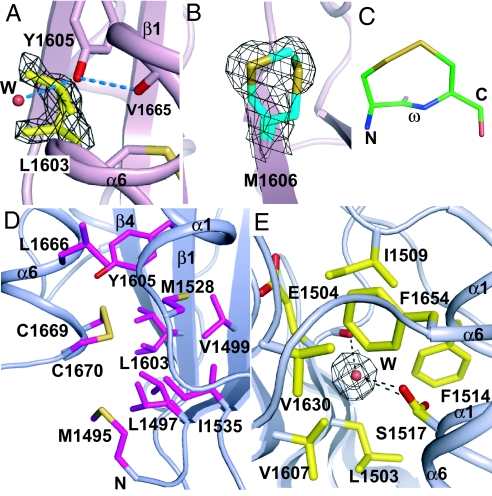
The ADAMTS13 cleavage site at Tyr1605 and Met1606 is in the middle of the β4-strand, deeply buried in the A2 hydrophobic core (Fig. 1 B and C). The side chain of Tyr1605 points toward the C terminus of the A2 domain, where it H-bonds to the backbone carbonyl oxygen of α6-helix residue Val-1665 (Fig. 3A). The Tyr1605 side chain also contacts the side chain of β4-strand residue Leu1603. Interestingly, the Leu1603 side chain adopts 2 markedly different conformations, which differ by 2 Å in Cγ atom position (Fig. 3A).
Fig. 3.
Structural specializations of A2. (A and B) The distinct rotamers of Leu1603 (A) and Met1606 (B). (C) The 8-membered ring formed by the vicinal disulfide. (D) The vicinal disulfide and its hydrophobic pocket. Key side chains are shown in stick and water molecules as spheres. (E) A water in a largely hydrophobic environment, with both Ser1517 rotamers shown. 2Fo-Fc densities around selected side chains and waters are contoured at 0.85 σ.
Cleavage site residue Met1606 locates on the opposite side of the β-sheet, underneath the α4-less loop. Met1606 also exhibits multiple side-chain rotamers, with alternative Met Cγ and Sδ atom positions 2 to 2.5 Å apart (Fig. 3B). Thus, β4-strand side chains on both sides of the β-sheet show poor packing. The β4-strand is also 1 residue shorter in A2 than in A1 and A3, because Pro1601 forms 1 less H-bond than the corresponding Ser residue in A1 and A3 (Fig. S3).
Unusual, C-Terminal, Vicinal Disulfide Bond.
In agreement with disulfide assignments in intact VWF (10), our structure shows that adjacent residues Cys1669 and Cys1670 are disulfide-bonded (Fig. 3 C and D). Together with the Cys1669–Cys1670 peptide backbone, the vicinal disulfide forms an 8-membered ring (Fig. 3C). The ring is strained, because the peptide bond is constrained to be nonplanar, with an ω angle of −152 ± 1°, in contrast to the ω angles in trans (±180°) and cis (0°) peptide bonds (Fig. S4). The vicinal disulfide forms an important part of the hydrophobic core of the A2 domain (Fig. 3D). It interacts with β4-strand residues Leu1603 (which has 2 rotamers) and Tyr1605 (at the cleavage site), α1-β2 loop residue Met1528, β2-strand residue Ile1535, and N-terminal residue Met1495. The importance of the vicinal disulfide in force resistance is discussed below.
Buried Waters.
Waters are buried in the A2 structure, including one in a mostly hydrophobic environment (Fig. 3E) and a network of 3 in a hydrophilic environment (Fig. 4B).
Fig. 4.
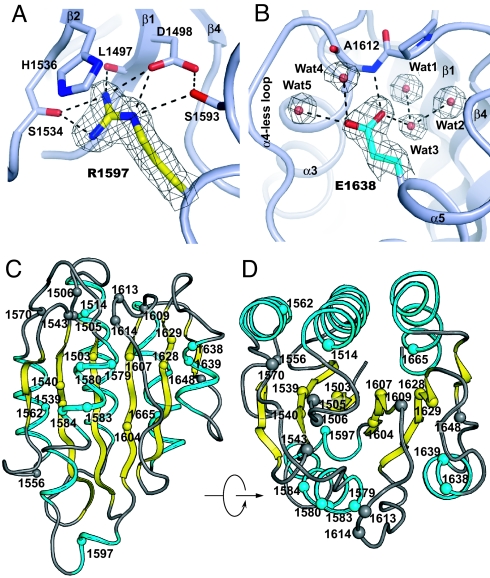
Type 2A VWD mutations. (A and B) The environments around Arg1597, commonly mutated to Trp (A) and Glu1638, mutated to Lys (B). Waters are spheres (only Waters 1, 2, and 3 are buried), H-bonds are dashed, and 2Fo-Fc densities are contoured at 1 σ. (C and D) VWD mutations, with each affected residue shown as a Cα sphere.
Type 2A VWD Mutations.
A subset of type 2A mutations in the A2 domain has been tested in vitro in pulse–chase biosynthesis experiments in transfected cells and further classified in 2 groups (11–13). Group II type 2A mutations do not affect secretion or multimer length before secretion; but after secretion in plasma, are trimmed by ADAMTS13 to markedly shorter lengths than healthy control VWF. The most commonly reported VWD mutation, R1597W (11, 14), as well as G1505E, I1628T, and E1638K belong to group II. Arg1597 is in a short 310 helix within the α3-β4 loop, the most flexible and extended loop in the A2 domain (Fig. 1 B–D). The Arg1597 side-chain guanido group stabilizes the orientation of this loop by stacking on His1536 and forming multiple H-bonds to the body of the domain (Fig. 4A). The Glu1638 side chain in the short α5-helix forms multiple H-bonds to the α4-less loop backbone and a network of buried water molecules (Fig. 4B). The E1638K mutation would destabilize packing of the α5-helix, and expose the buried waters to external solvent. The I1628T mutation is in the β5-strand. Compared with Ile, Thr lacks one methyl, and another methyl is substituted to a hydroxyl. Thus, I1628T introduces a small hole, and a polar atom, into the hydrophobic core. Gly1505 is buried just beneath the protein surface; substitution to larger residues requires structural rearrangements. The G1505E substitution is in group II, and G1505R is in group I.
Group I mutations impair secretion and may also affect multimer assembly. The S1506L mutation is in the same buried portion of the β1-α1 loop as the Gly1505 mutations. Ser1506 corresponds to the first Ser of the metal-binding Asp-Gly-Ser-X-Ser motif of integrin VWA or I domains; the first 3 residues of this motif are conserved in VWF A domains (Fig. 1E), although they do not bind metal. Glu1504 and Ser1506 are buried and H-bond to one another. Thus, the S1506L mutation disrupts side-chain H-bonds important in forming the β1-α1 loop. V1607D substitutes a hydrophobic, buried residue in the central β4-strand adjacent to the ADAMTS13 cleavage site for a charged residue. The L1540P mutation in the β2 strand abolishes a β-sheet H-bond. Thus, the Group I mutations are overall more structurally disruptive than those in Group II.
Of 24 different A2 domain residues that are mutated in type 2A VWD (www.shef.ac.uk/vwf), most have not been classified in group I or II. Many are buried in the hydrophobic core of the A2 domain (Fig. 4 C and D). Four Leu-to-Pro substitutions, L1503P, L1562P, L1580P, and L1639P locate to β-strands or α-helices and would destabilize A2 similarly to L1540P described above. Three mutations of Gly to Arg, G1579R, G1629R, and G1609R, affect Gly residues that are partially buried and would induce structural rearrangements. Similarly, S1543F, Q1556R, and V1604F substitute buried residues for larger residues. Conversely, F1514C represents a buried position mutated to a smaller residue. The most interesting ungrouped mutation is D1614G, which abolishes the ability of α4-less residue Asp1614 to N-cap the α3-helix.
Discussion
A2 as a Shear Bolt and VWD Mutations.
The structure of the A2 domain has important implications for A2 domain unfolding and how hydrodynamic force and ADAMTS13 cleavage regulate the size and, hence, hemostatic potency of VWF concatamers in vivo. Furthermore, the structure illuminates the basis for VWD, an important inherited disease. Type 2 VWD mutations are defined as affecting the function, rather than only the quantity of VWF in the circulation. The Group II subset of type 2A VWD substitutions (11) include mild packing defects in the protein interior, and loss of hydrogen bonds that stabilize loops, α-helices, or buried water molecules. These perturbations result in cleavage by ADAMTS13 to smaller, less active VWF multimers, and thus can be inferred to enable unfolding of the A2 domain at lower elongational forces.
The ADAMTS13 cleavage site is buried at the center of the β-sheet in the hydrophobic core of the A2 domain. Thus, the structure unequivocally demonstrates that unfolding of the A2 domain at least up to the β4-strand is required before cleavage by ADAMTS13. In an intact VWF multimer, elongational force will be transmitted from the A1 and the A3 domains through O-glycosylated linkers to the N and C termini of the A2 domain (Fig. 1A). Unfolding will proceed from the C rather than the N terminus of A2, because the C-terminal structural elements can be pried away one at a time from the end of the domain, whereas the β1-strand at the N terminus is buttressed by β-strands on either side in the core of the domain. Unfolding from the C terminus up to the α3–β4 loop, the long, extended loop that together with the α4-less loop has the highest B factors in the A2 domain, should be sufficient for cleavage by ADAMTS13, because this region corresponds to a 76-residue peptide substrate commonly used in VWF assays (15, 16).
The A2 domain provides surprising insights into the structural specializations of a domain that has evolved to be a force sensor. The A2 domain is the only domain within VWF that is not protected from unfolding by medium or long-range disulfide bonds (Fig. 1A). Based on this, and the known dependence of VWF cleavage by ADAMTS13 on hydrodynamic shear force, we conceptualize the A2 domain as a shear bolt domain. A shear bolt has a groove cut in its shaft. At this break-off groove, the bolt is designed to separate into 2 pieces above a threshold force, to protect other parts of a machine from accidental damage. Similarly, the α4-less loop, packing defects around the central β4-strand, and buried water molecules may endow the A2 domain with desirable characteristics as a force sensor, i.e., to unfold over a narrow, rather than wide, range of forces.
Specialized Structural Features of a Force-Sensing Domain.
The α4-less loop sets A2 apart from all other known VWA domains. The VWA fold superfamily is widely distributed in prokaryotes and eukaryotes. Structurally characterized VWA domains include those in integrins on cell surfaces, in complement components in the bloodstream, and in vesicle trafficking and DNA repair proteins inside the cell (17). Relationship among many of these domains is only detectable by structural similarity and not by sequence homology. Among 16 diverse, structurally characterized VWA domains, only VWF A2 lacks the α4-helix; furthermore, these 16 domains share all other 11 structural elements (6 β-strands and 5 α-helices). Moreover, the VWF A1 and A3 domains contain α4-helices, despite their sequence homology to and relatively more recent divergence from the A2 domain. In biology, form evolves to adapt to function. Thus, the available evidence strongly suggests that the lack of an α4-helix in the A2 domain is a unique evolutionary adaptation among VWA domains to the function of A2 as a shear bolt domain.
The B-factors in the α4-less loop and the α3-β4 loop are higher than any other region of the A2 domain; furthermore, positive density near the α4-less loop hints at a minor, alternative backbone conformation. In solution, these regions are likely to be dynamic and to explore different conformations that could seed unfolding. α-Helices have extensive H-bond networks that stabilize their secondary structure; in contrast, the α4-less loop does not, and has a substantially lesser number of residues that stabilize its association with neighboring structural elements. The α4-less loop may thus function to lower the force required for unfolding of the A2 domain.
The α4-less loop could alternatively function to delay refolding. Refolding of the A2 domain occurs relatively slowly, with a lifetime of 2 s (9). Molecules tumble in shear flow, and exposure to peak elongational force in shear flow occurs over a much shorter lifetime; therefore, slow refolding appears important to give ADAMTS13 time to cleave unfolded A2 domains (9). α-Helices form in the absence of other structural elements; however, there are no local interactions within the α4-less loop that could guide its formation in the absence of nearby structural elements. Therefore, a plausible alternative/additional function of the α4-less loop would be to slow refolding compared with VWA domains that contain α4-helices.
Other features of the A2 domain may contribute to its shear sensor function, including loose packing at its β4-strand and buried waters. To examine whether these are specializations of the A2 domain, we examined A1 and A3 domain structures solved at similarly high resolutions (18, 19). Inspection of their electron density shows substantially tighter packing of their β4-strands, with only Val1806 of A3 showing evidence for multiple side-chain rotamers. In contrast, in the A2 β4-strand, Leu1603, Met1606, and Thr1608 have alternative side-chain rotamers, and the alternative side-chain rotamers of Leu1603 and Met1606 differ substantially in position from one another. One reason for the looseness of Met1606 in A2 is that it is neighbored by Val1502 in the β1-strand, instead of the larger Leu residue found at this position in A1 and A3 domains (Fig. 1E and Fig. S3). Close packing of other residues around Leu1603 appears hindered by the large cleavage-site Tyr1605 side chain, which is unique to A2, and extends over Leu1603 toward the α6-helix. Another interesting specialization of the A2 domain is a buried water molecule in an environment that is largely hydrophobic, except for the Ser1517 side chain and β1-strand backbone, to which this water H-bonds (Fig. 3E). The buried Ser1517 residue is replaced by hydrophobic residues in the A1 and A3 domains.
Vicinal Disulfide Rigidity and Resistance to Unfolding.
Disulfide bonds between vicinal cysteines are rare in proteins and, when present, often are functionally important (20). Vicinal disulfides are not found in any other known VWA domains. The unusual 8-membered ring formed by the main chain and side chains of vicinal disulfide-bonded cysteines imparts rigidity (Fig. S4), which is important in resisting force.
Because C-terminal residue Ser1671 interacts only with Cys1670, all elongational force acting on the C terminus will be transmitted to Cys1670. Cys1669 and Cys1670 are the last 2 residues of the α6-helix; because of the rigid connection between them, these 2 residues will together bear the applied force. The enlargement of the force-resisting unit allows elongational force to be resisted simultaneously by the α-helical H-bonds of the Cys1669 and Cys1670 amides with the Val1665 and Leu1666 carbonyls, respectively, and by the hydrophobic and van der Waals interactions of the fused Cys1669–Cys1670 side chains with the hydrophobic pocket formed by the side chains of Leu1497, Met1528, Ile1535, Leu1603, Tyr1605, and Leu1666. The vicinal disulfide at the C terminus of the α6-helix is suggested to be an important specialization that functions to set a high energy barrier for the earliest step in A2 force-induced unfolding. It is further suggested to act like a plug for the hydrophobic core that, when pulled out, destabilizes the core and initiates unfolding.
The α4-less loop and the poor packing around the β4-strand may function to lower the barrier for force-induced unfolding, once the vicinal disulfide barrier has been breached. Upon removal of the vicinal disulfide from its hydrophobic pocket, water will be admitted to the hydrophobic core of the domain, a crucial step in protein unfolding. Cleavage site residue Tyr1605 provides a direct pathway between the vicinal disulfide and the β4-strand. Its side chain interacts with the vicinal disulfide and H-bonds to α6-helix residue Val1665 and solvent. Thus, after the initial breach, motion of the Tyr1605 side chain and its solvation may provide a pathway for transmission of disorder to the β4-strand, which, together with the packing defects around the β4-strand and the α4-less loop, may enable concerted unfolding of the C-terminal half of the A2 domain, exposing the cleavage site residues in the β4-strand to ADAMTS13. A similar argument holds in the reverse direction during refolding, so that refolding of the C-terminal portion of the domain may also be highly cooperative.
Cis-Pro and Refolding in an Extracellular Environment in the Absence of Prolyl Isomerases.
Cis-Pro1645 in the A2 domain (Fig. 2C) may play an important regulatory role in A2 domain refolding in the extracellular environment. Intracellular cis-trans proline isomerization acts as an on–off switch in some signaling proteins (21). Cis-Pro1645 is an A2 specialization, because a Pro found at a similar position in the A1 and A3 domains (Fig. 1E) is trans (18, 19). The A2 cis-peptide is stabilized by a hydrogen bond to C-capping, α4-less residue Arg1618 (Fig. 1C) in the folded state, but in the unfolded state could undergo cis-trans isomerization. Proline isomerization occurs with a rate on the order of 0.01 s−1 and is rate-limiting for protein folding (22). Conversion of the unfolded A2 domain to a state in which it was unlikely to refold over short time periods (time scale of 60 s), and subsequent recovery of refolding ability (time scale of 500 s), are consistent with cis-trans and trans-cis isomerization, respectively (9).
In vivo in the vasculature, conversion to a trans Trp1644-Pro1645 peptide bond would greatly delay A2 domain refolding, and would serve as a marker for the A2 domain in VWF multimers that had been unfolded for unusually long periods of time or been exposed to unusually high elongational forces, which can accelerate cis to trans peptide conversion (23). VWF free in the circulation is only briefly exposed to moderate forces, and refolding is rapid compared with the estimated time for cleavage by ADAMTS13 (9). However, VWF bound to endothelium or collagen at a site of hemorrhage binds multiple platelets, which exert much higher forces over substantially longer periods of time. Thus, conversion to a trans Trp1644-Pro1645 peptide bond would delay refolding and expedite cleavage by ADAMTS13 of VWF during vessel repair.
Summary.
The A2 structure reveals very interesting specializations including the lack of an α4-helix, poor packing around its central β4-strand, a cis-Pro, and a C-terminal, vicinal disulfide bond that is buried in a hydrophobic pocket. These specializations have important implications for the function of the A2 domain as a shear sensor domain within the ultralarge, vascular VWF protein, and for its cleavage by ADAMTS13.
Materials and Methods
Protein Expression and Purification.
A DNA segment encoding the murine Ig κ chain signal peptide, human VWF A2 (Met1495 to Ser1671 with prepro-VWF numbering), and a PHHHHHH sequence was cloned into the EcoR I and NotI sites of plasmid pLEXm (24). HEK293S GNT1− cells (25) were transiently transfected by using calcium phosphate. Culture supernatants (500 mL) were harvested after 5 days. Purification was performed with Ni-NTA affinity chromatography followed by Mono Q 4.6/100 (GE Healthcare) chromatography in 20 mM Tris (pH 8.0) with a 0 to 0.5 M NaCl gradient. Protein was concentrated to 8.4 mg/mL in 20 mM Tris·HCl (pH 8.0) and stored at −80 °C.
Crystallization and Data Collection.
Clusters of thin needles appeared in 0.1 M Bis-Tris (pH 7.2), 0.2 M lithium sulfate, 25% PEG3350. After microseeding with crushed needle clusters, crystals suitable for data collection appeared in 2 or 3 days at 277 K. Cryoprotection was by a quick dip in reservoir solution with added solid sucrose and 15% glycerol, followed by flash freezing. Data were collected at 100 K at NE-CAT ID24E microfocus beamline, by using ADSC quantum 315 detector (Advanced Photon Source, Argonne National Laboratory, Argonne, IL). Integration and scaling were done by using XDS and XSCALE (26). The crystal belongs to space group P21, with 2 molecules per asymmetric unit. The Matthew's coefficient is 2.8, and solvent content is 38%. Statistics of data collection are shown in Table 1.
Structure Determination and Refinement.
Crystal structure of human vWF A3 domain (PDB ID code 1FE8) was truncated to Cβ atoms. The output was used as search model in Molrep (27). Both rotation and translation search gave convincing results. The model was subjected to simulated annealing in CNS (28) at 1,500 K. Model building was with Coot (29). Further refinement with refmac5 used NCS restraints and TLS refinement (30). Coordinates are deposited as PDB ID code 3GXB
Supplementary Material
Acknowledgments.
We thank Drs. J. Evan Sadler and Wendy Thomas for reviewing the manuscript. This work was supported by National Institutes of Health Grant HL48675.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3GXB).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903679106/DCSupplemental.
References
- 1.Mayans O, et al. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 2.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Ruggeri ZM, Mendolicchio GL. Adhesion mechanisms in platelet function. Circ Res. 2007;100:1673–1685. doi: 10.1161/01.RES.0000267878.97021.ab. [DOI] [PubMed] [Google Scholar]
- 4.Batlle J, et al. Proteolytic degradation of von Willebrand factor after DDAVP administration in normal individuals. Blood. 1987;70:173–176. [PubMed] [Google Scholar]
- 5.Dong JF, et al. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 6.Tsai HM. Shear stress and von Willebrand factor in health and disease. Semin Thromb Hemost. 2003;29:479–488. doi: 10.1055/s-2003-44556. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins PV, Pasi KJ, Perkins SJ. Molecular modeling of ligand and mutation sites of the type A domains of human von Willebrand factor and their relevance to von Willebrand's disease. Blood. 1998;91:2032–2044. [PubMed] [Google Scholar]
- 8.Sutherland JJ, O'Brien LA, Lillicrap D, Weaver DF. Molecular modeling of the von Willebrand factor A2 Domain and the effects of associated type 2A von Willebrand disease mutations. J Mol Model. 2004;10:259–270. doi: 10.1007/s00894-004-0194-9. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein, von Willebrand Factor. Science. 2009 doi: 10.1126/science.1170905. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti T, Rosselet SJ, Titani K, Walsh KA. Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry. 1987;26:8099–8109. doi: 10.1021/bi00399a013. [DOI] [PubMed] [Google Scholar]
- 11.Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. Impaired intracellular transport produced by a subset of type IIA von Willebrand disease mutations. J Biol Chem. 1992;267:4424–4430. [PubMed] [Google Scholar]
- 12.Lyons SE, Cooney KA, Bockenstedt P, Ginsburg D. Characterization of Leu777Pro and Ile865Thr type IIA von Willebrand disease mutations. Blood. 1994;83:1551–1557. [PubMed] [Google Scholar]
- 13.Ribba AS, Voorberg J, Meyer D, Pannekoek H, Pietu G. Characterization of recombinant von Willebrand factor corresponding to mutations in type IIA and type IIB von Willebrand disease. J Biol Chem. 1992;267:23209–23215. [PubMed] [Google Scholar]
- 14.Ginsburg D, et al. Molecular basis of human von Willebrand disease: Analysis of platelet von Willebrand factor mRNA. Proc Natl Acad Sci USA. 1989;86:3723–3727. doi: 10.1073/pnas.86.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanardelli S, et al. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281:1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 16.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci USA. 2006;103:19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Springer TA. Complement and the multifaceted functions of VWA and integrin I domains. Structure (London) 2006;14:1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huizinga EG, Martijn van der Plas R, Kroon J, Sixma JJ. Crystal structure of the A3 domain of human von Willebrand factor: imlications for collagen binding. Structure (London) 1997;5:1147–1156. doi: 10.1016/s0969-2126(97)00266-9. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda K, et al. Structural basis of von Willebrand factor activation by the snake toxin botrocetin. Structure (London) 2002;10:943–950. doi: 10.1016/s0969-2126(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 20.Ruggles EL, Deker PB, Hondal RJ. Synthesis, redox properties, and conformational analysis of vicinal disulfide ring mimics. Tetrahedron. 2009;65:1257–1267. doi: 10.1016/j.tet.2008.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreotti AH. Native state proline isomerization: An intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 22.Schmid FX. Prolyl isomerase: Enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–142. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 23.Valiaev A, Lim DW, Oas TG, Chilkoti A, Zauscher S. Force-induced prolyl cis-trans isomerization in elastin-like polypeptides. J Am Chem Soc. 2007;129:6491–6497. doi: 10.1021/ja070147r. [DOI] [PubMed] [Google Scholar]
- 24.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 25.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 27.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 28.Brunger AT, et al. Crystallography and NMR System: A new software suite for macromolecular structure determination. Acta Crystallograllgr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.