Abstract
Conflict/reconciliation between mitochondria and nuclei in plants is manifested by the fate of pollen (viable or nonviable) in the cytoplasmic male sterility (CMS)/fertility restoration (Rf) system. Through positional cloning, we identified a nuclear candidate gene, RETROGRADE-REGULATED MALE STERILITY (RMS) for Rf17, a fertility restorer gene for Chinese wild rice (CW)-type CMS in rice (Oryza sativa L.). RNA interference-mediated gene silencing of RMS restored fertility to a CMS plant, whereas its overexpression in the fertility restorer line induced pollen abortion. The mRNA expression level of RMS in mature anthers depended on cytoplasmic genotype, suggesting that RMS is a candidate gene to be regulated via retrograde signaling. We found that a reduced-expression allele of the RMS gene restored fertility in haploid pollen, whereas a normal-expression allele caused pollen to die in the CW-type CMS. RMS encodes a mitochondrial protein, 178 aa in length, of unknown function, unlike the majority of other Rf genes cloned thus far, which encode pentatricopeptide repeat proteins. The unique features of RMS provide novel insights into retrograde signaling and CMS.
Keywords: cytoplasmic male sterility, fertility restoration, mitochondria, Oryza sativa, retrograde signaling
Cytoplasmic male sterility (CMS), which is a maternally inherited male sterility trait, is observed in more than 150 higher plant species. CMS is a useful system for commercial F1 hybrid breeding programs. CMS also is a focus of nuclear–mitochondrial research in plants, because aberrant mitochondrial genomic organization causes dysfunction in pollen development (1, 2). Fertility restoration (Rf) often is governed by a nuclear-encoded Rf gene. The first Rf gene cloned was Rf2a from maize, encoding a protein with aldehyde dehydrogenase activity (3, 4). Except for maize Rf2a, Rf genes in various plant species recently have been found to encode pentatricopeptide repeat (PPR) proteins (5–9). It has been proposed that a common function of PPR proteins is organelle posttranscriptional regulation, exemplified by RNA processing and editing (10, 11). The first PPR-encoding Rf gene reported was the petunia Rf-PPR592, which contains 11 continuous PPR motifs (5). Rf-PPR592 eliminates the CMS mitochondrial-specific protein PCF and resides within a high-molecular-weight protein complex associated with the mitochondrial membrane (12). PPRs also are responsible for fertility restoration in Brassica napus (6–8); in this case, a PPR protein encoded by Rfo reduces the abundance of CMS-associated ORF138 protein (6). In rice, Rf1a (13–16) and Rf1b (16) for Boro-Taichung (BT)-type CMS also have been identified as encoding PPR proteins. Rf1a promotes the processing of the BT-CMS-specific mitochondrial operon B-atp6-orf79 transcripts (13, 16), whereas Rf1b decreases the abundance of dicistronic transcripts of B-atp6-orf79 (16). Because Rf1a and Rf1b are located close together on the same chromosome and have highly conserved amino acid sequences, these 2 Rfs may be recently duplicated homologous genes. At least 9 Rf1a homologous sequences have been found around the Rf1a locus (16), and such Rf homologous sequences also are present around the respective Rf loci in petunia and B. napus CMS (5, 7, 8). These findings suggest the complex co-evolution of the Rf locus and mitochondrial CMS-associated gene. Other modes of Rfs within the same species must be identified to understand fully the co-evolution of these 2 genes. Currently, however, there are no reports in which 2 or more Rfs of independent origin have been cloned from the same plant species. In addition, the PPR-encoding Rf genes cloned to date all function dominantly against their nonfunctional rf allele. Their common working scheme is that the PPR proteins are carried into the mitochondria, preventing the accumulation of CMS-associated gene products. Further, no loss-of-function type Rf has been cloned, although CMS is caused by nuclear–mitochondrial incompatibility.
In rice, more than 20 independent CMS cytoplasms have been described. Accordingly, this plant species is ideal for identifying novel aspects of CMS. Although the CW-type cytoplasm was the first CMS system discovered in rice, the molecular components involved in the induction of male sterility and fertility restoration have not been described. Male fertility of rice plants carrying CW-type male sterility-inducing cytoplasm is restored gametophytically in haploid pollen by a single nuclear gene, Rf17 (17). Although plants carrying the CW-type CMS cytoplasm develop morphologically normal pollen, the pollen fails to germinate on the stigma after anthesis and lacks germination ability (17). In this study, we performed positional cloning of Rf17 and found that the reduced expression of a gene designated RETROGRADE-REGULATED MALE STERILITY (RMS) is sufficient to explain fertility restoration. The identification of RMS provided more information about functionally recessive Rfs. RMS was found not to encode a protein containing a pentatricopeptide repeat but to encode a protein of unknown function with a segment partially similar to acyl-carrier protein synthase (ACPS). The relationship between mitochondrial retrograde signaling and CMS is discussed also.
Results
Suppression of ORF11 Restored Fertility to a CMS Plant.
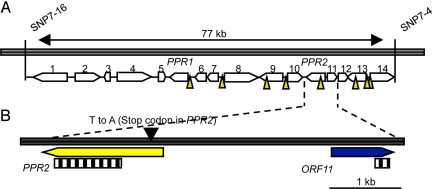
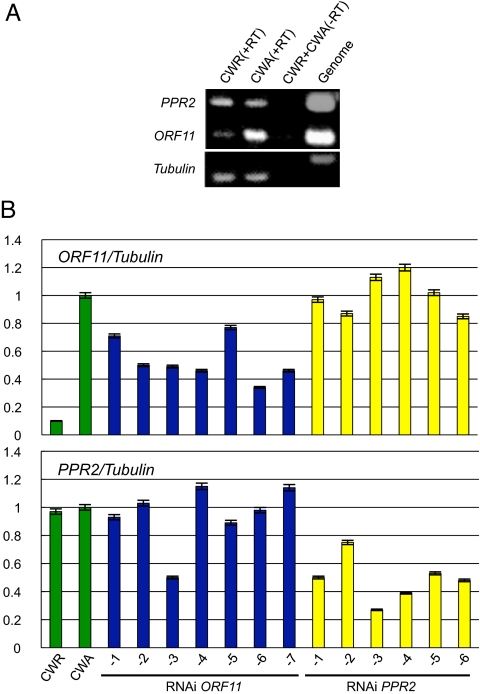
We previously described the determination of the coarse position of Rf17 (17). Using 4032 BC1F1 individuals and 9184 F2 individuals derived from a cross of the CW fertility restorer line (CWR) and the nonrestoring Kasalath/Koshihikari chromosome segment substitution line CSSL209, we identified Rf17 on the long arm of chromosome 4, in the 77-kb region within 2 cleaved amplified polymorphic sequence (CAPS) markers, SNP 7–16 and SNP7–4, and on a BAC clone, OsJNBa0022H21. The region contained 16 predicted genes, including 2 PPR genes (designated as PPR1 and PPR2 in Fig. 1A), a retrotransposon (ORF8 in Fig. 1A), and a transposase gene (ORF9 in Fig. 1A). By sequencing the BAC clone of the corresponding region screened from the CWR BAC library, a total of 25 SNPs and insertion/deletions (InDels) were found in the 77-kb region compared with the nonrestoring Nipponbare allele (Fig. 1A). In our previous study, we found that fertility restoration of CW-type CMS is controlled in a gametophytic manner (17), but whether Rf functions dominantly or recessively was unknown. The only nonsynonymous mutation identified was in the PPR gene Os04g0475800 (PPR2) of CWR, in which a T-to-A transition resulted in a premature stop codon (Fig. 1B). To gain more information about the predicted genes in this region, we monitored the expression of these 14 genes (excluding transposable elements) by RT-PCR. ORF11 (Os04g0475900) was the only gene determined to be differentially regulated in the CWR and the CW-CMS line (CWA) in mature anthers [Fig. 2A, supporting information (SI) Fig. S1] and was down-regulated in CWR. ORF11 down-regulation was thought to be caused by the SNP in PPR2, which could have been included in a promoter region of ORF11. PPR2 showed similar expression levels in these 2 lines. The result was confirmed by quantitative real-time RT-PCR, and the calculated mRNA abundance of ORF11 in CWA was ≈10-fold more than that in CWR (Fig. 2B). Thus, we considered that either a premature stop codon in a PPR gene or down-regulation of an unknown gene, ORF11, was involved in restoring fertility.
Fig. 1.
Mapping of the Rf17 locus. (A) The Rf17 locus on chromosome 4 mapped between DNA markers SNP7–16 and SNP7–4. Locations of 16 predicted genes are indicated by numbers 1 through 14. Genes encoding the PPR motif were designated PPR1 and PPR2. Arrowheads indicate the positions of 25 SNPs and InDels. SNPs or Indels with duplicate positions were omitted for clarity. (B) Detailed view of the 5.0-kb region spanning PPR2 and ORF11. Striped bars in each gene indicate the sequences chosen as the RNAi trigger region.
Fig. 2.
mRNA expression of PPR2 and ORF11. (A) Expression of PPR2 and ORF11 in the mature anthers of CW-type fertility-restorer line (CWR) and CW-type CMS line (CWA), as determined by RT-PCR. A mixture of CWR and CWA RNA without reverse transcription was used as the negative control (Third Lane). Transcript levels of tubulin were used as the internal control. (B) Transcript abundance of PPR2 and ORF11 in mature anthers of CWR and CWA ORF11 RNAi transgenic lines and PPR2 RNAi transgenic lines, as determined by quantitative real-time PCR. Relative transcript abundance was normalized to the levels of tubulin alpha-1 chain is shown as relative mRNA expression values against untransformed CWA (untransformed CWA = 1, Second Lane).
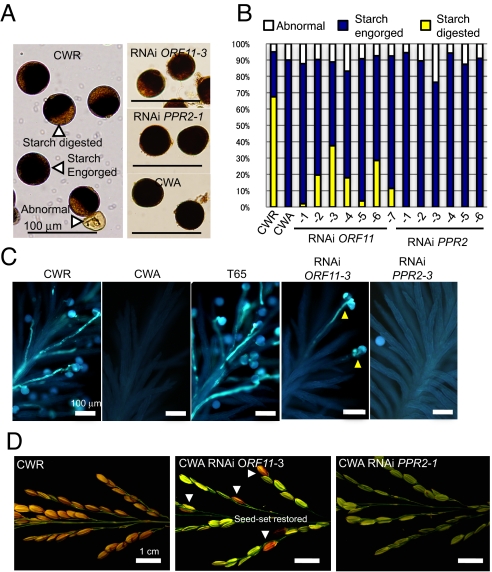
To examine these possibilities further, we introduced RNAi constructs carrying specific sequences of each gene as a trigger region into a CWA plant to down-regulate PPR2 or ORF11 (Fig. 1B). From the 15 independent transgenic lines for each construct, we obtained 7 plants with ORF11 expression reduced to 30% to 77% of the untransformed CWA, and 6 plants with PPR2 expression reduced to 27% to 75% (Fig. 2B). CWA plants exhibited complete seed-setting sterility, without any defects in pollen development until maturation that could be seen by light microscopy (17). Pollen dysfunction in the CWA line was observed first ≈ 2 h before anthesis after maturation. The CMS plants showed no starch digestion, as seen in wild-type plants. Starch digestion occurs when flowers are ready to open, possibly providing a fusible carbohydrate energy source for pollen germination on the stigma (18) Thus, we counted the starch-digested pollen of transgenic plants 2 to 3 h before anthesis (Fig. 3 A and B). In the male-fertile CWR line, 68% of the pollen exhibited a starch-digested phenotype before anthesis, and 27% of the pollen exhibited a starch-filled, engorged phenotype (Fig. 3B), whereas pollen in the CWA showed no starch digestion, and all of the pollen remained engorged with starch (Fig. 3B). We also examined the pollen of RNAi-mediated ORF11 or PPR2 knockdown transgenic plants and found that ORF11-knockdown plants produced starch-digested pollen ranging from 2% to 37.5% (Fig. 3B). On the other hand, the PPR2-knockdown plant phenotypes were unchanged from CWA, showing no starch-digested pollen at all. Thus, we concluded that suppression of ORF11 is sufficient for restoring starch digestion in pollen of a CMS plant.
Fig. 3.
The effects of ORF11 suppression on pollen germination and seed setting. (A) Pollen was collected 2 h before anthesis and categorized into 3 groups: starch-digested, starch-engorged, and abnormal pollen (CWR, Left). (B) Ratio of starch-digested, starch-engorged, and abnormal pollen in CWR and CWA ORF11 RNAi transgenic lines and PPR2 RNAi transgenic lines. (C) Stigmas 6 h after anthesis stained with aniline blue. Arrowheads in RNAi ORF11-3 panel indicate the traces of pollen germinations. (D) Seed setting observed on RNAi ORF11-3.
The effects of ORF11 mRNA suppression on pollen germination were evaluated by counting the germinating pollen on stigmas 6 h after anthesis (Fig. 3C and Table S1). Stigmas in CWR and Taichung 65 (T65) plants showed penetration of more than 100 pollen tubes 6 h after anthesis (Fig. 3C and Table S1). The frequency of germinating pollen in each stigma was 77% and 81%, respectively, and pollen tube germination was observed in all the florets examined (Table S1). In contrast, pollen germination never was observed in the stigmas of CWA plants after anthesis, although pollen grains without tube emission occasionally were found stuck in the stigmas (Fig. 3C and Table S1). The mature pollen of CWA was morphologically normal and was able to bind to stigmas under normal growth conditions. Few pollen grains remained at the time of the pollen tube observation, however, because the aniline-blue staining procedure required washing of the stigmas with alkaline buffer, and pollen grains not emitting tubes were wiped off. In the stigmas of RNAi ORF11-2 and -3, approximately 1/4 of the florets observed held pollen germination events (Table S1). Zero to 5 pollen grains emitted a tube within a floret, which accounted for 0.4 and 0.5 germinating pollen grains per stigma, on average (Fig. 3C and Table S1). As expected from the absence of the pollen starch degradation (Fig. 3), stigmas of RNAi PPR2-1 and -3 lines lacked pollen tubes (Table S1).
Although the seed-setting rate of CWR plants was greater than 74%, the seed-setting percentages of RNAi ORF11 lines Nos. 2, 3, 4, and 6 were only 2% to 5% (Fig. 3D), possibly because of incomplete suppression of ORF11 as compared with that of CWR (Fig. 2 A and B). Nevertheless, we confirmed that the suppression of ORF11 restored pollen fertility, because 4 independent lines with 37% to 47% suppression of ORF11 mRNA retained the seed-setting ability.
Overexpression of ORF11 Induces Pollen Lethality in CWR.
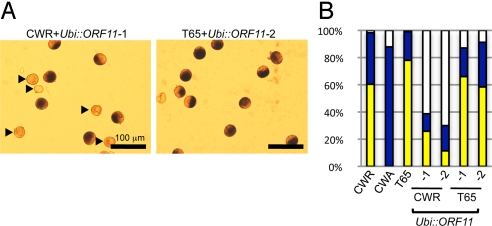
To investigate further, we overexpressed the 602-bp region including the full-length 537-bp ORF of ORF11 in a CWR background under the control of the maize ubiquitin promoter (Fig. S2 and Fig. 4). Of 10 independent transgenic lines, 2 lines carrying 1 copy of Ubi::ORF11 were chosen for further analysis. Approximately half of the pollen in CWR transgenic plants carrying 1 copy of Ubi::ORF11 hemizygously was shrunken and had reduced starch accumulation (Fig. 4 A and B); this phenotype was not observed in T65 transgenic plants carrying Ubi::ORF11. Thus, we concluded that the overexpression of ORF11 caused defects only in a CW-cytoplasm background. The pollen phenotypes of ORF11 overexpressing CWR lines seemed to be more deleterious than those of CWA lines (Fig. 3A), possibly because of the pleiotropic effect of ORF11 overexpression.
Fig. 4.
The effects of ORF11 overexpression in CWR and Taichung 65 (T65). (A) Pollen 2 h before anthesis. Arrowheads indicate the shrunken pollen observed in CWR+Ubi::ORF11 lines. (B) Pollen was categorized into 3 groups as in Fig. 3. The ratio of starch-digested, starch-engorged, and abnormal pollen in each line is shown.
ORF11 Encodes an ACPS-Like Domain Containing Protein.
ORF11 was predicted to encode a protein 178 aa long with an ACPS-like domain in amino acids 139–156 (Fig. 5A). ACPSs are directly involved in the elongation of acetyl-CoA to 18:0-ACP (19). Although the conserved part of the ACPS sequence was present in ORF11, it is not likely that ORF11 functions as a general ACPS, because the ACPS-like domain is shorter than ACPS catalytic domains, which normally are 70–90 aa long; furthermore, the conserved lysine residue among species is replaced by alanine (amino acid 140 in Fig. S3). BLASTP and BLASTN searches revealed no similar proteins in other organisms. Although we ran 4 different protein-targeting prediction programs (iPSORT, Mitoprot, Predotar, and TargetP v1.1), these tools did not produce a consistent prediction regarding the subcellular localization of ORF11. Thus, the GFP was fused to ORF11 and stably introduced into rice. GFP fluorescence in the protoplasts from the transgenic calli co-localized with the fluorescence of Mito Tracker Red, which indicated that ORF11 was targeted to mitochondria (Fig. S4).
Fig. 5.
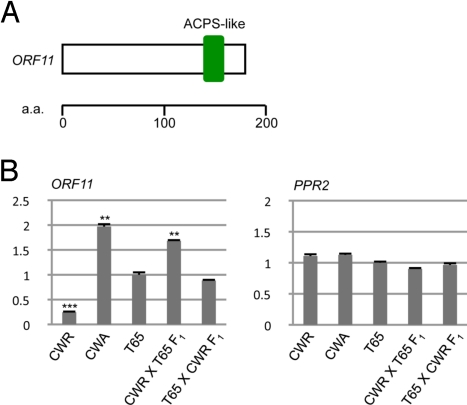
ORF11 expression in reciprocal hybrids. (A) Schematic structure of ORF11. (B) Relative expression of ORF11 and PPR2 in mature anthers of CWR and CWA, Taichung 65 (T65), and reciprocal F1 hybrids of CWR (female) X T65 (male), and T65 (female) X CWR (male). Relative transcript abundance was normalized to the levels of tubulin alpha-1 chain and is shown as relative mRNA expression values against untransformed T65 (T65 = 1).
ORF11 mRNA Expression Is Dependent on the Cytoplasmic Genotype.
The mRNA expression level of ORF11 in mature anthers in CWA was approximately double that of T65 (Fig. 5B), whereas PPR2 mRNA expression levels were unchanged. The nuclear backgrounds of T65 and CWA were identical on the 157 simple sequence repeat (SSR) markers across chromosomes (see Materials and Methods for details), but their mitochondrial sequences were different (Fig. S5). Therefore, we concluded that mRNA expression of ORF11 depends on the cytoplasmic genotype and, given the nature of CMS, ORF11 gene expression could depend on the mitochondrial genotype. To confirm the relevance of the cytoplasmic genotype to the mRNA expression level of ORF11, we compared the expression level of ORF11 in the F1 plants obtained from the reciprocal crosses between CWR and T65 plants. In comparison with T65 plants, significant up-regulation of ORF11 was observed in the F1 plants carrying the CW cytoplasm (CWR X T65 F1), whereas slight down-regulation of ORF11 was observed in the F1 plants carrying the T65 cytoplasm (T65 X CWR F1) (Fig. 5B). A wealth of recent studies support the cytoplasmic control of nuclear gene expression and retrograde signaling in broad range of Eukaryote species (20–22). It is likely that the up-regulation of ORF11 under CW cytoplasm could be controlled by a retrograde signaling pathway. Because the suppression of ORF11 restored pollen fertility (Fig. 3) and overexpression caused deleterious effects in pollen carrying the CW cytoplasm background (Fig. 4), we designated ORF11 RETROGRADE-REGULATED MALE STERILITY (RMS).
We also were curious whether the difference in mRNA expression level of RMS between CWR and CWA was controlled via a cis-element. As we reported in the prior section, the only nucleotide mutation in the RMS gene region was the SNP 2,286 bp upstream of the start codon, which could be included in the genic region of PPR2 (Fig. 1B). We constructed a reporter assay experiment using the monomeric red fluorescent protein (mRFP) gene, fusing mRFP downstream of the RMS promoter and coding regions of the CWR-type allele (carrying T at the −2,286th nucleotide) or the CWA-type allele (carrying A at the −2,286th nucleotide). mRFP reporter constructs were stably introduced into T65 plants, and 3 independent transgenic lines carrying 1 copy of the transgene were chosen for the analysis. As a result, in plants carrying CWA_RMS promoter::mRFP hemizygously, ≈ 25% to 33% of the pollen emitted the RFP fluorescence (Fig. S6). On the other hand, the frequency of pollen fluorescence of the plants carrying CWR_RMS promoter::mRFP was equivalent to that of the plants with the empty vector carrying solely 35S::HPT (Fig. S6). Thus, we concluded that the SNP at 2,286-bp upstream in the promoter region of RMS might be important for its suppression in the Rf17 allele.
Discussion
We performed positional cloning of Rf17 and succeeded in delimiting Rf17 to 2 candidates, PPR2 and ORF11 (RMS). By suppressing RMS, CW-CMS plants reverted to seed setting, although the molecular function of the putative mitochondrial RMS protein remains unknown. RMS possesses a partial ACPS domain that does not seem to function as general ACPS when compared with other proteins containing an ACPS domain (Fig. S3). In CW-CMS plants with depressed levels of PPR2 mRNA expression, there was no recovery of either seed setting or pollen fertility. Overexpression of RMS apparently was lethal to pollen in the CW cytoplasm background. We conclude that the CW-type CMS system of rice results from RMS up-regulation and that CWR retains male fertility because RMS expression may be reduced by the SNP in its promoter region, resulting in restored compatibility between the nucleus and mitochondria. We therefore propose that the CWR allele expressing relatively low levels of RMS mRNA is Rf17 and that the CWA allele expressing high levels of RMS mRNA is rf17.
RMS does not encode a PPR-containing protein, although, except for Rf2a for maize T-CMS, all the other Rf genes so far reported encode PPR proteins (3–9, 13–16). In addition to Rf1 for BT-CMS rice, our study cloned a second Rf gene of independent origin in the same plant species. Our RNAi experiment does not, however, completely rule out the possibility that PPR2 is a factor that restores the relationship between the CW cytoplasm and the nucleus. It is possible that RNAi-mediated suppression of RMS has epistatic effects on the neighboring gene, PPR2. Furthermore, PPR2 contains an AUG codon downstream of the premature stop codon in the CWR allele, which could produce a protein of 330 aa with 8 PPR motifs that might function as the fertility-restorer protein. Antibodies to PPR2 are required to determine if such a truncated protein is produced. Nevertheless, based on our current results, it is unlikely that PPR2, rather than ORF11, is the restorer gene. Additional mutants would be necessary to assess further the function of PPR2 in CW-CMS. Unfortunately, we were unable to find mutants with Tos17 or T-DNA inserted in PPR2 or RMS.
To the best of our knowledge, the only loss-of-function fertility restorer locus reported to date is restorer-of-fertility lethal 1 (rfl1) for maize S-CMS (23). The dominant Rfl1 allele is related positively to the accumulation of the mitochondrial A subunit of ATP synthase (ATPA), and ATPA could interact with the mitochondrial orf355-orf77 gene product, which is associated with CMS. rfl1 homozygosity is lethal, however, because of the lack of mitochondrial ATPA accumulation (23). A significant aspect of our findings is that a reduced-expression allele of RMS restored fertility in haploid pollen, whereas a normal-expression allele caused pollen lethality in the CW-type CMS. Although there were no indications of RMS functions other than the partial ACPS-like domain, we speculate that some metabolic alteration in mitochondria restores pollen fertility, similar to the mechanism in the maize Rf2 system (3, 4). We do not consider RMS, unlike other PPR-encoding Rf genes previously identified (5–9, 13–16), to be the protein responsible for posttranscriptional RNA modification of the mitochondrial CMS-associated gene expression; rather, loss-of-function of the RMS gene may provide a bypass to fertility restoration.
Retrograde regulation of nuclear genes by organelle status has been reported recently in animals, yeast (20), and in plants (21, 22, 24). It is assumed that this gene regulation is required for the coordinated expression of nucleus and organelle genes in plants, because protein complexes participating in the respiratory chain in mitochondria are composed of chimeric structures of nuclear and mitochondrial-derived proteins (25). Although virtually nothing is understood about mitochondrial retrograde regulation in plants, a key component in plastid retrograde signaling, GUN1, has been cloned and shown to encode a PPR protein (26). GUN1 mediates plastid signals to ABI4, a transcription factor containing an AP2 (APETALLA2) domain. ABI4 binds to G-box elements in a retrogradely regulated LHCB promoter, negatively regulating its expression (26). Our results indicate that up-regulation of RMS occurs upon introduction of the CW-type cytoplasm, suggesting that RMS is a candidate gene to be regulated via retrograde signaling. Because overexpression of RMS results in pollen sterility (Fig. 4), it is possible that retrograde signaling is directly involved in the occurrence of CW-CMS. This issue could be clarified by isolating a factor that promotes the expression of RMS and by analyzing its function in relation to mitochondrial signaling. Our present research suggests how CMS occurs and may help reveal the relationship between mitochondria and CMS and mitochondrial–nuclear retrograde signaling.
We also showed that the SNP at 2,286 bp upstream of RMS might be involved in the suppression of RMS in CWR by reporter assay experiments (Fig. S6). Recently, a 12-kb upstream SNP was shown to be involved in the cis-regulatory system of the spatial expression pattern of OsRPL, a gene responsible for seed shattering in rice (27). Expression of RMS is likely to be regulated by the SNP, although we have not detected any known cis-regulatory elements in this region.
CMS has been used widely for hybrid rice breeding in Southeastern Asia. In China, hybrid rice, which has an average 15% to 20% yield advantage over inbred strains, is planted on ≈16 million hectares—more than half of China's total rice area of 28 million hectares (28). The CW-CMS and its restorer gene have not been used for hybrid rice production, however. As discussed by Komori and Imaseki (29) and Sattari et al. (30), most commercial rice hybrids are based on a single CMS source, the wild-abortive (WA) cytoplasm. To avoid the potential threat of genetic vulnerability of rice hybrids, the development of several CMS/Rf systems is desirable. Introduction of loss-of-function type Rf genes to hybrid breeding programs will broaden the combination of F1 pairs and contribute to improve yields.
Materials and Methods
Plant Materials.
The BC3F1 CMS line was derived from a cross of Oryza japonica cultivar Reimei and Oryza rufipogon Griff., W1 strain (31). The T65 nuclear background CMS line, CWA with [cms-CW] rf17rf17, was obtained by backcrossing T65 6 times with the Reimei CMS line. The genotype of T65 is [normal] rf17rf17. The fertility-restorer line, CWR, was obtained by backcrossing T65 5 times with a restorer line described in (31) and has a genotype of [cms-CW] Rf17Rf17. We carried out the marker-assisted selection of CWA and CWR to obtain efficiently lines with nuclear genomes substituted by T65, using 157 SSR markers covering all the chromosomes (Table S2). The nuclear genomic substitutions of CWA and CWR used in this study were assessed by the 157 SSR markers. As a result, we obtained CWA and BTA lines completely isogenic to T65. CWR had almost the same genome except for the Rf17 region of chromosome 4, the region within SSR markers RM7535 and RM3276. BC1F1 and F2 mapping populations were derived from the cross of CWR and the nonfertility restoring Kasalath/Koshihikari chromosome segment substitution line CSSL209 (32). Most of chromosome 4, except for the 15-cM telomeric region of the long arm, was replaced by Kasalath.
Genetic Mapping Experiments.
For positional cloning, recombinants were screened from 4032 BC1F1 and 9184 F2 progeny. The 21 PCR markers used in this study are listed in Table S2. Through mapping, 2 recombinants were obtained for each of the cleaved amplified polymorphic markers, SNP7–16 and SNP7–4, which were ≈77 kb apart. Construction of the BAC library and sequencing were performed by the previously described methods (33).
Production of Transgenic Rice Lines.
ORF11 (accession no. Os04g0475900 in the Rice Annotation Project DataBase, RAP-DB and PPR2 (accession no. Os04g0475800) RNAi constructs overdriven by the maize ubiquitin promoter were constructed using the pANDA vector provided by D. Miki and K. Shimamoto, following their protocols (34). A gene-specific region of ORF11 was PCR cloned by primer set 5′-CACCAGCGTTGAAGAGTTTGGGGA-3′ and 5′- CGAGCTCCAACATACTGGCT-3′. 5′-CACCTGAAGAGTGCAAACCCTCTG-3′ and 5′-TTCGCAGACTCTCAACAAGG-3′ were used for PPR2. Nucleotides CACC were added to the original ORF11 and PPR2 sequences for cloning into the pENTER D-TOPO vector (Invitrogen). For overexpressing ORF11, the 602-bp region including the full-length 537-bp translational region and the 5′UTR region was PCR-cloned using the primer pairs 5′-CACCGGATCCCTCGCTGAGGTGCTCTCCCTCT-3′ and 5′-GGATCCCTAGGTGGCTAAACTTGGCCAGC-3 and was fused to the maize ubiquitin promoter. The resulting RNAi and overexpressing constructs were introduced into Agrobacterium tumefaciens EHA105 and were further introduced into plants by the previously described method (35).
ORF11 (RMS) promoter and coding regions from each CWR and CWA were PCR-cloned using primer pairs 5′-GGATCCCCTTCAAGCCTTCATGAAATGCTCC-3′ and 5′-GGATCCGGTGGCTAAACTTGGCCAGC-3′. The 4,603-bp regions were fused in upstream of the mRFP gene, as described in our previous study (36). The constructs were stably introduced to T65 plants as described in the previous paragraph.
Subcellular Localization of ORF11-GFP Fusion Protein.
The stop codon-deleted ORF11 was fused to the N-terminus of GFP and was driven by the maize ubiquitin promoter to produce T65 plants stably expressing the ORF11-GFP fusion protein. Calli from the transgenic lines were treated with 0.05% (wt/vol) pectolyase Y-23 (Kyowa Chemical Products), 0.05% (wt/vol) cellulase RS (Yakult Pharmaceutical Ind. Co.), 0.01% (wt/vol) calcium chloride, and 0.9% (wt/vol) mannitol-containing buffer (pH 5.6) and were subsequently stained with Mito Tracker Red (Invitrogen) according to the manufacturer's protocols. Protoplasts were washed with 0.01% calcium chloride and 0.9% mannitol-containing buffer and were observed under fluorescent microscopy for GFP fluorescence.
mRNA Expression Analysis.
Total RNA from mature anthers was extracted using RNeasy (Qiagen) according to the manufacturer's instructions. DNA elimination and cDNA synthesis were performed using the QuantiTect Reverse Transcription kit (Qiagen). RT-PCR for genes predicted in the Rf region was performed using exTaq DNA polymerase (TaKaRa-Bio). PCR cycles were as follows: 94°C for 3 min, followed by 23, 27, and 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s, and final extension at 72°C for 3 min. Band intensities at a nonsaturating condition were compared. Quantitative RT-PCR for ORF11 and PPR2 was performed for 3 biologic replicates, using SYBR Premix Ex Taq (TaKaRa-Bio) and Thermal Cycler Dice Real Time System TP800 (TaKaRa-Bio). The expression level of tubulin alpha-1 chain (accession no. Os07g0574800) was used as the internal control. Specific primers used for each gene are listed in Table S3.
Pollen Observation.
Spikelets were harvested 2 to 3 h before anthesis and fixed in 3:1 ethanol:acetic acid (vol/vol). After staining with 2% iodine-potassium iodide, pollen was classified into 3 groups: starch-digested, starch-engorged, and abnormal pollen, and grains were counted under light microscopy. At least 400 pollen grains were counted per spikelet, and 5 spikelets were counted for each line. Pollen germination on the stigmas was observed by the method previously described (17).
Supplementary Material
Acknowledgments.
We thank Drs. Takashi Matsumoto, Harumi Ishikubo, and Hiroyuki Kanamori (Society for Techno-innovation of Agriculture, Forestry, and Fisheries, Japan) for constructing the BAC library and subsequent screening and sequencing. Seeds of Kasalath/Koshihikari chromosome segment substitution line CSSL209 were distributed by the Rice Genome Resource Center, Japan. We are grateful to Dr. D. Miki and Dr. K. Shimamoto at Nara Institute of Science and Technology, Japan, for kindly providing us pANDA vector. This study was supported in part by Grants QT-2001 and QT-2007 from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated Research Project for Plants, Insects and Animals Using Genome Technology), and by Grant-in-Aid 18075002 for Special Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan. S.F. is the recipient of Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The sequence reported in this paper has been deposited in the DNA Data Bank of Japan (accession no. AB481199).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901860106/DCSupplemental.
References
- 1.Hanson M-R, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16:S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chase C-D. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends in Genetics. 2006;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Cui X, Wise R-P, Schnable P-S. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science. 1996;272:1334–1336. doi: 10.1126/science.272.5266.1334. [DOI] [PubMed] [Google Scholar]
- 4.Liu F, et al. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell. 2001;13:1063–1078. doi: 10.1105/tpc.13.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentolila S, Alfonso A, Hanson M-R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koizuka N, et al. Genetic characterization of pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 2003;34:407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown G-G, et al. The radish Rfo restorer gene Ogura cytoplasmic male sterility encode a protein with multiple pentatricopeptide repeats. Plant J. 2003;35:262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- 8.Desloire S, et al. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Reports. 2003;4:588–594. doi: 10.1038/sj.embor.embor848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein R-R, et al. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L. ) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor Appl Genet. 2005;111:994–1101. doi: 10.1007/s00122-005-2011-y. [DOI] [PubMed] [Google Scholar]
- 10.Small I, Peeters N. The PPR motif—a TRP-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 11.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillman J-D, Bentolila A, Hanson M-R. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007;49:217–227. doi: 10.1111/j.1365-313X.2006.02953.x. [DOI] [PubMed] [Google Scholar]
- 13.Kazama T, Toriyama K. A pentatricopeptide repeat-containing gene that promote the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 2003;544:99–102. doi: 10.1016/s0014-5793(03)00480-0. [DOI] [PubMed] [Google Scholar]
- 14.Komori T, et al. Map-based cloning of a fertility restorer gene, Rf-1 in rice (Oryza sativa L. ) Plant J. 2004;37:315–325. doi: 10.1046/j.1365-313x.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- 15.Akagi H, et al. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor Appl Genet. 2004;108:1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii S, Toriyama K. Molecular mapping of the fertility restorer gene for rice ms-CW-type cytoplasmic male sterility of rice. Theor Appl Genet. 2005;111:696–701. doi: 10.1007/s00122-005-2054-0. [DOI] [PubMed] [Google Scholar]
- 18.Koike S, Satake T. Sterility caused by cooling treatment at the flowering stage in rice plants. Japanese Journal of Crop Science. 1987;56:666–672. [Google Scholar]
- 19.Nishida I. In: Molecular Biology and Biotechnology of Plant Organelles. Daniell H, Chase C, editors. The Netherlands: Springer; 2004. pp. 543–564. [Google Scholar]
- 20.Butow R-A, Avadhani N-G. Mitochondrial signaling: The retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 21.Nott A, Jung H-S, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 22.Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Current Opinion in Plant Biology. 2007;10:600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Wen L, et al. A nuclear restorer-of fertility mutation disrupts accumulation of mitochondrial ATP synthase subunit a in developing pollen of S male sterile maize. Genetics. 2003;265:771–779. doi: 10.1093/genetics/165.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zubko M-K. Mitochondrial turning fork in nuclear homeotic functions. Trends in Plant Science. 2004;9:61–64. doi: 10.1016/j.tplants.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Heazlewood J-L, Howell K-A, Whelan J, Millar H. Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 2003;132:230–242. doi: 10.1104/pp.102.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 27.Konishi, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 28.Barclay A. A hybrid history. Rice Today. 2007;6:22–25. [Google Scholar]
- 29.Komori T, Imaseki H. Transgenic rice hybrids that carry the Rf-1 gene at multiple loci show improved fertility at low temperature. Plant Cell Environ. 2005;28:425–431. [Google Scholar]
- 30.Sattari M, et al. Development and use of a two-gene marker-aided selection system for fertility restorer genes in rice. Euphytica. 2007;153:35–42. [Google Scholar]
- 31.Toriyama K, Hinata K. Anther culture application to breeding of a restorer for a male-sterile cytoplasm of wild rice [ms-CW] Japanese Journal of Breeding. 1987;37:469–473. [Google Scholar]
- 32.Ebitani T, et al. Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breeding Science. 2005;55:65–73. [Google Scholar]
- 33.Baba T, et al. Construction and characterization of rice genomic libraries: PAC library of japonica variety, Nipponbare and BAC Library of indica variety, Kasalath. Bulletin of the National Institute of Agrobiological Research. 2000;14:41–49. [Google Scholar]
- 34.Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- 35.Yokoi S, Tsuchiya T, Toriyama K, Hinata K. Tapetum-specific expression of the Osg6B promoter-ß-glucuronidase gene in transgenic rice. Plant Cell Rep. 1997;16:363–367. doi: 10.1007/BF01146774. [DOI] [PubMed] [Google Scholar]
- 36.Fujii S, Yamada M, Toriyama K. Cytoplasmic male sterility-related protein kinase, OsNek3, is regulated downstream of mitochondrial protein phosphatase 2C, DCW11. Plant Cell Physiol. 2009;50:828–837. doi: 10.1093/pcp/pcp026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.